Figure 20.1
Proposed mechanism for HLA-B*5701 abacavir hypersensitivity
Australia was one of the earliest countries to introduce routine HLA-B*5701 screening and results from the prospective study that was conducted between 2002 and 2005 demonstrated a significant reduction of ABC-HSR from 8 to 2 % [11, 12]. Additional studies with similar results prompted one of the largest pharmacogenetics clinical trials ever conducted, the Prospective Randomized Evaluation of DNA Screening in a Clinical Trial (PREDICT-1) [13]. This double-blind, randomized trial was also the first study to address the clinical utility of a pharmacogenetic marker to reduce toxicity of a drug. PREDICT-1 enrolled 1,956 patients, who were predominantly Caucasian, from 19 different countries. Patients were randomized into two groups. The control group received standard of care, meaning no screening and administration of abacavir without restriction. The second group received prospective HLA typing and abacavir was withheld from patients who tested positive for HLA-B*5701. One aspect that made this study unique was that all suspected cases of clinical ABC-HSR were confirmed by patch testing. The results obtained were important because prescreening eliminated all cases of patch-confirmed, immunologically mediated, HLA-related abacavir hypersensitivity and yielded 100 % negative predictive value.
Prior to PREDICT-1, reliance on clinical diagnosis had led to inaccurate estimates of the negative predictive value of HLA-B*5701 testing, casting doubts on the utility of prospective screening. This study provided robust evidence for preventive screening in Caucasians, but the lack of ethnic diversity in study participants limited generalization to other ethnic populations. The frequency of the HLA-B*5701 allele varies globally, with a high frequency of 8 % in Caucasians and only 2.5 % in African Americans [14]. The Study for Hypersensitivity to Abacavir and Pharmacogenetic Evaluation (SHAPE) confirmed the reduced prevalence of ABC-HSR in African Americans, but presented clear evidence that screening had applicability across races [15].
Further studies evaluated the cost-effectiveness of pretreatment screening and considered variables such as the frequency of HLA-B*5701 carriers, and the sensitivity and specificity of assays for detection of the HLA-B*5701 allele, as well as the cost of preventive screening, alternative therapies, and treatment of ABC-HSR. The conclusion was that pretreatment screening was a cost-effective approach and the evidence supported adoption of widespread implementation of prescreen testing [16, 17].
Clinical Utility
Clinical utility of prospective screening for HLA-B*5701 to prevent abacavir hypersensitivity reaction is well established. Genetic screening has now been widely adopted in most Western countries, particularly Australia, Ireland, the United Kingdom, and the USA. In 2007, guidelines from the US Department of Health and Human Services that address antiretroviral treatment recommended screening for HLA-B*5701 prior to initiation of abacavir therapy [18]. In 2008, the US Food and Drug Administration also revised the product label and added a black box warning for all abacavir-containing formulations to reflect the new guidelines for HLA-B*5701 screening.
Available Assays
One hurdle for widespread implementation of screening was the availability of a simple, reliable, and inexpensive assay to detect HLA-B*5701 carriers. Because of the level of resolution needed to discriminate between the many closely related alleles present at each locus, sequencing is the gold standard for HLA haplotyping. Generally, HLA-B*5701 haplotyping by sequencing is available only in highly specialized laboratories that perform tissue matching in the setting of organ transplantation. The expense, labor, and expertise required for the sequencing method are barriers to widespread availability of testing. In an effort to overcome these challenges, simplified molecular assays have been developed and some groups have even explored alternative, surrogate markers for HLA-B*5701. Rs2395029(G) allele in the HCP5 region of the HLA complex is in linkage disequilibrium with HLA-B*5701 and has been proposed as a substitute for HLA-B*5701 testing [19, 20]. Genotyping for the rs2395029 T/G SNP is technically less challenging than genotyping for HLA-B*5701, as this necessitates reliable discrimination between closely related alleles, and it is less expensive than sequencing methods; however, as discussed below, this marker does not always correlate with the presence of an HLA-B*5701 allele.
For direct detection of HLA-B*5701, allele-specific PCR methods with gel electrophoresis or melting curve analysis are available commercially [21, 22]. Flow cytometry-based typing is cost effective and also utilized by diagnostic laboratories. A Taqman allelic discrimination assay for the HCP5 rs2395029 T/G SNP has also been described [19]. Sequencing for HLA-B*5701 remains an alternative.
Interpretation of Results
The presence of one HLA-B*5701 allele is sufficient to predispose to ABC-HSR, because HLA expression is codominant. As reviewed above, HLA is extremely polymorphic; therefore, most individuals will be heterozygous at this locus and carry only one copy of the HLA-B*5701 allele. Laboratories may offer HLA-B*5701 allele detection but not all assays directly detect HLA-B*5701. Assays that rely on the detection of the HCP5 rs2395029(G) allele that is in linkage disequilibrium with HLA-B*5701 and is an indirect test for HLA-B*5701 are available in the USA. In the initial study using this approach, 100 % sensitivity and 99.4 % specificity were observed [19]. However, there are now multiple reports that HLA-B*5701 and the HCP5 rs2395029(G) allele are not in complete linkage disequilibrium and patients can be positive for HLA-B*5701 but negative for HCP5 rs2395029 [23, 24].
Although HLA-B*5701 allotype has a negative predictive value approaching 100 %, a negative HLA-B*5701 result does not absolutely preclude the development of ABC-HSR. Therefore, careful clinical observation for signs and symptoms associated with a hypersensitivity reaction remain a vital component of patient care. Abacavir therapy should be withdrawn in individuals with symptoms of a hypersensitivity reaction regardless of the HLA-B*5701 genotype result. Skin patch testing can be used to immunologically confirm ABC-HSR, but has no usefulness in prospective screening. As is typical for most HLA associations with disease or ADRs, the positive predictive value is often low and has been estimated to be only 47 % in this case [13]. In other words, only about half of patients that carry an HLA-B*5701 allele would be expected to develop ABC-HSR when placed on abacavir therapy. Clearly, other genetic or environmental factors contribute to the risk of developing ABC-HSR in patients who are HLA-B*5701 positive.
Hepatitis C and IL28B Genotyping
Chronic hepatitis C (HCV) infection affects millions of individuals worldwide with a substantial risk for liver cirrhosis and hepatocellular carcinoma decades after infection. Although the incidence of HCV infection has decreased, the number of patients that will be diagnosed and must be treated is expected to increase for years to come. Current therapeutic regimens can produce a sustained viral response (SVR) and slow or prevent these long-term complications [25]. Current therapy is based on the combined use of interferon and ribavirin (RBV) for infections with HCV genotypes 2 through 6, and beginning in 2011, the combined use of interferon, RBV, and a protease inhibitor for HCV genotype 1 infections [26].
The first important biomarker for guidance of therapy was pre-therapeutic viral genotyping. Genotype 1 infections require longer treatment and have substantially lower rates of SVR. Identification of HCV genotypes 2 or 3 infection allows for a reduction in the length of interferon RBV treatment to 24 weeks compared to 48 weeks for other genotypes. More recently, measurement of the kinetics of viral clearance during therapy has been implemented as standard of care to predict the likelihood of SVR for all HCV genotypes. A rapid viral response (RVR) defined as a fall in HCV viral load to undetectable levels after 4 weeks of therapy strongly predicts SVR. Although RVR is less likely to be achieved with an HCV genotype 1 infection, achieving an RVR is the best predictor of SVR compared to HCV genotype or host physiologic markers. This observation suggests the existence of host factors which affect the likelihood of achieving an SVR. Until recently, no host genetic markers have been available to predict the likelihood of response or drug toxicity prior to therapy.
In late 2009, a series of papers was published by several groups that performed GWAS seeking biomarkers associated with HCV treatment outcome. Several SNPs were identified on chromosome 19 near the IL28B gene that were associated with both spontaneous and treatment-induced HCV clearance (Fig. 20.2) [27–31]. Two SNPs, rs12979860 and rs8099917, have been identified by multiple investigators as having the strongest predictive values for achieving an SVR in patient cohorts of varied ethnic background and have emerged as candidates for IL28B-associated genotyping assays. For the rs12979860 C/T SNP those with the CC genotype are two-fold more likely to achieve an SVR and 2.5 times more likely to clear HCV spontaneously, compared to CT heterozygotes or TT homozygotes [28, 29]. For rs8099917, the TT genotype predicts likelihood of an SVR, while TG heterozygotes and GG homozygotes have an increased risk of treatment failure [27].
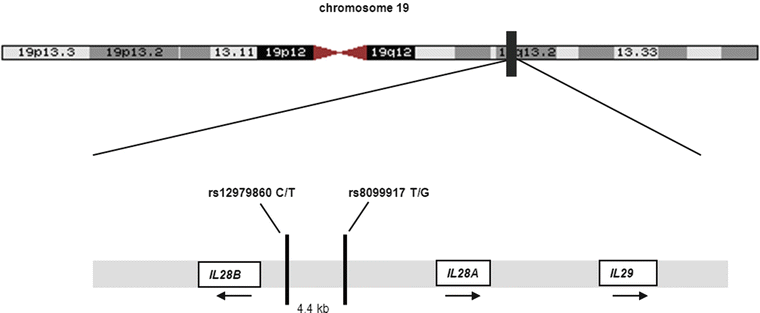

Figure 20.2
Location of SNPs in the IL28 region
These associations more powerfully predict spontaneous clearance and therapeutically driven SRV than all previously described host markers including gender, age, liver steatosis, and insulin resistance [32, 33]. In addition, they account for a significant component of the long-appreciated poor treatment outcomes for individuals of African descent. African-American populations have a much higher prevalence of the rs12979860 TT genotype compared to Caucasian or Hispanic populations. African Americans who carry the CC genotype fare better than those carrying CT or TT genotypes but still worse than Caucasian or Hispanic population with the same genotype, suggesting that IL28 explains much but not all of the long-observed differences in HCV spontaneous clearance and treatment response [29].
The mechanism by which IL28B genotype exerts its effects remains elusive. The two SNPs, rs 12979860 and rs 8099917, are in linkage disequilibrium, approximately 4 kb apart, and both are located upstream of IL28B. The IL28 gene encodes a type III lambda interferon. Lambda interferons, like alpha interferons, stimulate antiviral effects through the JAK-STAT signaling pathway, but use a different cell surface receptor that is preferentially expressed in hepatocytes. Lambda interferons can inhibit HCV growth in vitro, but the precise mechanism for clearance of HCV in vivo remains uncertain [34, 35]. Further complicating interpretation is that both SNPs associated with HCV response are located upstream of the IL28B gene and not in the gene itself. This finding suggests that these SNPs affect transcription of the gene. However, initial messenger RNA expression studies have yielded conflicting results. Although the underlying mechanism has not been clarified, the predictive power of these IL28B-associated SNPs for HCV clearance has been unequivocally substantiated.
Clinical Utility
The significance of the IL28B genotype was originally discovered in patients infected with HCV genotype 1 who were undergoing standard-of-care (SOC) treatment with interferon and RBV. Subsequent studies have investigated IL28B genotype in patients infected with HCV genotypes 1, 2, and 4. Although the results from these investigations support the overall conclusion that favorable IL28B genotype is predictive of likelihood of response to treatment, IL28B genotype has lower predictive value in patients infected with HCV genotypes other than 1. Clinical trials and one study have investigated the utility of IL28B genotyping in the era of direct-acting antiviral therapies. Data demonstrate that those with a favorable IL28B genotype are more likely to have abbreviated therapy under this new treatment regimen [36–38]. Individuals with the favorable IL28B genotype have higher SVR rates with triple therapy as compared to SOC; therefore, no recommendation can be made for one therapy over another based on IL28B genotype.
In summary, IL28B genotyping is a strong pretreatment predictor of treatment response to interferon and RBV therapy, as well as protease inhibitor triple therapy, in patients infected with HCV genotype 1. Predictive value is lower in patients infected with HCV genotypes 2 and 3 [26, 33]. Currently, insufficient evidence exists to recommend one therapy over another for HCV genotype 1 or to recommend a specific duration of therapy based on IL28B genotype alone. IL28B genotype should be considered, if information regarding the likelihood of response and probable duration of response is needed [26].
Available Assays
Both TaqMan allelic discrimination and dual-color fluorescence resonance energy (FRET) probe assays for IL28B genotyping have been published and are commercially available [39].
Interpretation of Results
Caution must be taken in interpreting IL28B genotype results. First, IL28B genotype is primarily a predictive marker for likelihood of response and duration of treatment, but cannot be used to recommend a particular therapy and does not directly dictate duration of treatment. Second, not all individuals with a favorable genotype will achieve a positive treatment outcome, and conversely not all individuals with a risk genotype are destined to fail therapy. Third, IL28B genotype assays are now commercially available that genotype for the rs12979860 or the rs8099917 SNP or both, and it is clinically important to know which SNP is the target. These two SNPs are in linkage disequilibrium with IL28B, but the degree (strength) of linkage disequilibrium between these two SNPs varies with ethnic background [29, 31, 39, 40]. The two SNPs provide interchangeable information in Caucasians, but not in African Americans. Because both SNPs are tag SNPs and neither is the causal variant, it is not well established which of the two SNPs is more reflective of observed response in patients who are discordant for these two SNPs.
Future Directions
HCV and ITPA
GWAS for HCV infection and therapy markers discovered that polymorphisms in the inosine triphosphatase (ITPA) gene were associated with hemoglobin concentrations in patients treated with RBV. Follow-up studies identified two variants, rs1127354 and rs7270101, that cause inosine triphosphatase (ITPAse) deficiency and protect patients from RBV-induced hemolytic anemia [41–45]. The rs1127354 C-to-A polymorphism is a missense mutation and the rs7270101 is an A-to-C splice mutation, both of which are significantly and independently associated with enzyme deficiency and protective effects. The biological mechanism is not well defined, but the protective effects have been confirmed. Genotyping assays for ITPA polymorphisms are available clinically on a limited basis.
Antiretroviral Therapy
One of the most promising areas of pharmacogenetics is antiretroviral therapy. The introduction of highly active antiretroviral therapy (HAART) in the mid-1990s for treatment of HIV infection has proven to be extremely effective and has virtually transformed HIV infection into a manageable, chronic disease. Because HIV is never cleared, long-term antiretroviral therapy (ART) is necessary for suppressing viral replication and is often complicated not only by emergence of viral resistance, but also by the development of severe toxic syndromes and unexpected health consequences. For example, extended use of ART correlates with an increased risk of developing accelerated atherogenesis and cardiovascular complications (events). Consequently, the link between genetic variations in lipid metabolism or transport genes and dyslipidemia has been examined by many investigators. Polymorphisms in APOC3, APOE, and APOA5 genes are believed to significantly contribute to increased triglyceride concentrations in patients on long-term ART, particularly those treated with ritonavir [46–49].
Other host-treatment associations of note include (1) HLA-DRB*0101 class II allele and nevirapine-associated hypersensitivity; (2) atazanavir- and indinavir-induced hyperbilirubinemia associated with UGT1A1 primarily but also P-gp (formerly MDR1) SNPs; (3) tenofovir renal proximal tubulopathy associated with multidrug-resistance protein (MRP2) transporter variation; and (4) efavirenz-associated neurotoxicity attributed to CYP2B6 differences [46, 47]. Genetic screening has been proposed for both UGT1A1 and CYP2B6 prior to atazanavir and efavirenz treatment, respectively. However, even for these well-established associations clinical utility of genetic testing remains controversial, because it is becoming increasingly clear that variation in more than one drug-metabolizing gene and multiple mechanisms may affect the pharmacokinetics of any one drug. The relative contribution of variation in all genes that impact metabolism of a drug has to be determined before utility of genotyping for one or multiple markers can be demonstrated. Currently, the majority of these associations are of research interest only due to the many limitations of pharmacogenetics, as discussed above. Perhaps the foremost limitations are the lack of widespread applicability and well-designed cost-effectiveness studies. Nevertheless, HLA-B*5701 is a great example that genetic associations can be successfully translated into clinical practice if the appropriate studies are conducted. There is considerable anticipation that personalized HAART pharmacogenetics will be possible in the near future.
Tuberculosis
Tuberculosis (TB) is an infectious disease with a long history. There is resurgence in the global morbidity and mortality caused by TB, primarily due to the emergence of antibiotic-resistant Mycobacterium strains and HIV coinfection. Globally, the prevalence of TB has been estimated at 30 %, with the highest burden in developing nations. SOC treatment for TB is a combination of multiple drugs: rifampicin, streptomycin/ethambutol, and pyrazinamide combined with isoniazid. Serious dose-dependent side effects associated with isoniazid therapy, mainly hepatotoxicity, were apparent as early as the 1970s. It is now well accepted that polymorphisms in NAT2 are responsible for the differential extent of N-acetylation of isoniazid and observed phenotypes. Genetic testing for this marker could reduce dose-dependent ADRs in slow metabolizers while increasing efficacy in fast metabolizers or acetylators. Appropriate patient dosing based on genetic testing likely would increase patient compliance with the added benefit of minimizing drug resistance. Drug resistance is a growing problem and has led to combination therapy as the mainstay for TB treatment. Although NAT2 testing is not recommended prior to prescribing isoniazid, pharmacogenetic information regarding the role of NAT2 in isoniazid metabolism is included in US drug labels. NAT2 genotyping or phenotyping is not widely available.
Malaria
Malaria is one of the deadliest infectious diseases known to man, with an enormous human and financial toll. Ironically, one of the earliest examples of an ADR associated with host genetics was the recognition that those who have glucose-6-phosphate dehydrogenase (G6PD) deficiency develop severe hemolytic anemia when administered primaquine, an antimalarial drug introduced in 1950 [50]. Importantly, G6PD deficiency is still relevant to developing efficacious antimalarial drugs, and as recently as 2008, complications due to G6PD deficiency caused withdrawal of chlorproguanil/dapsone therapy [51–53]. Antimalarial drugs have been available for decades and were initially effective and relatively inexpensive. However, monotherapy approaches to treatment led to widespread resistance. Hope was restored around 2000 with the development of artemisinin combination therapies (ACT). ACT is predicated on different classes of drugs with different modes of action for parasite elimination, impeding the emergence of drug-resistant parasites. At the moment, the three ACT options widely utilized for TB management are amodiaquine and artesunate, artemether-lumefantrine, and artesunate-mefloquine [54].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


