Virus Assembly
Eric Hunter
Virus assembly, a key step in the replication cycle of any virus, involves a process in which chemically distinct macromolecules are transported, often through different pathways, to a point within the cell where they are assembled into a nascent viral particle. A diversity of strategies and intracellular assembly sites are employed by members of the various virus families to ensure the efficient production of fully infectious virions. Nevertheless, a virus, irrespective of its molecular structure (membrane enveloped or nonenveloped) or the symmetry with which it assembles (icosahedral, spherical, or helical), must be able to take advantage of the intracellular transport pathways that exist within the cell if it is to achieve this goal. The end product of this selection process is assembly of each virus at a defined point within the cell. This chapter will focus on the cell biology of these intracellular targeting events, the intermolecular interactions that mediate targeting, and the assembly steps themselves.
Most viruses encode a very limited number of gene products. They therefore depend on the cell not only for biosynthesis of the macromolecules that constitute the virus particle but also for the pre-existing intracellular sorting mechanisms that the virus utilizes to achieve delivery of those macromolecules to the sites of virion assembly. Because these are the same sorting mechanisms that the cell uses to delineate its subcellular organelles, the viral macromolecules must possess targeting signals similar to those of the components of those organelles. For a virus such as adenovirus, which assembles its nonenveloped capsids in the nucleus, this means that, following translation in the cytoplasm, each of the structural proteins of the mature virus must have the necessary protein-targeting information to be efficiently routed through the nuclear membrane to the assembly site. The situation is more complicated for a membrane-enveloped virus, such as influenza virus. For this virus, which assembles and releases virions from the apical surface of the epithelial cells that it infects, there is a necessity to ensure that the surface glycoproteins of the virus are correctly sorted by the secretory pathway of the cell to apical membranes. In addition, nucleocapsids, assembled in the nucleus, must be transported into and through the cytoplasm to the same location. As will be discussed later, the intracellular site at which the final phase of assembly and budding of an enveloped virus takes place is most often defined by the accumulation of the viral glycoproteins at a specific point in the secretory pathway of the cell. This also implies that there is specific molecular recognition of the virally encoded, membrane-spanning envelope components by the cytoplasmic nucleocapsids for a productive budding process to occur. Thus, interactions between proteins of viral and cellular origin, between viral proteins and nucleic acids and lipids, and between the viral proteins themselves are at the heart of the assembly process.
Partitioning of Proteins within the Cell
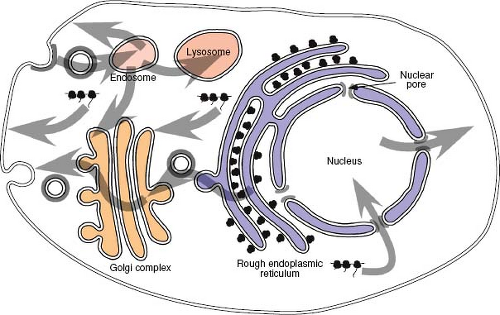
For an actively growing eukaryotic cell, there is a constant need to transport proteins and nucleic acids from their site of synthesis to the specific intracellular domains where they must function (Fig. 6.1). Proteins synthesized on cytosolic ribosomes, for example, must be transported to specific regions of the cytoplasm, to mitochondria and into the nucleus, whereas those synthesized on membrane-bound ribosomes will enter the secretory pathway and be targeted to specific organelles along the way. At the same time, messenger RNAs (mRNAs) and integral RNA components of the ribosome, or ribosomal RNAs (rRNAs), must be transported out of the nucleus. Proteins that participate in these intracellular trafficking processes have evolved to contain specific motifs that ensure the correct localization of the protein within the cell. Viruses have similarly evolved to take advantage of these pre-existing
pathways to accumulate, in specific locations, the necessary components for assembly of a nascent virus.
pathways to accumulate, in specific locations, the necessary components for assembly of a nascent virus.
Nuclear Import and Export of Proteins and Nucleic Acids
The Nuclear Pore Complex
The nucleus is segregated from the cytoplasm by an inner and outer membrane; thus, access to and egress from this subcellular domain is mediated by specialized structures termed nuclear pore complexes (NPCs).202 There are approximately 3,000 NPCs on the nuclear envelope of an animal cell, and each provides a proteinaceous channel between the nucleus and cytosol. The NPC itself is a very large structure, with a molecular mass exceeding 50 mDa that exhibits eightfold symmetry and is constructed of multiple copies of approximately 30 different proteins called nucleoporins (Nups). Negative stain and cryo-electron microscope reconstructions of these complexes reveal a 125-nm diameter core structure in which eight spokes in a radially symmetrical arrangement join to form three main rings surrounding a central channel of approximately 35 nm.3 Attached to both faces of the central framework are peripheral structures, cytoplasmic filaments, and a nuclear basket assembly, which interact with molecules that transit the NPC.58 The channel is filled with flexible, filamentous FG-Nups, which are characterized by regions of multiple Phe-Gly repeats and form a virtual gate restricting transport into and out of the nucleus.181 Small molecules and proteins may be able to passively diffuse through the NPC; however, it acts as a molecular sieve for macromolecules. Larger proteins and macromolecular assemblages must be actively moved through what is clearly a dynamic, malleable transporter structure that has the capacity to accommodate macromolecular complexes with diameters of up to nearly 35 nm.154
Nuclear Localization Signals
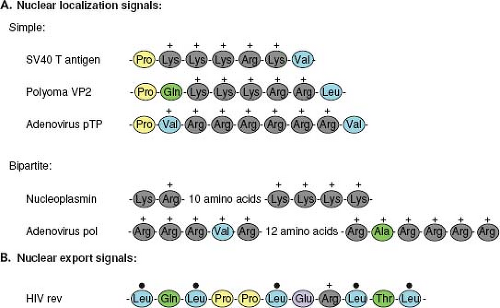
Proteins that are actively transported into or out of the nucleus are characterized by the presence of amino acid motifs that allow them to interact with the nuclear transport machinery. For import into the nucleus, these motifs are termed nuclear localization signals (NLS) and for export, nuclear export signals (NES). NLS motifs, such as that first identified in the SV40 T antigen,95 are not only necessary for nuclear localization of the proteins in which they are present but are also sufficient to actively direct large foreign proteins, such as β-galactosidase, into the nucleus. Although there is no conservation of sequence in different NLS motifs, they are generally short (<20 amino acids), rich in basic amino acids, and
frequently preceded by proline residues (Fig. 6.2A). Some NLS, such as that in the adenovirus DNA-binding protein, are bipartite and require two separate short clusters of basic residues to be functional.238
frequently preceded by proline residues (Fig. 6.2A). Some NLS, such as that in the adenovirus DNA-binding protein, are bipartite and require two separate short clusters of basic residues to be functional.238
To be exported from the nucleus, proteins contain an NES. This nuclear transport signal is also short (∼10 amino acids) and contains a pattern of conserved leucines134 (Fig. 6.2B). Some proteins, such as Rev of human immunodeficiency virus type 1 (HIV-1), possess both an NLS and an NES and appear to shuttle back and forth between the cytoplasm and the nucleus.124
Nuclear Transport Pathways: In and Out
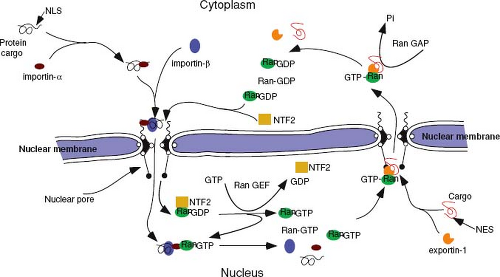
Nuclear import is a two-stage process. In the first stage, the newly synthesized NLS-containing protein interacts with cytosolic receptor proteins that then bind to phenylalanine-glycine (FG) repeat containing Nups (FG-Nups) that make up the filaments on the cytoplasmic side of the NPC – a process referred to as docking. This complex is then translocated, in an energy-independent process, through the nuclear pore into the nucleus, where the complex is disassembled, allowing the transported protein to become functional.
