Viral Replication Strategies
Sean Whelan
Introduction
Replication of genetic information is the single most distinctive characteristic of living organisms, and nowhere in the biosphere is replication accomplished with greater economy and apparent simplicity than among viruses. To achieve the expression, replication, and spread of their genes, different virus families have evolved diverse genetic strategies and replicative cycles to exploit the biology of their hosts. Despite their comparatively limited genetic repertoire, viruses encode the information necessary to rewire their hosts to become viral factories. The intimacy of this relationship and the co-evolution of virus and host continue to provide unique mechanistic insights into host biology at the molecular, cellular, organismal, and population levels. Understanding this interplay enriches our understanding of the biosphere in general and virus–host relationships in particular, but also creates opportunities for the rational development of antiviral drugs, and for domesticating viruses as expression vectors, live-attenuated vaccines, and pesticides. This chapter provides an overview of the replication strategies of the major virus families that infect vertebrates, attempting where possible to emphasize the general principles that guide and constrain virus replication and evolution.
Viral Genome Diversity and Replication Strategies
Perhaps the most striking aspect of viruses at the molecular level is the diversity of their genome structures and replication strategies. Unlike cellular genomes, which consist uniformly of double-stranded DNA (dsDNA), viral genomes provide examples of almost every structural variation imaginable. As shown in Table 5.1, different families of viruses have genomes made of either double-stranded (ds) or single-stranded (ss) DNA or RNA; of either positive, negative, or ambisense polarity; of either linear or circular topology; and comprising either single or multiple segments. Each variation has consequences for the pathways of genome replication, viral gene expression, and virion assembly. This diversity argues strongly that viruses had several different evolutionary origins and can be thought of in D. J. McGeoch’s evocative phrase as “mistletoe on the tree of life.” Accordingly, viral taxonomy above the family level is patchy, with only 22 of 87 families assigned to the six orders that are currently recognized.57 However, it is likely that more distant phylogenetic relationships will emerge as the number of genome sequences and protein structures increase, and as more powerful comparison algorithms become available.
Unique Biology of Virus Replication
As obligate intracellular parasites, all viruses depend heavily on functions provided by their host cells. This dependence, as well as the extensive metabolic overlap between host and parasite, limits the number of possible targets for antiviral therapy. Nevertheless, almost all viruses encode and express unique proteins, including enzymes, and many viruses exploit pathways of information transfer that are unknown elsewhere in the biosphere. This is particularly evident among the RNA
viruses, which are the only organisms that are known to store their genetic information in the form of RNA. They accomplish this by replicating their genomes via one of two unique biochemical pathways—either by RNA-dependent RNA synthesis (RNA replication), or, among the retroviruses, by RNA-dependent DNA synthesis (reverse transcription) followed by DNA replication and transcription. Both pathways require enzymatic activities that are not usually found in uninfected host cells and must therefore be encoded by the viral genome and expressed during infection. Furthermore, in some families of RNA-containing viruses those unique synthetic processes are required right at the start of the infectious cycle. This necessitates co-packaging of the corresponding polymerase and other associated enzymes with the viral genome during the assembly of viral particles in preparation for the next round of infection.
viruses, which are the only organisms that are known to store their genetic information in the form of RNA. They accomplish this by replicating their genomes via one of two unique biochemical pathways—either by RNA-dependent RNA synthesis (RNA replication), or, among the retroviruses, by RNA-dependent DNA synthesis (reverse transcription) followed by DNA replication and transcription. Both pathways require enzymatic activities that are not usually found in uninfected host cells and must therefore be encoded by the viral genome and expressed during infection. Furthermore, in some families of RNA-containing viruses those unique synthetic processes are required right at the start of the infectious cycle. This necessitates co-packaging of the corresponding polymerase and other associated enzymes with the viral genome during the assembly of viral particles in preparation for the next round of infection.
Table 5.1 Families and Genera of Viruses that Infect Vertebrates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
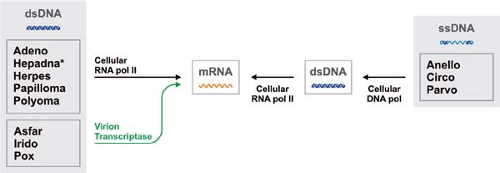
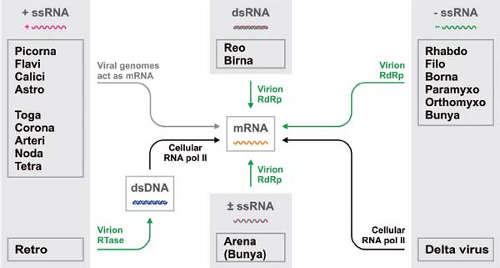
Whatever the structure and replication strategy of their genomes, all viruses must express their genes as functional messenger RNAs (mRNAs) early in infection in order to direct the cellular translational machinery to make viral proteins. The various genomic strategies employed by viruses can therefore be organized around a simple conceptual framework centered on viral mRNA (Figs. 5.1 and 5.2). By convention, mRNA is defined as positive-sense and its complement as negative-sense. The pathways leading from genome to message vary widely among the different virus families and form the basis of viral taxonomy. Although it is generally believed that viruses originated from cellular organisms, perhaps fairly recently in evolutionary times, it remains possible that some RNA viruses are descended directly from a primordial “RNA world” or “ribonucleoprotein world,” which may have predated the emergence of DNA and cells.
Subcellular Sites of Viral Replication
Most DNA viruses of eukaryotes transcribe and replicate their genomes and assemble progeny in the nucleus, the site of cellular DNA transcription and replication. The exceptions are the poxviruses, iridoviruses, and African swine fever virus, which replicate their DNA genomes partly or completely in the cytoplasm. In contrast, most RNA viruses replicate their genomes in the cytoplasm. However, in addition to the retroviruses that integrate DNA copies of their genomes into the host chromosomes, other notable exceptions to this generalization are the orthomyxoviruses, bornaviruses, and many plant-infecting rhabdoviruses, whose linear negative-sense RNA genomes replicate in the nucleus. The circular RNA genome of hepatitis delta virus (HDV), also replicates in the nucleus (Table 5.1). Each site of replication presents distinct opportunities and challenges in terms of which cellular components and pathways are available to be co-opted, and how the synthesis and trafficking of viral proteins, genome replication, virion assembly, and the release of progeny can be coordinated. For example, RNA splicing occurs only in the nucleus, so among the RNA viruses,
this mechanism of accessing more than one open-reading frame in a single transcript can be employed by only the retro-, orthomyxo-, and bornaviruses that transcribe there. It is remarkable that the paramyxoviruses that replicate in the cytoplasm have evolved a transcriptional editing mechanism that achieves a similar result.99 Irrespective of the site of replication (nuclear or cytoplasmic) the viral replication machinery itself is frequently compartmentalized within specific structures or viral-induced organelles. For example, herpesviruses form replication compartments within the nucleus at nuclear speckles,16,88 and many RNA viruses that replicate in the cytoplasm do so in association with membranes or an inclusion-like structure that contains the viral replication machinery.25
this mechanism of accessing more than one open-reading frame in a single transcript can be employed by only the retro-, orthomyxo-, and bornaviruses that transcribe there. It is remarkable that the paramyxoviruses that replicate in the cytoplasm have evolved a transcriptional editing mechanism that achieves a similar result.99 Irrespective of the site of replication (nuclear or cytoplasmic) the viral replication machinery itself is frequently compartmentalized within specific structures or viral-induced organelles. For example, herpesviruses form replication compartments within the nucleus at nuclear speckles,16,88 and many RNA viruses that replicate in the cytoplasm do so in association with membranes or an inclusion-like structure that contains the viral replication machinery.25
Evasion of Host Response to Infection
To ensure their survival, host organisms have evolved a variety of responses to combat viral infection. In turn, many viruses express specific gene products that act to circumvent one or more of those antiviral defense mechanisms. Examination of these measures and countermeasures provides a revealing glimpse into the heart of the host–parasite relationship as it plays out in nature. Host-defense mechanisms can be categorized as innate or adaptive. Among the former, which operate at the cellular level, are apoptosis (programmed cell suicide that limits the spread of infection, see Chapter 8), the induction and action of interferons in vertebrates (inducible cytokines that render cells resistant to infection by inducing a multifaceted antiviral state, see Chapter 8), and RNA interference in plants and invertebrates (a sequence-specific mechanism of RNA degradation, see Chapter 8). Adaptive immune mechanisms operate at the organismal level, and include the cell- and antibody-mediated immune response (see Chapter 9). Increasingly, specific restriction factors have been identified that limit the replication of subsets of viruses. Such factors include tripartite motif containing protein 5 (TRIM5), which appears to trigger the premature disassembly of the incoming human immunodeficiency virus type 1 (HIV-1) capsid to limit the establishment of infection; the apolipoprotein B mRNA editing, enzyme catalytic (APOBEC) family, which induces a biased hypermutation in RNA through its cytidine deaminase activity that converts C to U; and bone marrow stromal antigen 2 (Bst2)/Tetherin, which is incorporated into the membranes of some enveloped viruses resulting in a linking together of budding viral particles. Although a number of other cellular proteins have been termed “restriction factors,” they are distinguished by their dependence on induction by interferon and are therefore not considered here. In different viruses, mechanisms and gene products have been identified that inhibit apoptosis, intercept interferons or suppress their activities, obstruct RNA interference, either evade or suppress different arms of the adaptive immune response, or block intrinsic restriction factors.4,36,46,47,63,70,90,92,93,100
Viruses are sensed by the host in ways that appear to involve recognition of unique signatures present in viral genomes or gene products. Such signatures are termed pathogen-associated molecular patterns (or PAMPS), and are recognized by an array of host pathogen-recognition receptors (or PRRs). Those PRRs include the toll-like receptors (TLRs), which are membrane-associated molecules that sense invading pathogens directly at the plasma membrane or during endosomal transit. The retinoic acid inducible gene (RIG)-like receptors (RLRs), which are cytoplasmic RNA helicases that recognize the products of RNA viral nucleic-acid replication, and the absent in melanoma 2-like receptors that recognize cytoplasmic DNA. Although viral ligands have not been defined, the NOD-like receptors (NLRs), which sense bacterial peptidoglycan, appear to also detect some viruses. A striking example of such PRR function is the detection of off-pathway products of replication such as abortive initiation products, dsRNA, and defective viral genomes, which can serve as ligands for the cytoplasmic sensors RIG-I and melanoma differentiation association protein 5 (MDA-5) to engage in a signaling cascade that leads to the activation of interferon.40 The net result of interferon activation is both the blocking of infection within the cell, and the preactivation of defense mechanisms in neighboring cells to render them less susceptible to infection. The latter is accomplished by the transcription of interferon (IFN)–stimulated genes (ISG), which themselves act to block various steps in the replication cycle of DNA and RNA viruses93 (see Chapter 8). In turn, viruses themselves have evolved countermeasures to such host-defense mechanisms that act to block the induction of IFN itself, or to interfere with specific ISG function.22,43 The elaborate arms race between viruses and their hosts is described in more detail in Chapter 8 and Chapter 9, as well as within the specific chapters dealing with individual virus families. Molecular signatures of this arms race throughout evolution are also visible in the sequences of virus and host genes. Retroviruses provide a unique insight into this, since they integrate into the host genome. Evidence for integration of portions of other viral genomes into the host chromosome including RNA viruses such as bornavirus, and lymphocytic choriomeningitis virus, has also emerged.33,80
Error Prone Nature of RNA Replication
The polymerases that catalyze RNA replication and reverse transcription have minimal proofreading activities. The polymerase error rate of such RNA-dependent RNA polymerases (RdRp’s) and reverse transcriptases is approximately three orders of magnitude higher than that of DNA-dependent DNA polymerases, and approaches the reciprocal of their genome length.28,52,74 The net result is that the genomes of RNA viruses evolve at a much faster rate than those of their hosts. Biologically, RNA viruses therefore represent a swarm of sequences around a consensus sequence or master sequence.31,62 This molecular swarm provides a fertile source of phenotypic variants that can respond rapidly to changing selection pressures by shifting its composition. As a consequence, RNA viruses can evolve up to 1 million times faster than DNA-based organisms. The error prone nature of RNA virus replication is also critical for pathogenesis in infected hosts. The diversity of viral sequences regenerated following bottleneck transmission of HIV in humans,89 and experimental poliovirus infection of mice,84,101 provide striking examples of this in vivo. In the case of HIV, the resulting sequence variation achieved following transmission of a limited number of genomes is enormous and accounts for—among other phenotypes—the rapid escape of the virus from neutralizing antibody, and the escape from antiviral monotherapy.
Such rapid rates of evolution are not without cost for the RNA viruses, however, because higher polymerase error rates impose upper limits on genome size. The combination
of replicative error rate and genome size defines an “error threshold” above which a virus cannot maintain even the sequence integrity of its quasispecies.31 As a result, few RNA virus genomes contain more than 30 kilobases (kb) and most have between 5 and 15 kb. RNA genomes of this size are poised just below their error thresholds, and although their genetic diversity inevitably wastes individual progeny that carry deleterious mutations, the cost is offset by the potential for rapid evolutionary response to changing selective pressures. This positioning of RNA viruses—just below their error threshold—may also present an opportunity for antiviral development. Specifically, therapeutics that lead to an increase in error rate can shift the balance beyond the error threshold toward “error catastrophe.” Indeed evidence has accumulated that this is one such mechanism by which ribavirin, an adenosine analog, may inhibit the replication of some RNA viruses.21 The largest RNA virus genomes currently recognized are those of the coronaviruses, which approach a size of 30 kb. Strikingly, it appears that for coronaviruses the nonstructural protein nsp14 functions as an RNA exonuclease that may function as a proofreading mechanism that could help maintain genome integrity.27
of replicative error rate and genome size defines an “error threshold” above which a virus cannot maintain even the sequence integrity of its quasispecies.31 As a result, few RNA virus genomes contain more than 30 kilobases (kb) and most have between 5 and 15 kb. RNA genomes of this size are poised just below their error thresholds, and although their genetic diversity inevitably wastes individual progeny that carry deleterious mutations, the cost is offset by the potential for rapid evolutionary response to changing selective pressures. This positioning of RNA viruses—just below their error threshold—may also present an opportunity for antiviral development. Specifically, therapeutics that lead to an increase in error rate can shift the balance beyond the error threshold toward “error catastrophe.” Indeed evidence has accumulated that this is one such mechanism by which ribavirin, an adenosine analog, may inhibit the replication of some RNA viruses.21 The largest RNA virus genomes currently recognized are those of the coronaviruses, which approach a size of 30 kb. Strikingly, it appears that for coronaviruses the nonstructural protein nsp14 functions as an RNA exonuclease that may function as a proofreading mechanism that could help maintain genome integrity.27
Levels of Segmentation
Another distinctive feature of eukaryotic cells—besides their partitioning into nuclear and cytoplasmic compartments—has a profound influence on the biology of their viruses. On most mRNAs, eukaryotic ribosomes require a methylated mRNA cap structure at the 5′ end that plays a critical role in signaling the initiation of protein synthesis. As a result, eukaryotes typically conform to the “one mRNA one polypeptide chain” rule; with very few exceptions, each message operates as a single translational unit. Similarly, viral RdRp’s generally appear somewhat restricted in their ability to access internal promoter elements on RNA templates, and this creates a problem of how an RNA virus can derive several separate protein products from a single genome.
Through evolution, different RNA virus families have found three different solutions: fragmentation at the level of proteins, mRNAs, or genes, with some viruses using more than one of those solutions. For example, RNA viruses in the picorna- toga-, flavi-, and retrovirus families rely on extensive proteolytic processing of polyprotein precursors to derive their final protein products.29 Others (in the orders Mononegavirales and Nidovirales) depend on complex transcriptional mechanisms to produce several monocistronic mRNAs from a single RNA template.1,91 Still others (in the reo-, orthomyxo-, bunya-, and arenavirus families, among others) have solved the problem by fragmenting their genomes and assembling virions that contain multiple genome segments, each often representing a single gene.34,69,76 Among plant viruses, such RNA genome segments are often packaged into separate virions, necessitating co-infection by several virus particles to transmit infectivity,107 but the genome segments of animal viruses are typically co-packaged into single virions. In contrast, DNA viruses seldom use either genome segmentation or polyprotein processing. This is likely due to the relative ease with which monocistronic mRNAs can be transcribed from internal promoter elements of dsDNA, and the extensive use of differential splicing of nuclear transcripts to express promoter-distal open-reading frames.
Host Cell Components for Replication
Viruses depend on their host cells to support their replication, and this degree of dependency—to some extent—reflects their genome size. Although all viruses depend on the host translational machinery, large DNA viruses, such as mimivirus, may encode specific initiation factors that may provide a translational advantage for viral genes.18 Entry of viruses into cells usually requires specific host-cell factors, and can require co-opting of cellular endocytic pathways.71 The end point of entry is the release of the minimal viral replication machinery into the host-cell cytoplasm to initiate infection. How viruses establish infection in the hostile environment of the host cell remains one of the least understood steps of the viral replication cycle. The input genomes must either associate directly with ribosomes in the case of positive-strand RNA viruses, or be copied into mRNA, in the case of the negative-strand RNA viruses, dsRNA viruses, and DNA viruses. Because the particle-to-infectivity ratio of some viruses approaches 1:1, this process must be highly efficient despite its inherent challenges. Our knowledge of the subsequent viral rewiring of host-cell structures to establish replication compartments, traffic viral proteins and nucleic acids, and assemble viral particles is also far from complete, but has yielded a wealth of information into host biology as well as that of the viruses themselves. Indeed, study of viruses has contributed enormously to our understanding of promoters, transcriptional enhancers, the mRNA cap structure, RNA splicing, and mechanism of translation. Similarly, critical discoveries in host-cell transport and trafficking pathways including endocytosis, exocytosis, and secretory transport were achieved because of the ability to synchronize infections with viruses. Although systematic approaches including RNA interference (RNAi), proteomics, gene-knockout studies, and microarrays are helping to further transform our understanding of the virus–host interaction at the molecular level, we have yet to understand fully the complexities of the interactions of any virus with its host. Zoonotic viruses must strike a balance for optimal replication in often quite disparate hosts, likely adding further complexity to this intimate relationship. Striking examples of this are provided by members of the Flaviviridae, such as Dengue virus (which replicates in both its mosquito host and animals), and experimentally with many viruses including vesicular stomatitis virus (which replicates in virtually all eukaryotic cells in culture).
Structures and Organization of Viral Genomes
DNA versus RNA Genomes
Among families of viruses that infect vertebrates, those with RNA genomes outnumber those with DNA genomes by about 2 to 1 (Table 5.1); among viruses infecting plants the disparity is even greater. Indeed, no dsDNA viruses of plants are known except for those that like the hepadnaviruses of vertebrates, replicate via reverse transcription (see Chapter 68). This remarkable observation remains to be explained, but it may suggest that non–RT dsDNA viruses arose only after animals and plants diverged. Be that as it may, the prevalence of RNA viruses attests to the evolutionary success and versatility of RNA as genetic material for smaller genomes. As discussed
previously, the high error rates of RNA replication restrict RNA genome sizes to 30 kb or less, whereas proofreading and error repair ensure sufficiently accurate replication of DNA virus genomes as large as that of the 1200-kb megaviruses.3 In addition, the fact that DNA is more chemically stable than RNA likely explains why all known viruses of thermophilic hosts have dsDNA genomes.57
previously, the high error rates of RNA replication restrict RNA genome sizes to 30 kb or less, whereas proofreading and error repair ensure sufficiently accurate replication of DNA virus genomes as large as that of the 1200-kb megaviruses.3 In addition, the fact that DNA is more chemically stable than RNA likely explains why all known viruses of thermophilic hosts have dsDNA genomes.57
Single- and Double-Stranded Genomes
Although all viral genomes replicate via conventional Watson-Crick base pairing between complementary template and daughter strands, viruses that belong to different families encapsidate and transmit different molecular stages of the genome replication cycle. Families of ssRNA viruses outnumber families of dsRNA viruses by almost 10 to 1, roughly the inverse of the ratio between ssDNA and dsDNA viruses. In view of the greater chemical stability of double-stranded nucleic acids of both types, this difference calls for an explanation. Two possibilities seem plausible: First, dsRNA viruses must somehow circumvent the translational suppression that can result from the coexistence of equimolar amounts of the sense and antisense RNAs. How the dsRNA reoviruses solve this problem is addressed in Chapter 44. Second, dsRNA is widely recognized by the cells of higher eukaryotes as a signal for the induction of defense mechanisms that act to suppress viral replication, such as the IFN system in vertebrates (see also Chapter 8), gene silencing in plants, and RNAi in a variety of organisms.15,40,93,106 These effects probably suffice to explain the relative scarcity of dsRNA virus families.
For these same reasons, it is important even for ssRNA viruses to limit the accumulation of replicative intermediates that contain regions of dsRNA, and the strategies to ensure this differs between the positive- and negative-sense RNA viruses. All known positive-strand RNA viruses synthesize disproportionately low amounts of the negative-strand RNA—typically 1% to 5% of the levels of the positive-strand—and thereby minimize the potential for dsRNA accumulation. Moreover, because the replication of these viruses appears to universally occur in sequestered membranous compartments, there appears to be a physical separation of the replicative intermediates from the host-cell cytoplasm, likely reducing the chances of detection.25 In contrast, negative-strand RNA viruses, which need substantial amounts of both positive- and negative-sense RNAs to use as messages and progeny genomes, respectively, prevent the complementary RNAs from annealing to one another by encasing the genomic and antigenomic RNAs with a viral nucleocapsid protein.2,44 Here, RNA synthesis also appears confined at some stages of infection to specific subcellular compartments that may help serve to limit detection of viral products of RNA synthesis by the innate immune system.
Positive, Negative, and Ambisense Genomes
The differences between positive- and negative-strand RNA viruses extend beyond the polarity of the RNA assembled into virions. Positive-sense RNA genomes exchange their virion proteins for ribosomes and cellular RNA binding proteins at the onset of infection. Once synthesized and assembled the virus-specified RdRp and other nonstructural proteins replace the ribosomes to accomplish RNA replication. Virion structural proteins are reacquired during the assembly of progeny virions. In contrast, negative-strand RNA genomes and their antigenomic complements remain associated with their nucleocapsid proteins, both within the viral particles and throughout the viral replication cycle, even during RNA replication. These fundamentally different adaptations can be attributed to the fact that whereas positive-sense RNA genomes must satisfy criteria for translation that are dictated by the host cell, negative-sense RNA genomes must only satisfy the template requirements for the virus-specified RdRp because they are replicated but never translated. Although the precise mechanism by which the protein-coated templates of negative-strand RNA genomes are copied by their cognate polymerases is not fully understood, short naked RNAs that correspond to the terminal promoters can be copied by their viral polymerases.26,59,73 Such experimental evidence is consistent with a model for RNA synthesis in which the nucleocapsid protein is transiently displaced from the template RNA during copying of the genome.
The dsRNA virus genomes are intermediates between the two. The parental genome remains sequestered within a subviral particle during the synthesis of the unencapsidated positive-sense mRNA transcripts, which are replicated to produce progeny dsRNAs only after being assembled into subviral core particles.81 Although the core RdRp’s of each of these viruses are structurally as well as functionally analogous, the distinctions in the genomic structure likely place additional structural constraints on the viral polymerase complexes.
Linear and Circular Genomes
Genome replication not only requires an acceptable error rate as described previously, but must also avoid the systematic deletion or addition of nucleotides. Genome termini are particularly troublesome in this respect, a fact that has been dubbed “the end problem.” For DNA replication, the end problem is exacerbated by the fact that DNA polymerases cannot initiate the synthesis of daughter strands and must therefore use primers, thus creating additional complications of replicating the primer-binding site(s). Among several known solutions, the most economical and widespread in nature is to eliminate the ends altogether by covalently circularizing the genomic DNA, as occurs in the genomes of prokaryotes. Polyoma-, papilloma-, circo-, and anellovirus genomes follow this model, and the dsDNA genomes of herpes and hepadnaviruses, although linear, in virions are covalently circularized before replication. Poxviruses and asfiviruses also have linear dsDNA genomes, but in these cases the individual complementary strands are covalently continuous at the termini of the duplex, which provides another solution to the end problem. A similar close-ended duplex DNA is generated during the replication of the ssDNA genomes of parvoviruses (Chapter 57). Terminal redundancy (iridoviruses), inverted terminal repeats (adenoviruses), and the use of protein primers that do not occlude the binding site (adenoviruses and hepadnaviruses) represent the other ways that DNA viruses have evolved to ensure accurate and complete replication of their genome termini.
Unlike DNA polymerases, most RNA polymerases do not require primers, so RNA genomes are less susceptible to the end problem. Accordingly, most RNA genomes are linear molecules. Covalently closed circular RNAs are found only in HDV in animals (Table 5.1) as well as among the viroids and other subviral RNA pathogens that infect plants. Nevertheless the termini of linear RNA genomes are vulnerable to degradation,
and their replication is likely to be particularly error prone. Consequently, every family of RNA viruses has features designed to preserve the termini of the genome.6 For example, many positive-strand RNA viruses have a 5′ cap structure and 3′ polyadenylate tail that serve to protect eukaryotic RNAs against degradation, and a similar role is likely played by the VPg that is covalently linked to the 5′ end of the picornavirus genomes,64 and by the stable RNA secondary structures present at the 3′ end of the flaviviral RNA and other genomes. The 3′ ends of many plant virus RNAs form clover leaf structures that resemble transfer RNAs (tRNAs) so closely that they are recognized by the cellular tRNA charging and modifying enzymes.30 In addition to playing protective roles, terminal modifications of positive-sense RNAs may also serve to bring their ends together by binding to interacting cellular proteins such as the poly(A) binding protein and cap-binding complex, thereby forming noncovalent functionally circular complexes that may promote repetitive translation by ribosomes and repetitive replication by RdRp’s.49,102
and their replication is likely to be particularly error prone. Consequently, every family of RNA viruses has features designed to preserve the termini of the genome.6 For example, many positive-strand RNA viruses have a 5′ cap structure and 3′ polyadenylate tail that serve to protect eukaryotic RNAs against degradation, and a similar role is likely played by the VPg that is covalently linked to the 5′ end of the picornavirus genomes,64 and by the stable RNA secondary structures present at the 3′ end of the flaviviral RNA and other genomes. The 3′ ends of many plant virus RNAs form clover leaf structures that resemble transfer RNAs (tRNAs) so closely that they are recognized by the cellular tRNA charging and modifying enzymes.30 In addition to playing protective roles, terminal modifications of positive-sense RNAs may also serve to bring their ends together by binding to interacting cellular proteins such as the poly(A) binding protein and cap-binding complex, thereby forming noncovalent functionally circular complexes that may promote repetitive translation by ribosomes and repetitive replication by RdRp’s.49,102
Unlike the genomes of positive-sense RNA viruses, negative-sense and ambisense RNA virus genomes rarely carry covalent terminal modifications. Those RNA genomes show some degree of terminal sequence complementarity that is thought to lead to the formation of a panhandle type of structure that, in the case of the segmented viruses, favors RNA replication. Because the templates are encapsidated by the viral nucleocapsid protein, it is not clear how the RNA bases can engage in base-pairing interactions between the termini. However, complementarity between the genomic termini favors replication and likely promotes polymerase transfer during RNA synthesis to ensure efficient reinitiation of replication. In other solutions to the end problem among the RNA viruses, retroviral genomes are terminally redundant and have direct repeats of 12 to 235 nucleotides at each end that maintain and restore the integrity of the termini during reverse transcription and virus replication (see Chapter 47).
Segmented and Nonsegmented Genomes
As discussed previously, segmentation of RNA genomes is one way to facilitate the production of multiple gene products in eukaryotic cells, but it also means that the various segments must each contain appropriate cis-acting signals to mediate their expression, replication, and assembly into virions. In some virus families whose members have segmented genomes (e.g., the orthomyxoviruses and some reoviruses), these signals comprise conserved sequences at the RNA termini, but in others (e.g., the bipartite nodaviruses and tetraviruses) sequence conservation between the segments is minimal. In these latter cases, the specificity of RNA replication and assembly is presumably dictated by conserved RNA secondary or tertiary structures. Moreover, segmentation of the viral genome requires a level of coordination to ensure that the correct amounts of viral gene products are expressed and to ensure the packaging of multiple genome segments to form infectious virus particles. How such coordination is achieved is not understood. Furthermore, in the case of the negative-sense, ambisense, and dsRNA viruses that have segmented genomes, a mechanism is required to ensure that the polymerase is packaged into the virus particle so that the incoming segments can be transcribed into mRNA. For the dsRNA viruses the polymerase is an integral structural component of the core transcribing particle ensuring that the polymerase and capping machinery are present within the incoming particle. In the case of the arenavirus, Machupo, this is a function of a small viral protein Z, which locks the polymerase on the promoter in an inactive form.60
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree