Type of analysis
Typically tested specimens
Specimen identification
Unknown specimen
Specimen confirmed to be from the known individual(s)
Histologic floater
Macrodissected floater tissue
Main tissue on slide separated from floater tissue
If available, additional specimen confirmed to have tissue from the known individual helps confirm the origin of the main tissue in the block
Pathogenic donor cells in a transplant recipient
Post-transplant tissue with suspected donor cells present
Specimen with only recipient cells (e.g., pre-transplant blood, tissue, or buccal cells)
Specimen with only donor cells
Maternal contamination of a prenatal diagnostic specimen
Prenatal specimen or products of conception
Maternal specimen
Gestational trophoblastic disease
Gestational specimen with suspicious features, microdissected to enrich for gestational cells (villi)
Maternal specimen (blood or carefully microdissected decidua)
Paternal specimen (optional)
For specimen identification testing, the “unknown” DNA from the specimen is compared to “known” DNA from a specimen verified to be from the individual, such as a blood sample or a tissue block from a previous procedure. For mislabeled specimens, additional documentation of specimen handling and processing is useful. In contrast, for the identification of suspected floaters that are not consistent with the remainder of the specimen section, “unknown” DNA from microdissected floater tissue will be tested and compared to “known” DNA from an area of tissue containing tissue consistent with what is expected for the specimen. In this scenario it may also be prudent to analyze a verified alternative sample from the same individual as the current specimen in question as a further confirmation that the “known” DNA did, in fact, originate from the individual whose sample was contaminated by the floater. Confirmation of the actual source of the floater is unnecessary in most cases. In a case of two possibly switched samples, both are “unknown” and can be compared with two “known” samples from the two potential source individuals.
Analysis of maternal cell contamination of a fetal specimen requires a known maternal blood sample in addition to the “unknown” fetal sample. Similarly, diagnosis of a hydatidiform mole requires analysis of both chorionic villi and a known maternal sample. In the typical post-transplant scenario, analysis of the “unknown” tumor specimen is compared with the analysis of “known” specimens from the donor and recipient.
These are only general examples, some of which are illustrated in the cases described later. In fact, identity testing for sample identification is useful in a host of situations, each requiring careful specimen selection on a case by case basis to ensure that the tested specimens reflect the best choices from the available specimens, and that the results of testing those specimens will enable resolution of the clinical question.
Tissue Processing
While specimen handling and processing are major issues for certain types of molecular genetic testing, the use of smaller polymorphic markers allows identity testing to be performed on specimens that would be less than ideal for other clinical molecular tests. Frequently, identity testing for sample identification involves the use of FFPE tissue specimens, which is the routine method for preserving tissue for histopathology in nearly all histology laboratories. Commercial reagents simplify extraction of adequate DNA for identity testing from FFPE tissue sections. However, not all fixatives or chemical treatments used in the histology laboratory are compatible with PCR amplification. Specimens exposed to fixatives containing heavy metals (for example B5 fixative) are not recommended due to inhibition of enzymatic reactions in PCR. Similarly, decalcification solutions are usually acidic as are some fixatives (e.g., Carnoy’s, Zenker’s, and Bouin’s fixatives), which degrade the DNA and render the specimens unsuitable for PCR amplification. Therefore, it is very important to review the tissue processing during selection of tissue blocks for identity testing to avoid incompatible treatments. In extraordinary cases when the only specimen available is suboptimal but the testing will have a significant impact on patient management, testing of the compromised specimen can be considered. In such cases, if there is some amplification of the DNA, it may only be for loci with the smallest PCR target sizes. In some situations, sufficient information may be obtained to provide a limited interpretation, at the risk of over-interpreting potential artifacts that can occur at low DNA concentrations. If critical, testing using SNPs or mitochondrial DNA can be considered since those methods are more tolerant of degraded specimens.
Some staining methods may produce added damage to the nucleic acids. Most hematoxylin and eosin (H&E) stained tissue sections are acceptable [6, 41]. However, because reagents for histochemistry or immunohistochemistry may substantially degrade the nucleic acids or inhibit PCR and the effects of such potential interferences may not have been thoroughly examined during assay validation, due caution should accompany interpretation of STR analysis of previously stained material.
Selection of Tissue for Testing
Identity testing of tissue specimens begins with review of all available material by an anatomic pathologist. After assessing the quantity and distribution of the available material, the best method of isolating DNA from the submitted material is chosen. Available methods range from extraction of DNA from an entire tissue section to extracting DNA from small, isolated areas within a tissue section by macrodissection or microdissection. Regardless of the final method used, the goal of tissue selection is to obtain the appropriate specimens for comparison.
Testing All Tissue Within a Block
When the identity of all of the tissue in a block is in question, several (e.g., three) 10 μm thick section rolls from the FFPE tissue block can be cut and placed directly into microcentrifuge tubes for DNA extraction. This saves time and eliminates the effort of preparing a slide and then scraping the tissue off the slide for DNA extraction.
Testing Areas of Interest Within a Single Block
When different portions of tissue in the same block potentially originate from different sources, macrodissection or microdissection of the tissue from representative slides (see below) is appropriate to separate the tissues for subsequent comparison. Macrodissection after appropriate training and practice can be regularly employed using readily available tools [42]. Microdissection techniques permit more precise selection of cells for testing, but require more expertise and expensive equipment.
When the only slide containing the area of interest has been previously stained and coverslipped, xylenes can be used to remove the coverslip (with a risk of losing the desired cells) for DNA extraction from the stained tissue [6, 41]. Importantly, the scenario of tissue present on only a single level is suggestive of a true “floater” and may not merit further testing because tissue that is truly a part of the specimen is typically present on more than one level of a block.
Macrodissection
For macrodissection, an anatomic pathologist first identifies and outlines representative cellular, nucleated, non-necrotic areas of interest on a stained slide using a permanent marking pen (see Fig. 57.1a). Then, unstained 4–10 μm sections are cut and placed onto glass slides (e.g., 5–15 slides are prepared). The first slide, “cut off the top,” and the last slide “cut off the bottom” are stained with H&E to demonstrate the distribution of the areas of interest across the intervening unstained slides. Identifying the distinct areas of interest is necessary when the tissue on a slide is not homogenous. Regions representing “floaters,” benign, malignant, or other relevant tissues and cells on a slide are delineated with a diamond etching pen (on the non-tissue-containing surface of the slide) or a permanent marking pen on a coverslipped slide (Fig. 57.1a, b). Alternatively, a permanent marking pen can be used to mark areas of interest directly on a faced FFPE block, and this marked area can be manually separated from the remainder of the block. The marked slides are used as guides for identifying and marking the areas of interest on the unstained slides that will be used for dissection and extraction of DNA (Fig. 57.1c). Macrodissection is done by eye or using a low power dissecting microscope and the equivalent circled tissue regions are scraped off the unstained slides (Fig. 57.1d).
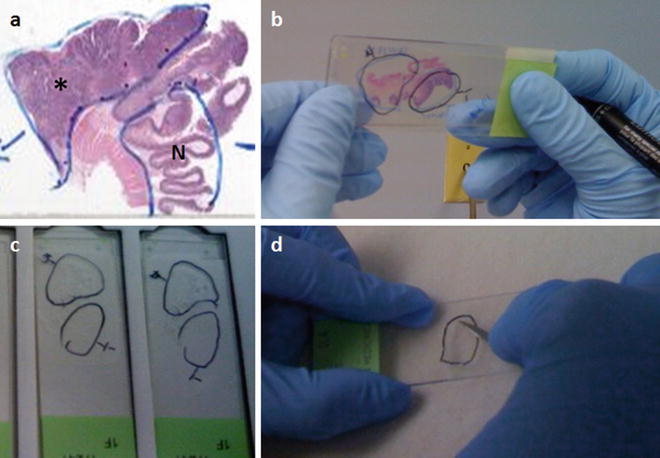

Figure 57.1
Macrodissection can be used to enrich for tumor and/or normal tissue from histologic tissue sections for identity testing, depending on the question being asked. Regions of interest on an H&E stained slide are indicated by an anatomic pathologist by circling the tissue areas of interest using a permanent marking pen, as shown in (a) (*, tumor; N, normal). This functions as a guide slide for marking regions of interest on unstained slides of sections adjacent to the tissue on the guide slide (b). A final H&E stained slide cut after the unstained sections is used to confirm that the tissue of interest is present on all the unstained slides. The marked tissue from the unstained slides (c) can then be individually scraped with a scalpel to isolate the tissue of interest (d)
Microdissection
Diamond etching pens and permanent marking pens are useful for delineating distinct areas on a slide, and in most cases of identity testing they are satisfactory for marking relevant areas to be collected for DNA extraction and comparison. When greater precision is required, microdissection techniques (i.e., use of a higher power microscope to assist dissection) may be used to isolate discrete cells or clusters of cells from a complex tissue section.
Laser Capture Microdissection
Laser capture microdissection (LCM) permits reliable procurement of pure cell populations from tissue sections. The principle advantage of LCM is the ability to isolate clusters of cells or even single cells of interest as a pure population free from contaminating stromal, inflammatory, and other surrounding cell types. Its precision can also complicate analysis by limiting the choice of analysis methods to those that are amplification-based because the amount of material isolated often is minute. Due to the increased expertise and cost associated with the LCM systems, many laboratories are unable to use LCM routinely for identity testing.
LCM systems such as the ArcturusXT™ LCM System (Life Technologies, Grand Island, NY), Leica LMD6500 and LMD7000 (Leica Microsystems Inc., Buffalo Grove, IL), or PALM MicroBeam (Carl Zeiss AG, Oberkochen, Germany) are available. Each system has unique technologies and techniques for separating the selected areas of interest from tissue sections. In general, the systems include an inverted microscope, an infrared laser, control systems for the laser and the microscope stage, and a digital imaging system to permit the user to view and capture images of the microdissected fragments [43]. The systems differ mostly in terms of the slides and other consumables they require to separate and collect the cells.
Chemical Microdissection
An alternative to LCM is The PinPoint Slide DNA Isolation System™ (Zymo Research, Orange, CA) [17]. With this system a solution is applied to the microscopic area of interest. The solution dries into a thin film that captures the underlying cells. The film is lifted with a scalpel and transferred into a tube for DNA extraction. This method is more time consuming than macrodissection of marked slides and requires a specific kit, but because the area of interest is precisely selected by an anatomic pathologist, the possibility that an incorrect area will be used for analysis is reduced.
Interpretation of Results
Specimen Identification
Sample identification determines whether two samples originated from the same or different individuals. Comparison of a tissue or other specimen with an identified specimen from the potential source patient, such as peripheral blood, can be used to verify the source of the specimen in question. Interpretation of the results involves comparison of the alleles (STR or sequence polymorphisms) between the samples. The identification of non-matching alleles, preferably in two or more loci, provides evidence to exclude a potential individual as the source of the sample, or at least to conclude that both samples did not originate from the same individual. The possibility of microsatellite instability, loss of heterozygosity, or chromosomal abnormalities should be considered when malignant tissues are being compared to normal samples using STR analysis, as described further below. If an exact allele match is observed between two samples, then the likelihood that they are from the same person is high, and the matching probability can be determined from published data or manufacturer data for the loci used. The following case is an example of identity testing for sample identification.
Case 1
Two blocks of tissue (Unknown A and Unknown B) obtained from two different individuals were suspected of being switched and mislabeled. An alternate, confirmed sample from each individual (Confirmed A and Confirmed B) was available. DNA was extracted from all four specimens and each was amplified at 12 STR loci. Comparison of the genotypes indicated that the unknown samples had been switched (Table 57.2). Blocks Unknown A and Unknown B were relabeled to accurately reflect their true source, thereby avoiding the diagnoses of the two individuals being switched.
Table 57.2
Case 1 genotypes confirming that specimens were switched
Alleles (repeats) | ||||
|---|---|---|---|---|
Locus | Confirmed A | Confirmed B | Unknown A | Unknown B |
D3S1358 | 17,18 | 15,16 | 15,16 | 17,18 |
TH01 | 6,9.3 | 7,9 | 7,9 | 6,9.3 |
D21S11 | 28,30 | 30,32.2 | 30,32.2 | 28,30 |
D18S51 | 14,16 | 14,15 | 14,15 | 14,16 |
PentaE | 7,11 | 11,16 | 11,16 | 7,11 |
D5S818 | 12,13 | 11,13 | 11,13 | 12,13 |
D13S317 | 9 | 10,11 | 10,11 | 9 |
D7S820 | 12,13 | 10,11 | 10,11 | 12,13 |
D16S539 | 11 | 9,10 | 9,10 | 11 |
CSF1PO | 10,11 | 11 | 11 | 10,11 |
vWA | 14,17 | 15,18 | 15,18 | 14,17 |
TPOX | 8 | 8,11 | 8,11 | 8 |
A “confirmed” specimen is partially a subjective designation due to the possibility of specimen misidentification. As such, in interpretive reports language such as “…block number 1 is identified as originating from patient ‘1’ and block number 2 is identified as originating from patient ‘2’…” makes it clear that even for specimens with presumably confirmed identity there remains the possibility of preanalytical errors resulting in a specimen misidentification.
Analysis of Floaters
To assess the origin of an extraneous, frequently small, tissue fragment (floater) in a tissue section, the genotype of the majority of the tissue on the slide is compared with that of the floater. As with other forms of identity testing, differences in two or more markers should be identified in order to exclude the patient as the source of the floater. Generally it is not necessary or cost-effective to identify the source of the extraneous tissue; however, if desired, possible source cases with similar histology that were processed during the same time period can be tested. As always, histopathologic review of the case to verify the nature of the submitted material and to select the appropriate tissues for analysis is essential before initiating identity testing. The histopathologic review also informs the analysis, as macrodissection is frequently imperfect and a minor component of the predominant tissue’s genotype may be detected in what is intended to be only floater tissue. When mixed genotyping result are obtained, correlation with the histologic picture usually will show intermingled “known” and “floater” cells in the specimen with the mixed genotype result.
Case 2
An isolated fragment of adenocarcinoma was identified in a single block of prostate needle biopsies. The remaining five blocks were completely benign. A “floater” was suspected and the possible source was another prostate biopsy case processed immediately prior to the case with the suspected floater. The adenocarcinoma floater was macrodissected, as well as the majority tissue from the benign case with the floater and the prior case with adenocarcinoma. The three tissue DNAs were tested at 12 STR loci and the results confirmed that the tissue fragment was a “floater” (Table 57.3). As expected, testing of the amelogenin locus confirmed that both samples were from male patients.
Table 57.3
Case 2 genotypes confirming the presence of a floater
Alleles (repeats) | |||
|---|---|---|---|
Locus | Suspected floater | Suspected source of floater | Benign block containing floater |
vWA | 11,18 | 11,18 | 16,17 |
TH01 | 10 | 10 | 6 |
TPOX | 8,11 | 8,11 | 9,11 |
CSF1PO | 11,12 | 11,12 | 10 |
LPL | 11,12 | 11,12 | 10,12 |
F13B
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

| |||