The best-characterized protein import receptor is importin-α (also named karyopherin-α). After binding its NLS-containing cargo, importin-α interacts with importin-β, which then mediates docking with the NPC.98 Most nuclear transport receptors belong to one large family of proteins (karyopherins), all of which share homology with importin-β (also named karyopherin-β). Members of this family have been classified as importins or exportins on the basis of the direction that they carry their cargo. Importins and exportins are regulated by the small guanosine triphosphatease (GTPase), Ran, which is highly enriched in the nucleus in its GTP-bound form. Importins recognize their substrates in the cytoplasm and transport them through nuclear pores into the nucleus. In the nucleoplasm, RanGTP binds to importins, inducing the release of their cargoes. In contrast, exportins interact with their substrates only in the nucleus in the presence of RanGTP and release them after GTP hydrolysis in the cytoplasm, causing disassembly of the export complex (Fig. 6.3).204 Thus, the directionality of transport is regulated by whether Ran is complexed with guanosine diphosphate (GDP) or GTP.
Active transport of large molecules in either direction across the nuclear pore involves interaction with the FG-Nups. The FG repeats in these filamentous proteins provide binding sites for the nuclear transport receptors as well as other molecules, such as nuclear transport factor 2 (NTF2), involved in this process. The exact mechanism by which the importin-cargo complex is carried through the nuclear pore is not known. It is likely, however, that the process involves a series of docking and release cycles with the FG-Nup proteins that make up the transporter machinery within the nuclear pore channel.199,202
The Secretory Pathway of the Cell
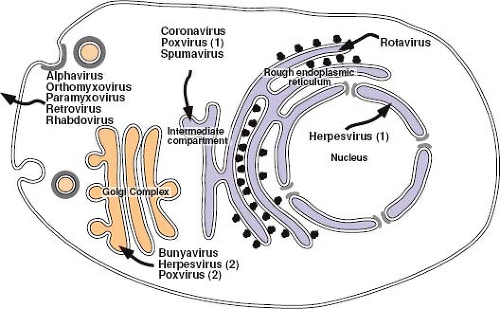
Proteins of both viral and cellular origin that are destined for the outer membrane of the cell travel along a highly conserved route known as the secretory pathway. This complex series of membrane-bound subcellular compartments, through which proteins pass sequentially, includes the endoplasmic reticulum (ER), an intermediate membrane compartment, and the cis-, medial-, and trans-compartments of the Golgi apparatus
(see Fig. 6.1). Proteins that traverse this pathway, such as the envelope glycoproteins of viruses, enter via the ER. Insertion of proteins into the ER occurs during translation through a process termed translocation. The ER network of tubules and sacs defines a unique environment in which protein modification and folding can occur isolated from the cytoplasm. Because polyribosomes, in the process of translating proteins that are translocating into the secretory pathway, are bound tightly to the ER membrane, regions containing them are known as the rough ER.
(see Fig. 6.1). Proteins that traverse this pathway, such as the envelope glycoproteins of viruses, enter via the ER. Insertion of proteins into the ER occurs during translation through a process termed translocation. The ER network of tubules and sacs defines a unique environment in which protein modification and folding can occur isolated from the cytoplasm. Because polyribosomes, in the process of translating proteins that are translocating into the secretory pathway, are bound tightly to the ER membrane, regions containing them are known as the rough ER.
