Figure 34.1
Genetic pathways for urothelial carcinoma tumorigenesis. Noninvasive papillary tumors are characterized by early activating mutations of the FGFR3 gene, inactivating mutations or epigenetic alterations of the P16 gene, and a diploid or near-diploid DNA content. Carcinoma in situ (CIS) and invasive tumors are characterized by early inactivating mutations within the TP53 and P16 genes, chromosomal instability (CIN), and an aneuploid DNA content. A small proportion of papillary tumors may acquire TP53 alterations or alterations of other unknown genes that cause invasive potential of these tumors
Table 34.1
Pathologic staging of primary bladder urothelial carcinoma
Stage | Description |
|---|---|
pTa | Noninvasive papillary |
pTis | Carcinoma in situ |
pT1 | Invasion into lamina propria |
pT2 | Invasion into muscularis propria |
pT3 | Invasion through the muscularis propria and into bladder adventitia |
pT4 | Invasion into surrounding organs (e.g., colon) |
Molecular Basis of Disease
At the chromosomal level, the majority of low-grade papillary tumors are diploid or near-diploid, while the majority of high-grade papillary UC, CIS, and invasive UC (pTa tumors) are aneuploid. Based on array-based comparative genomic hybridization and fluorescence in situ hybridization (FISH) studies, noninvasive, low-grade pTa papillary UC have relatively few chromosomal abnormalities except for loss of all or part of chromosome 9, while CIS, high-grade pTa, and invasive UC (pT1 tumors) have a high number of chromosomal gains and losses [1, 2]. The pT1 tumors also have loss of all or part of chromosome 9 but have numerous additional chromosomal abnormalities, which include whole or partial chromosomal losses and gains. Frequent sites of allelic imbalance (AI) in UC include 3p, 4p, 8p, 9p, 9q, 11p, 13q, 17p, and 18q based on microsatellite analysis (MA) [3–5]. Regions with high rates of AI are the sites of known or putative tumor suppressor genes. Many of the regions that show high rates of AI correspond to the areas of chromosomal gains and losses detected by aCGH.
Two important molecular genetic alterations that contribute to UC tumorigenesis are mutational and epigenetic alterations that inactivate the P16 and TP53 tumor suppressor genes. P16 loss is one of the earliest events in the development of both papillary and flat/invasive UC [1, 5–7]. Mutations that inactivate TP53 are found primarily in CIS and invasive UC and not low-grade papillary tumors and in part may be responsible for the aggressive behavior of these tumors [8, 9]. According to the Catalogue of Somatic Mutations in Cancer (COSMIC) database, other oncogenes and tumor suppressor genes mutated in decreasing order of frequency in UC include FGFR3, PIK3CA, CDKN2A, HRAS, KRAS, PTEN, AKT1, APC, CTNB1, and NRAS (http://www.sanger.ac.uk/genetics/CGP/cosmic/).
Defective DNA mismatch repair (MMR) is manifested as MSI at >30 % of microsatellite markers examined and in most cases is associated with a loss of expression of one of the DNA MMR proteins, hMSH2, hMLHl, hMSH6, or hPMS2. MMR is rarely observed in UC of the bladder but is found in approximately 20–30 % of upper urinary tract UC [10, 11]. The finding of defective MMR in an upper tract UC should prompt an investigation into the possibility that the patient may have hereditary nonpolyposis colorectal cancer (HNPCC) and a germline mutation of one of the DNA MMR genes.
Chromosomal instability (CIN) is present in invasive UC and CIS. It is likely that genes that maintain genomic stability are inactivated early during invasive UC tumorigenesis. CIN drives tumorigenesis and tumor progression by accelerating the mutation rate in tumor cells [12]. The genes responsible for CIN in invasive UC are not known, and the role of TP53 inactivation in CIN has been a matter of debate. pTa tumors show little evidence of CIN but, as noted above, tend to be diploid or near-diploid tumors with relatively few chromosomal alterations. Chromosome 9 and P16 alterations play a major role in the formation of low-grade pTa tumors. In addition, most low-grade papillary UC and urothelial papillomas have missense mutations of the fibroblast growth factor 3 (FGFR3) gene, while mutations of this gene are less common in invasive UC and CIS [13].
Taken together, two genetic pathways lead to the development of UC [1, 7]. One pathway leads to the formation of noninvasive papillary UC and the other to the development of CIS/invasive UC (Fig. 34.1). The pathway for noninvasive papillary UC is characterized by the presence of FGFR3 mutations and/or chromosome 9 alterations and P16 inactivation. The pathway for invasive UC is characterized by early alterations in the TP53 and P16 genes, late alterations of other tumor suppressor genes and oncogenes, chromosomal instability, and aneuploidy. The genetic differences between noninvasive papillary and CIS/invasive tumors likely explain the markedly different behavior and prognosis of these tumors [14].
Clinical Utility of Testing
In general, clinical molecular tests for solid tumors can be categorized as being useful for predicting predisposition to developing tumor, aiding in a diagnosis of the tumor type, detecting the presence of tumor, predicting prognosis, or guiding therapy. Examples of assays that are currently being used or investigated for each of these indications for UC are presented below.
Available Assays
The most clinically useful clinical molecular tests, as described below, are the following:
MSI analysis and DNA MMR protein immunohistochemistry (IHC) of upper urinary tract UC to assess for Lynch syndrome (LS)
FISH for UC detection
FGFR3 mutation analysis for UC detection
Numerous assays with a variety of clinical purposes for UC have been investigated but have not yet transitioned into the clinical use. TP53 mutations are common in UC, especially in high-grade UC [15]. Assays that assess for TP53 status could potentially be used to assess prognosis and detect tumor recurrence. Some studies have shown that TP53 overexpression detected by IHC analysis of formalin-fixed, paraffin-embedded tumors is associated with worse prognosis and higher risk of muscle invasion [8, 16], while others have not [17]. IHC analysis of bladder tumors for TP53 expression has not been widely utilized by urologists or pathologists. A few studies have shown that the antiapoptotic protein survivin may be a sensitive and specific marker for the detection of recurrent UC [18], but blinded prospective studies are needed to further evaluate the clinical utility of this assay. Alterations in certain genes, such as glutathione S-transferase M1 and N-acetyltransferase that encode proteins that metabolize carcinogens, may increase an individual’s risk of developing bladder cancer, especially among smokers, but assays for these alterations also have not been used clinically.
Interpretation of Results
MSI Analysis and DNA MMR Protein IHC of Upper Tract UC to Assess for LS
Defective DNA MMR is rarely observed in UC of the bladder but is found in approximately 20–30 % of upper tract UC [10, 11]. Patients with early onset upper tract UC or an upper tract UC and a family history of LS-related tumors should be evaluated for LS. This evaluation can consist of assessing the tumor for defective DNA MMR with MSI testing and/or DNA MMR IHC. Patients whose tumors exhibit high-level MSI (MSI-H phenotype) have defective DNA MMR and almost always show loss of expression of one or more of the DNA MMR proteins by IHC. Most histopathology laboratories perform immunostains for four DNA MMR proteins: MLH1, PMS2, MSH2, and MSH6. The most common pattern of protein expression loss in tumors that exhibit defective DNA MMR is loss of MLH1 and PMS2 with retention of staining for MSH2 and MSH6. This pattern of expression is most often due to epigenetic silencing of the MLH1 gene through promoter hypermethylation. However, some patients with MLH1 and PMS2 loss have a germline mutation in the MLH1 gene and consequently have HNPCC. Less common IHC staining patterns are loss of MSH2 and MSH6 with retention of MLH1 and PMS2, loss of MSH6 alone, or loss of PMS2 alone. These three patterns are strongly associated with the presence of a germline mutation in the MSH2, MSH6, and PMS2 genes, respectively. Patients with genetically proven LS are at risk of developing various tumors such as colorectal cancer, endometrial cancer, upper tract UC, gastric cancer, and sebaceous skin tumors and should undergo regular surveillance for these tumors.
Fluorescence In Situ Hybridization for UC Detection
Urine cytology has been the primary laboratory method for diagnosing and monitoring UC for the past 50 years. Urine cytology has excellent specificity but poor sensitivity for the detection of UC [19]. The problem with false-negative urine cytology test results, if combined with a negative cystoscopy, is that clinical surveillance regimens recommend rescreening in 3 months, allowing an undetected tumor to progress to a higher, potentially incurable state before it is detected. This is of particular concern for grade 3 UC, which routinely progress if not removed or treated. The suboptimal sensitivity of urine cytology has prompted the development of new tests with improved sensitivity for UC detection.
Most UC are characterized by numerical and structural chromosomal abnormalities and a marked degree of CIN with variation in the chromosomal abnormalities found from cell to cell. The finding of aneusomy (i.e., abnormal chromosome copy number) and CIN in a population of cells by FISH is strongly correlated with the presence of malignancy. UroVysion (Abbott Molecular, Des Plaines, IL) is a FISH assay that has been developed for the detection of UC in urine. This assay utilizes four FISH probes, CEP3, CEP7, CEP17, and LSI 9p21, that are labeled with red, green, aqua, and yellow fluorophores, respectively [2, 19]. UroVysion received FDA approval in 2001 for monitoring UC patients for tumor recurrence and FDA approval in 2005 for assessing patients with hematuria (gross or microscopic) for bladder cancer. Representative examples of patients with FISH-positive and FISH-negative findings are shown in Fig. 34.2.
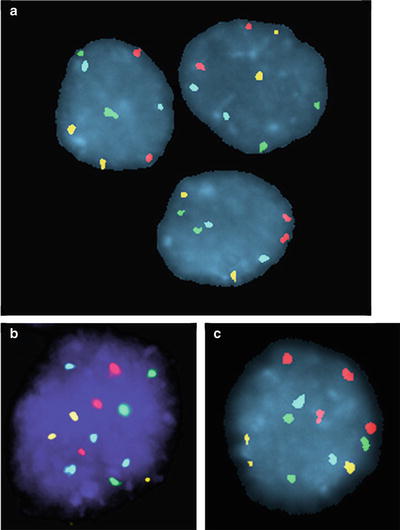

Figure 34.2




Representative examples of FISH results for nonneoplastic urothelial cells (panel a) and UC cells (panels b and c) using the UroVysion FISH assay (Abbott Molecular, Des Plaines, IL). Nonneoplastic cells generally have two signals for each of the four probes, though occasional nonneoplastic cells show only one signal for one or more of the probes due to random overlap of signals or imperfect hybridization efficiency. UC cells generally have gains for two or more of the probes (i.e., polysomy) of the UroVysion probe set. The finding of just a few cells with polysomy has high specificity for the presence of malignancy. Panel b shows UC cells with a gains of all four probe signals, CEP3(red), CEP7(green), CEP17 (aqua) and LSI 9p21 (yellow). Panel c shows UC cells with gain of CEP3 (red) and CEP7 (green) probe signals.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