Translocation
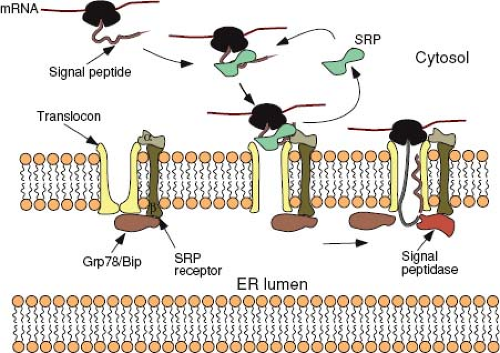
Translating ribosomes are directed to the ER membrane by a short sequence in the nascent polypeptide known as the signal sequence. In most proteins, this 15 to 30 amino acid sequence, which contains a core of hydrophobic amino acids, is located at the N-terminus of the protein. Shortly after the signal peptide emerges from the ribosome, it is bound by a ribonucleoprotein (RNP) complex known as the signal recognition particle (SRP). Binding of SRP transiently arrests any further translation and directs the ribosome to an SRP-receptor on the ER (Fig. 6.4). Both SRP and its receptor have GTP-binding components, and the presence of this nucleotide is essential for efficient targeting.187
Following the initial docking of the translationally arrested complex, the ribosome becomes tightly associated with the ER membrane via the translocon—a gated, aqueous, protein channel that spans the ER membrane. Concomitant with this process, the SRP and its receptor are released, the signal peptide is introduced into the channel, and translation resumes. The components of the translocon have been identified through biochemical approaches in mammalian cells and genetic approaches in yeast and are comprised of a heterotrimeric complex known as the Sec61p (composed of Sec61 alpha, beta, and gamma chains). Based on the crystal structure of the archaebacterial Sec61 homolog, SecYβγ, this complex forms a 40 Å × 40 Å structure with a pore-like central cavity and a single potential lateral opening to allow transition of membrane-spanning domains into the lipid bilayer.195,239 Additional proteins (e.g., the translocating chain-association membrane or TRAM protein) are necessary for optimal translocation. It is unlikely that the central pore of the translocon ever allows free diffusion between the cytosol and the lumen of the ER, because these compartments are chemically distinct. A constriction in the channel appears to be plugged by a short helix, and widening of the constriction as well as displacement of the plug have been linked to conformational changes induced by SRP binding to the complex.219 GRP78 (BiP), a member of the Hsp70 family of chaperone proteins located in the lumen of the ER, plays multiple roles in gating the channel and facilitating translocation. The chaperone is then poised to facilitate the folding of the nascent polypeptide chain as it emerges from the pore, although
this binding is not essential for translocation to proceed. The signal peptide, in those proteins with a transient N-terminal sequence, is cleaved from the rest of the polypeptide by a complex of five proteins called the signal peptidase shortly after it enters the lumen of the ER (see Fig. 6.4).
this binding is not essential for translocation to proceed. The signal peptide, in those proteins with a transient N-terminal sequence, is cleaved from the rest of the polypeptide by a complex of five proteins called the signal peptidase shortly after it enters the lumen of the ER (see Fig. 6.4).
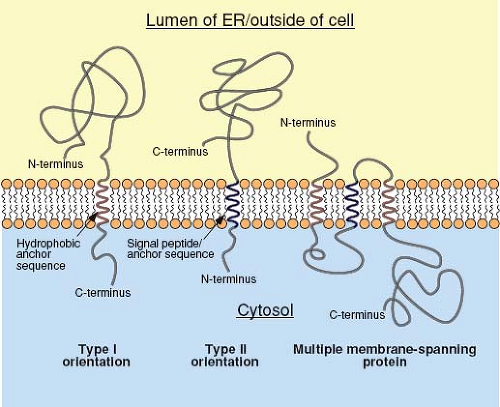
For secreted proteins, translocation of the polypeptide through the translocon continues until the entire protein is present in the lumen of the ER. In contrast, integral membrane proteins, such as the envelope glycoproteins of viruses, contain a stop-transfer, membrane-anchor sequence that is generally located toward the C-terminus of the protein. Following the translation of this short (∼25 amino acids) and mostly hydrophobic sequence, the translocon undergoes a conformational change that allows the membrane-spanning domain to be associated directly with the lipid bilayer. The product of this process is a type I integral membrane protein (Fig. 6.5), such as the influenza virus hemagglutinin (HA), in which the N-terminal ectodomain is in the lumen of the ER and the C-terminus is in the cytoplasm. In some proteins, such as the influenza virus neuraminidase (NA), a longer N-terminal signal peptide also functions as the membrane anchor. In this case, the signal peptide is not cleaved from the polypeptide chain and the sequences C-terminal to it are translocated into the ER lumen, resulting in a type II orientation. Multiple membrane-spanning proteins, such as the M protein of the coronaviruses, appear to possess hydrophobic sequences that are alternatively recognized as signal and stop-transfer sequences, and translocation of such proteins may involve multiple Sec61 heterotrimers.195
Posttranslational Modifications
Protein Folding and Quality Control
Proteins enter the lumen of the ER in an unfolded state and the process of folding into a transport-competent conformation is facilitated by interactions with molecular chaperones and folding enzymes located there. This collection of proteins includes BiP, calnexin (Cnx), calreticulin (Crt), GRP94, and protein disulfide isomerase (PDI). In addition to assisting in folding the nascent molecules, these proteins retain incompletely folded molecules in the lumen and act as a quality control system for the secretory pathway.20 Oligomeric proteins, such as the receptor/fusion proteins of enveloped viruses, also assemble into their quaternary conformation in this compartment. For many of these molecules, oligomerization appears to be a prerequisite for transport out of the ER.
Glycosylation
Most proteins that traverse the secretory pathway are modified by the addition of oligosaccharide side chains either to the amino group of asparagines (N-linked glycosylation) or through the hydroxyl group of serines or threonines (O-linked glycosylation). N-linked moieties are added co-translationally in the lumen of the ER, where mannose-rich
oligosaccharides are transferred by oligosaccharyltransferase from a lipid (dolichol) carrier to asparagine residues present in NXS/T motifs (where X is any amino acid but proline) within the protein. Trimming of terminal glucose and mannose residues from the branched oligosaccharide occurs in the ER and is closely linked with the Cnx- and Crt-mediated quality control process.20 Further trimming of mannose residues followed by addition of other sugars (N-acetylglucosamine, galactose, fucose, and sialic acid), to yield complex oligosaccharide structures, occurs in the Golgi complex. O-linked oligosaccharides are also added in this organelle.
oligosaccharides are transferred by oligosaccharyltransferase from a lipid (dolichol) carrier to asparagine residues present in NXS/T motifs (where X is any amino acid but proline) within the protein. Trimming of terminal glucose and mannose residues from the branched oligosaccharide occurs in the ER and is closely linked with the Cnx- and Crt-mediated quality control process.20 Further trimming of mannose residues followed by addition of other sugars (N-acetylglucosamine, galactose, fucose, and sialic acid), to yield complex oligosaccharide structures, occurs in the Golgi complex. O-linked oligosaccharides are also added in this organelle.
Transport Through the Secretory Pathway
Transport of soluble and membrane-spanning proteins from one compartment of the secretory pathway to the next is mediated by the formation of coated membrane vesicles that travel to and fuse with the target organelle. Thus, once the process of protein folding and quality control has been completed in the ER, proteins are sequestered into these transport vesicles prior to transit to the Golgi complex. The processes of cargo protein selection, budding, targeting, and fusion are probably all mediated by specific protein constituents that define the different transport vesicles involved in shuttling proteins between components of the secretory pathway.86,210 In the case of ER-to Golgi-transport, the vesicles have coat protein complex II (COPII),43 whereas retrograde transport of vesicles from the Golgi to the ER, as well as anterograde transport through the Golgi, is mediated by COPI coats.149 Budding is initiated at specialized regions of the ER (transitional ER) when a small myristoylated protein (SAR1) is converted to the GTP-bound form, allowing it to bind to the membrane and recruit coat proteins (Sec23, Sec24, Sec13, and Sec31) in a stoichiometric manner.43 Formation of the coat itself induces membrane curvature and vesicle budding. Sorting signals displayed on the cytosolic surfaces of transmembrane protein cargo direct it into COPII vesicles, in some instances through physical association of the Sec23/Sec24 components in a sorting signal-dependent manner.100 Soluble proteins and transmembrane proteins lacking COPII sorting signals depend on a diversity of transmembrane adaptor proteins that can link them to the budding machinery. These include the endoplasmic reticulum–Golgi intermediate compartment-53 (ERGIC-53) and p24 family
of receptor proteins, as well as a set of multiple membrane-spanning ER vesicle (Erv14p, 26p, and 29p) proteins that can facilitate concentration of soluble proteins in COPII vesicles.43
of receptor proteins, as well as a set of multiple membrane-spanning ER vesicle (Erv14p, 26p, and 29p) proteins that can facilitate concentration of soluble proteins in COPII vesicles.43
Membrane receptors that mediate docking of the transport vesicle with the target organelle are also incorporated into the coat and appear to define the specificity with which the cargo protein is delivered. Rab-GTPases and tethering proteins appear to play an important role in defining the initial vesicle-target interactions,79,210 whereas soluble N-ethylmaleimide-sensitive factor (Nsf) attachment protein receptors (SNAREs) are generally accepted to mediate the final stage of vesicle docking and the subsequent membrane fusion events that are critical to transport.16,79 A vesicle-specific SNARE (v-SNARE) interacts with a target-membrane–specific SNARE (t-SNARE) complex (generally comprised of three peptides) during this process. Two additional proteins, the Nsf and soluble Nsf attachment proteins (SNAPs), act to disassemble the complex following fusion, allowing the SNARE components to be recycled.79,86
Protein Localization
Subcellular localization of proteins within the secretory pathway appears to be determined by a combination of sorting/targeting signals that mediate interactions with the coat complex for inclusion in a transport vesicle and retention signals that localize the protein to a specific compartment within the secretory pathway. Localization is enhanced by the interplay of anterograde and retrograde transport that allows retrieval of proteins inadvertently transported beyond their target location. The classical example of this is the KDEL peptide sequence found on soluble proteins that are localized to the lumen of the ER.131 Proteins containing this sequence are efficiently retrieved from the cis-Golgi by the KDEL receptor, which is incorporated into COPI vesicles for trafficking back to the ER. Similarly, membrane-spanning proteins localized to the ER have, at the C-terminus of the cytoplasmic domain, a dilysine (KKXX) COPI-binding motif, which ensures their efficient retrieval from the Golgi complex.84 This type of motif is utilized by the primate foamy viruses to concentrate the envelope glycoprotein complex (gp80/gp48) in the ER/intermediate compartment (IC), where virus budding occurs.66 For integral membrane proteins, retention signals often appear to be associated with the membrane-spanning domain(s) of the protein, as is the case for the coronavirus M protein, and may reflect preferred association with specific lipid compositions of the membrane within a particular component of the secretory pathway.
The Golgi Complex
The Golgi complex represents a unique organelle within the secretory pathway in that it is comprised of a series of membrane-bound compartments that are the sites for specific biochemical modifications to proteins and oligosaccharides, as well as locations where specific protein-sorting decisions are made. Proteins transported from the ER enter the Golgi complex via the cis-Golgi network and, after traversing the cis-, medial-, and trans-cisternae, exit via the trans-Golgi network.179 Each of the compartments provides a spatially distinct site for maintaining an ordered set of enzymes involved in the process of oligosaccharide maturation. They are also the sites at which proteins undergo O-linked glycosylation, through the addition, at certain serines and threonines, of monomeric sugar residues. It is in the trans-Golgi cisternae and trans-Golgi network where viral glycoprotein precursors, such as the Env polyprotein of the retroviruses, are cleaved to their mature forms through the action of members of the furin family of proteinases—enzymes that normally function to process cellular substrates such as polypeptide hormone precursors. This cleavage event is critical for the generation of a biologically functional glycoprotein and thus for virus infectivity.
Intracellular Targeting and Assembly of Virion Components
Viruses can be nominally divided into two groups based on the presence or absence of a lipid bilayer envelope. The nonenveloped viruses can assemble in the cytoplasm or nucleus and generally, for those that propagate in animal cells, exhibit icosahedral symmetry (Chapter 3). For these viruses, the viral structural proteins and genomic nucleic acid must be targeted to or retained at the subcellular domain at which assembly occurs. Enveloped viruses, by their very nature, must acquire a lipid bilayer from one of the cell’s membranes during the process of assembly. In some viruses, such as the herpesviruses and some retroviruses, this envelopment step takes place after the assembly of an intact capsid shell, whereas for others the processes of envelopment and capsid assembly occur concomitantly. Some viruses undergo transient envelopment and in some cases re-envelopment during the process of assembly.
For nonenveloped viruses, the tightly assembled structure of the icosahedral shell forms a protective coat that prevents degradation of the genome by environmental factors. For enveloped viruses, the integrity of the nucleocapsid structure is less critical because the membrane provides a barrier to external degradative enzymes.
Assembly of Nonenveloped Viruses in the Nucleus
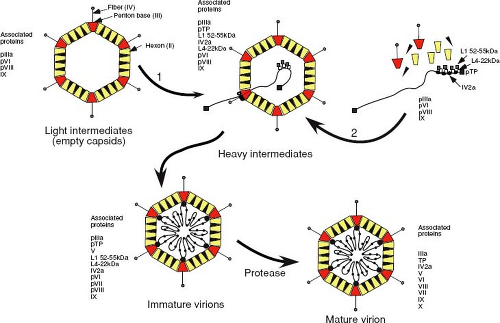
Adenoviruses are nonenveloped icosahedral viruses, 70 to 100 nm in diameter, that have a protein shell surrounding a DNA core. The protein shell (capsid) is composed of 252 capsomeres, of which 240 are hexons and 12 are pentons. Each penton consists of a five-subunit base (polypeptide III) and a trimeric fiber (polypeptide IV) that extends out and away from the shell. The hexon capsomeres are comprised of trimers of three tightly associated molecules of polypeptide II (Fig. 6.6).
For a nonenveloped virus such as adenovirus, which replicates exclusively in the nucleus, there is a strong dependence on nuclear targeting/transport pathways to export newly synthesized mRNAs out of the nucleus and to import structural proteins back into the nucleus. Nuclear import of the major capsid protein—hexon or polypeptide II—depends on the involvement of a second adenovirus protein, the pVI (precursor) polypeptide, which acts as a nucleocytoplasmic shuttling adapter and provides the necessary NLS for transporting the hexon into the nucleus.229 Trimer formation, in turn, depends on yet another virus-encoded, chaperone-like protein—L4 100K—which transiently binds to the newly synthesized hexon monomer and mediates its association with two additional monomers.24 Thus, the most abundant structural protein of the adenovirus capsid needs to interact with two additional virus-encoded factors to attain the correct tertiary structure and subcellular location for assembly.
The two other proteins that form the 12 vertices of the capsid—penton and fiber—appear to assemble independently in the cytoplasm. Mutations in the C-terminus of the penton that block assembly into pentamers do not prevent transport into the nucleus,96 indicating that each monomer has an active NLS. In contrast, the fiber must form trimers to be efficiently transported to the nucleus,146 even though this protein has an active NLS located at its N-terminus. It seems likely that the penton base and fibers are transported independently into the nucleus and assemble into the intact penton at the site of assembly.
Although two distinct pathways for adenovirus capsid assembly have been postulated from a large body of work in this area, it now appears that an empty procapsid is first formed around scaffolding proteins, in a manner similar to that observed with DNA phages. Results from experiments that combined kinetic labeling with temperature-sensitive replication mutants and additional site-directed mutants have yielded the assembly scheme shown in Figure 6.6. Several viral products appear to act as scaffolding proteins, around which shells are assembled and that facilitate the encapsidation process. These proteins are present in the intermediate capsid-like particles but are absent once DNA has been encapsidated.39 Disruption of adenovirus capsids with denaturants results in the release of groups of nine hexons that are associated with the faces of the icosahedron and lack the peripentonal hexons. Under acidic conditions, these nanomers can reassemble to form icosahedral shells that lack the 12 vertices, which would normally be composed of the penton and the five peripentonal hexons, raising the possibility that hexon nanomers are intermediates of adenovirus capsid assembly. Following procapsid assembly, DNA and associated core proteins are then subsequently packaged into these empty shells to yield immature virions that then undergo proteolytic maturation. Recent studies have demonstrated that the approximately six to eight copies of the pIV2a protein, which binds to the packaging sequence, are present at a single apex of the mature virion.29 This, and the fact that the pIV2a protein contains motifs (the Walker A and B boxes) associated with the binding and hydrolysis of adenosine 5´triphosphate (ATP), suggests that it may represent the ATP-hydrolyzing component required to drive DNA through a portal structure in the procapsid. Consistent with this, mutations that either prevent synthesis of pIV2a or prevent binding
of ATP to the protein block genomic packaging and result in the assembly of empty procapsids.150
of ATP to the protein block genomic packaging and result in the assembly of empty procapsids.150
Assembly of Enveloped Viruses in the Nucleus
Two enveloped animal virus families—the herpesviruses and the orthomyxoviruses—utilize cell components that are located within the nucleus in their replication and initiate their assembly within that compartment. In addition to importing into the nucleus the necessary components for assembly, these viruses must also export large nucleoprotein complexes back out into the cytoplasm.
In some respects, the herpesviruses represent a hybrid between a nonenveloped virus, such as adenovirus, and a more conventional enveloped virus, such as a retrovirus, in that they utilize the compartments of the secretory pathway to transport large capsid structures from the nucleus to the outside of the cell. Members of this group of large viruses assemble, within the nucleus, an icosahedral capsid shell that is 160 Å thick and 1,250 Å in diameter. The major component of this protein shell is pUL19, which forms both the pentameric and hexameric capsomeres necessary to assemble the icosahedral structure. Associated with the outer surface of the hexamers is the abundant small protein pUL35. Two additional proteins—pUL38 and pUL18—in a 1:2 ratio, form heterotrimeric triplexes that fit between and link together adjacent capsomeres.140 Scaffolding proteins are essential for herpes simplex virus type 1 (HSV-1) capsid assembly/maturation; in their absence, incomplete and aberrantly shaped capsids are assembled. As with adenovirus, the major capsid protein lacks a nuclear targeting signal, and its transport into the nucleus requires an interaction with either the scaffolding protein (pUL26.5) or the triplex protein pUL38, which presumably provide the necessary NLS. pUL18 similarly requires pUL38 for nuclear localization, whereas pUL35 appears to be directed there via its interaction with pUL19.170
Studies of virus-infected cells together with in vitro assembly studies have provided valuable insights into the assembly process.123,142 These studies point to a pathway in which pUL19, pUL38, and pUL18 assemble around a scaffold to form an icosahedral but predominantly spherical procapsid—the B-capsids identified by electron microscopy. Although not required for capsid formation, in its presence the portal complex apparently initiates capsid formation leading to its incorporation into the nascent capsid.141 Cleavage of the scaffolding protein at a site near its C-terminus by the viral protease removes a 25 amino acid sequence that is necessary for binding to pUL19. The resulting disassociation and release of scaffold allows for packaging of the viral DNA genome and induction of maturation of the capsid into a more angular icosahedral structure, the previously identified C-capsids. DNA enters the procapsid through a unique vertex composed of the portal protein pUL6, which is assembled into rings composed of 12 subunits to form the portal complex.143 The pUL6 portal resembles the connector or portal complexes employed for DNA encapsidation by double-stranded DNA bacteriophages such as φ29, T4, and P22. In the absence of an active proteinase, the scaffolding proteins remain associated with the protein shell, preventing packaging of the viral DNA, and the procapsid is unable to mature.144
Unlike adenoviruses, which accumulate in the nucleus and are released on lysis of the cell, herpesviruses exit the nucleus by budding into the lumen of the nuclear membrane. This process depends on the products of two highly conserved genes—UL31 and UL34—that encode a phosphoprotein and a type II membrane protein, respectively. Nuclear localization of pUL31 depends on its interaction with pUL34.97 Although both proteins are present in the primary enveloped virions present in the lumen of the nuclear membrane, they are absent from mature virions, consistent with a model in which herpesvirus virions are first enveloped at the inner nuclear membrane, de-envelop by budding through the outer nuclear membrane and are re-enveloped by Golgi membranes in the cytoplasm. The complexities of this interaction and subsequent steps in assembly will be discussed later in this chapter.
For influenza virus to take advantage of its unusual capacity to “steal” the capped 5´ ends of host cell mRNAs to initiate its own mRNA synthesis, transcription and viral RNA replication must occur in the nucleus (Chapters 5, 40 and 41). Genomic (minus sense) RNAs are replicated by a different mechanism to yield templates for mRNA synthesis as well as progeny viral genomes. The eight viral RNA segments of this virus are packaged into individual RNPs containing the four proteins of the transcriptase complex (PB1, PB2, PA, and NP, each containing a functional NLS sequence) and the nuclear export protein (NEP, previously NS2), but are not exported into the cytosol until late in infection when the viral matrix protein begins to be synthesized.113 Under conditions where matrix synthesis is inhibited, the viral ribonucleoproteins (vRNPs) accumulate in the nucleus, tightly associated with the nuclear matrix. The block to export can be relieved by expression of matrix from an independent vector.22 It has also been shown that the vRNPs remain in the nucleus of cells if NEP is not encoded by the virus. NEP does not interact directly with vRNPs; rather, it mediates (RanGTP-dependent) formation of a bridge between the cellular export receptor Crm1 and the N-terminal domain of M1, which in turn binds to the vRNP via its C-terminal domain. This daisy-chain complex of (Crm1–RanGTP)–NEP–M1–vRNP is likely what mediates the export of vRNP across the nuclear envelope.2,137 Matrix association with the vRNPs also appears to be important for preventing their re-entry into the nucleus, because conditions such as acidification or mutations that promote dissociation of M1 allow the RNPs to be reimported.228 Thus, the matrix protein of influenza virus is a key modulator of vRNP transport into and out of the nucleus.
Assembly of Viruses in the Cytoplasm
Targeting and import of proteins into the nucleus or secretory pathway involves well-characterized motifs on the proteins involved; therefore, the processes by which proteins are targeted to destinations within the cytoplasm remains for the most part obscure. Nevertheless, most viruses, even those that are nonenveloped, initiate or complete their assembly in association with membranes of the secretory or endocytic pathways, although the intracellular pathways that function to transport their capsid components and genomes to these sites have not been defined. Reoviruses are the only animal viruses that appear to complete their assembly entirely in the cytoplasm without the involvement of membranes. Genome replication and virus assembly both occur in specialized areas of the cytoplasm known as virus factories or viroplasms. The virus nonstructural proteins NSP2 and NSP5 appear to play a critical role in establishing these sites of virus replication and can form morphologically similar structures when expressed in the absence of other viral proteins.158 It is likely that they are responsible for recruitment of the other viral proteins and viral
nucleic acid to these sites, thereby avoiding the complexities of transporting virion components to multiple separate cytoplasmic assembly sites following translation.
nucleic acid to these sites, thereby avoiding the complexities of transporting virion components to multiple separate cytoplasmic assembly sites following translation.
Intracytoplasmic Transport and Assembly of Retroviral Capsids
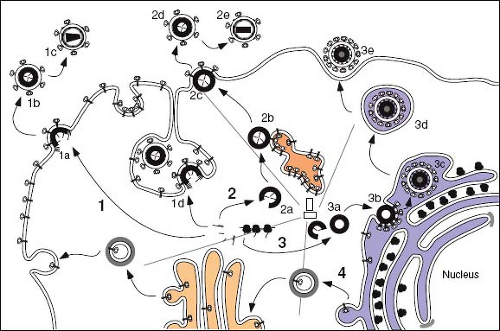
Retroviruses are enveloped viruses that, for the most part, complete their assembly by budding through the plasma membrane of the infected cell. For these viruses, the immature capsid of the virus is assembled from polyprotein precursors that must be transported through the cytoplasm to the inner leaflet of the membrane. The viral glycoproteins, on the other hand, must be transported through the secretory pathway of the cell to the cell surface, where they co-localize with the nascent, membrane-extruding capsid (Fig. 6.7). All replication competent retroviruses contain four genes that encode the structural and enzymatic components of the virion. These are gag (capsid protein), pro (aspartyl proteinase), pol (reverse transcriptase and integrase enzymes) and env (envelope glycoprotein) (Chapter 47). However, the product of the gag gene has been shown to possess the necessary structural information to mediate intracellular transport, to direct self-assembly into the capsid shell, and to catalyze the process of membrane extrusion known as budding.183 For most retroviruses, the nascent Gag polyproteins are transported to the plasma membrane, where assembly of the capsid shell and envelopment occur simultaneously (see Fig. 6.7, Pathway 1). Viruses that undergo this type C form of morphogenesis include members of the alpha- and
gammaretroviruses. Lentiviruses and deltaretroviruses assemble their capsids in a similar fashion in most cell types. In the second morphogenic class of retroviruses, the type B/D class, the Gag precursors are targeted first to an intracytoplasmic site, where capsid assembly occurs. These assembled immature capsids are then transported to the plasma membrane, where they undergo budding and envelopment (see Fig. 6.7, Pathway 2). Viruses that undergo this process of assembly and release include members of the betaretroviruses. Members of the spumavirus family also assemble immature capsids in the cytoplasm but are targeted to the ER or ERGIC for envelopment (see Fig. 6.7, Pathway 3).
gammaretroviruses. Lentiviruses and deltaretroviruses assemble their capsids in a similar fashion in most cell types. In the second morphogenic class of retroviruses, the type B/D class, the Gag precursors are targeted first to an intracytoplasmic site, where capsid assembly occurs. These assembled immature capsids are then transported to the plasma membrane, where they undergo budding and envelopment (see Fig. 6.7, Pathway 2). Viruses that undergo this process of assembly and release include members of the betaretroviruses. Members of the spumavirus family also assemble immature capsids in the cytoplasm but are targeted to the ER or ERGIC for envelopment (see Fig. 6.7, Pathway 3).
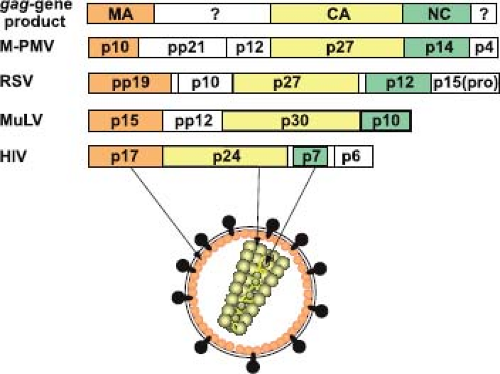
Whereas the size and protein content of the precursor varies between different retroviral families, at least three gag-encoded proteins are found in all retroviruses: the matrix protein (MA), the capsid protein (CA), and the nucleocapsid protein (NC). In addition to these functionally conserved domains, the Gag precursor can, depending on the virus encoding it, contain additional peptide sequences (Fig. 6.8) whose functions in virus assembly and future cycles of infection are only now being resolved.
The detailed mechanisms by which the capsid precursor proteins are directed to the site of assembly are only now starting to be elucidated in molecular detail; however, the process is mediated primarily by the MA domain of the Gag precursor. In most retroviruses, the matrix protein contains two elements involved in plasma membrane targeting. The first of these is an N-terminal myristic acid, which is thought to insert into the hydrophobic lipid bilayer. The second is a surface patch of basic amino acids that are hypothesized to mediate the initial interaction of Gag with the negatively charged, phospholipid head groups of the membrane. Mutations that interfere with either myristoylation or the charged residues can abrogate plasma membrane targeting, and in some instances, the mutated Gag precursors are targeted to internal membranes.57
In the betaretroviruses, where capsid assembly and virus budding are discrete events, a genetic dissection of this process has shown that Gag-containing precursors express a dominant sorting signal (the cytoplasmic targeting/retention signal [CTRS]) that targets the proteins to the initial assembly site.28 Recent studies suggest that the CTRS interacts with Tctex-1, a light chain of the microtubule-associated dynein motor, and directs nascent Gag proteins and translating polysomes to the centriolar region of the cell, where capsid assembly occurs.194,222 A point mutation within the Mason-Pfizer monkey virus (M-PMV) CTRS domain can abrogate intracytoplasmic targeting and results in the type C–like transport of precursors to the plasma membrane, where efficient capsid assembly occurs.81 Efficient transport of wild-type capsids out of the assembly site depends on both the presence of the M-PMV Env protein and endosomal trafficking, and appears to reflect a requirement for Gag-Env interactions at the pericentriolar recycling endosome.193 Interestingly, studies in murine leukemia virus (MuLV), in which the viral RNA was tagged for visualization by fluorescence microscopy, have suggested that prebudding complexes of Env, Gag, and RNA associate with late endosomes and are routed in this way to the plasma membrane.8 It is likely that similar preassembly complexes also participate in the intracellular transport of HIV-1 Gag proteins, because their transport is modulated by the cellular adaptins AP-1, AP-2, and AP-3, and intracellular interactions with Env direct Gag assembly to specific plasma membrane regions of the cell.5,49 Although in macrophages HIV-1 Gag was initially thought to be targeted to a late endosomal compartment for intracellular assembly and budding, more recent studies suggest that these compartments are derived from deep invaginations of the plasma membrane5,11 (see Fig. 6.7, Pathway 1d). Targeting of the plasma membrane by HIV-1 Gag for assembly in part reflects specific MA domain recognition of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], which is enriched there. Depletion of this lipid component by overexpression of the cognate phosphatase (5-phosphatase IV) redirects HIV-1 assembly away from the plasma membrane to internal membranes.5
Irrespective of the assembly site, Gag precursor proteins must associate in a reproducible fashion to assemble into the nascent capsid. Mutational analyses of gag genes, as well as in vitro assembly studies, have shown that the CA and NC domains of Gag play a critical role in assembly; MA is dispensable for this process.36,57 NC binding to RNA may act to nucleate the capsid assembly process, whereas CA forms a symmetrical hexameric network of proteins. In vitro, the CA protein alone can assemble into tubes that exhibit local sixfold symmetry (hexamers), and cryo-electron tomography studies of immature HIV-1 capsids have revealed a similar but distinct arrangement of the CA domain of Gag. A striking observation in the context of HIV-1 is that released immature virus particles have an incomplete protein shell, with the ordered
Gag lattice covering, on average, only two-thirds of the membrane surface.19,231
Gag lattice covering, on average, only two-thirds of the membrane surface.19,231
For most retroviruses, capsid assembly drives the process of membrane extrusion known as budding. As we will discuss later, for an infectious virus to be formed, the Gag precursors (or for other viruses, NPs) and surface glycoproteins of the virus must be targeted to the same region of the same membrane. In this way, during virus budding, a proper complement of glycoproteins can be incorporated into the nascent virion.
Assembly of Enveloped Viruses at Cellular Membranes
For most enveloped viruses, the location within the cell at which envelopment takes place is determined by the targeting to or retention of the viral glycoproteins at that site. Indeed, with the exception of the retroviruses, the efficiency of virus budding and particle release highly depends on the presence of the envelope glycoprotein(s), and in its absence, few particles are produced. Because the glycoproteins define the site of virus budding, specific interactions between the viral NP and the glycoproteins, sometimes mediated by a matrix protein, must take place to ensure that the genome of the virus is incorporated. For each of the viruses that assemble at intermediate points within the secretory pathway, fully assembled viruses must traverse the remainder of the pathway to be released from the cell (Fig. 6.9). Glycoproteins on these released viruses have complex oligosaccharides and are most likely modified by the Golgi-localized enzymes on the way to the cell surface.
Assembly at the Endoplasmic Reticulum–Golgi Intermediate Compartment
The coronaviruses are positive-stranded RNA viruses with large (30-kb) genomes packaged in a helical nucleocapsid. The nucleocapsid acquires its envelope by budding into the lumen of the ERGIC, a pre-Golgi compartment of the secretory pathway. Coronaviruses invariably encode three envelope proteins. The spike protein (S), which determines the host range of the virus, is a type I glycoprotein that forms the distinct bulbous peplomers of the virus. Expressed independently of the other glycoproteins, infectious bronchitis virus (IBV) S is transported to the plasma membrane, although it does contain both ER retention and endocytosis signals that can redirect it to the ERGIC.106 The most abundant virion protein is the membrane (M) glycoprotein. M spans the lipid bilayer three times, exposing a short N-terminal domain outside the virus and a long C-terminus inside the virion. Because of its abundance and because M is transported to the Golgi complex but not to the surface, it was initially thought to define the site at which this family of viruses was enveloped. Studies, however, have shown that the small envelope protein (E), which is only
a minor component of virions, is the key to defining the site and nature of coronavirus envelopment.77 E is a hydrophobic type I membrane protein that localizes to the ERGIC and induces the formation of tubular, convoluted membrane structures characteristic of virus infection.34,166 The E protein of IBV appears to be retained in the ERGIC by a novel ER retrieval signal (RDKLYS-COOH) and localizes M to this site through intermolecular interactions.102 Co-expression of E, M, and S results in the assembly and release from cells of virus-like particles (VLPs) containing these three viral membrane proteins. The enveloped particles produced by this system form a homogeneous population of spherical particles indistinguishable from authentic virions in size and shape.221 Only M and E are required for efficient particle formation, and expression of E alone can mediate particle release.110 The S glycoprotein is thus dispensable for virus particle assembly but is retained in the Golgi by the M protein, which appears to direct its assembly into virions. M protein also directs the incorporation of nucleocapsids containing the genome-length RNA into virions, and nucleocapsids associated with newly synthesized M protein have been localized to the budding site in the ERGIC.135
a minor component of virions, is the key to defining the site and nature of coronavirus envelopment.77 E is a hydrophobic type I membrane protein that localizes to the ERGIC and induces the formation of tubular, convoluted membrane structures characteristic of virus infection.34,166 The E protein of IBV appears to be retained in the ERGIC by a novel ER retrieval signal (RDKLYS-COOH) and localizes M to this site through intermolecular interactions.102 Co-expression of E, M, and S results in the assembly and release from cells of virus-like particles (VLPs) containing these three viral membrane proteins. The enveloped particles produced by this system form a homogeneous population of spherical particles indistinguishable from authentic virions in size and shape.221 Only M and E are required for efficient particle formation, and expression of E alone can mediate particle release.110 The S glycoprotein is thus dispensable for virus particle assembly but is retained in the Golgi by the M protein, which appears to direct its assembly into virions. M protein also directs the incorporation of nucleocapsids containing the genome-length RNA into virions, and nucleocapsids associated with newly synthesized M protein have been localized to the budding site in the ERGIC.135
Although most retroviruses are enveloped at the plasma membrane of the cell, for most members of the spumavirus genus, this occurs on internal membranes. In human fibrosarcoma cells, primate foamy virus (FV) Gag and Env appear to co-localize predominantly in the trans-Golgi region of the cell;235 however, in other cells, immature capsids appear to bud into the ERGIC region of the secretory pathway. This is consistent with the observation that expression of the primate FV Env complex, gp80/gp48, in the absence of other structural proteins, results in its localization to the ER. A di-lysine ER retrieval motif at the C-terminus of gp48 is responsible for this localization, and mutation of either lysine results in efficient transport of the protein to the plasma membrane. Although a greater fraction of capsids bud from the plasma membrane in mutant virus–infected cells, envelopment of capsids at the ER is still observed.66 A second distinguishing feature of foamy viruses is the dependence of capsid envelopment on Env expression.6 As with the betaretroviruses, assembly of immature FV capsids is targeted to the pericentriolar region of the cell by a CTRS located around an arginine residue at position 50 in Gag.235 They are then transported to the ER (or plasma membrane) for envelopment. In the absence of FV Env, these preassembled capsids do not associate with membranes or initiate budding. Recent experiments have shown that it is the posttranslationally cleaved (148 amino acids long), membrane-spanning, signal peptide domain of the FV Env that mediates capsid membrane association/envelopment; they have also shown that this protein is incorporated into the virus in the process.103
Assembly in the Golgi Complex
Bunyaviruses are negative-stranded, enveloped viruses with a segmented genome that assembles in tube-like virus factories that are built around the Golgi complex and are connected to mitochondria and rough ER226 (see Fig. 6.9). By physically juxtaposing viral RNA replication and assembly, these factories appear to allow accumulation of RNPs that can associate with viral glycoproteins and bud into the lumen of swollen Golgi stacks. The glycoprotein spikes of the best-characterized member of this family, Uukuniemi virus, are comprised of two type I glycoproteins—Gn (previously G1) and Gc (G2)—that determine the site of virus budding. Gn and Gc are co-translationally cleaved from a single precursor protein by signal peptidase, which cleaves after the internal signal sequence that mediates translocation of Gc. The two proteins have been shown to fold with distinctly different kinetics, but once properly folded, they form a Gn-Gc heterodimer that is transported to the Golgi complex. Gc expressed in the absence of Gn is retained in the ER, whereas Gn expressed alone is targeted to the Golgi.71 The Uukuniemi virus Golgi localization signal of Gn has been mapped, through analysis of mutations and glycoprotein chimeras, to the membrane proximal half of the 98 amino acid long cytoplasmic tail of the protein. Glycoprotein retention in this case appears, therefore, to depend on interactions between the cytoplasmic tail of Gn in the Gn-Gc heterodimer with components residing on the cytoplasmic side of the Golgi membrane. However, although all bunyavirus Gn-Gc complexes accumulate in the Golgi, the exact location and nature of the signal(s) that ensure this do appear to differ among the genera.226
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree