Virus: RTI and CAP
Bacteria: RTI and CAP
Bacteria: VAP and HAP
Adenovirus (ADV)
Bordetella pertussis and Bordetella parapertussis
Acinetobacter spp.,
Human bocavirus (HBoV)a
Chlamydophila pneumoniae
Corynebacterium spp.
Coronavirus (CoV) (OC43, 229E, NL63, HKU-1)
SARS, MERS
Haemophilus influenzae
Enterobacteriaceae (MDRO)
Enterovirus (EV)
Legionella pneumophila
Legionella pneumophila
Human metapneumovirus (HPMV)
Moraxella cattharalis
Pseudomonas spp.
Influenza A (seasonal H1 [A/H1], seasonal H3 [A/H3], (H1N1)pdm09 [A/2009 H1])
Mycoplasma pneumoniae
Stenotrophomonas maltophilia
Influenza B
Neisseria meningitidis
Staphylococcus aureus (MSSA and MRSA)
Parainfluenza virus 1, 2, 3, 4 (PIV)
Staphylococcus aureus (MSSA and MRSA)
Human rhinovirus (HRV)
Streptococcus pneumoniae
Respiratory syncytial virus (RSV)
Since 2008 several clusters of respiratory illness associated with human enterovirus 68 (HEV68) were reported in Asia, Europe, and the USA (MMWR [29]). HEV68, a unique enterovirus with similar biologic features to HRV, was associated with RTIs ranging from relatively mild illness to severe illness requiring intensive care and mechanical ventilation. Severe disease was particularly pronounced in children (MMWR [29]). Although human bocavirus (HBoV) has been implicated as a cause of RTI, in the majority of cases HBoV is detected in conjunction with other viral pathogens and the clinical significance of HBoV is still debated [105, 200]. HBoV is more commonly found in children; however, HBoV can affect persons of all ages. Underlying disease, such as cancer, is associated with severe infections requiring hospitalization.
Generally the CoVs (NL-63, OC43, HKU-1, 229E) cause mild, self-limiting URTIs such as the common cold. However, in the last decade, two CoVs—Severe Acute Respiratory Syndrome (SARS-CoV) and the Middle East Respiratory Syndrome (MERS-CoV) have emerged with a potential pandemic threat due to person-to-person communicability [33]. Both viruses can cause severe lower respiratory tract disease with extrapulmonary involvement and are associated with high case-fatality rates. Although SARS-CoV is currently not circulating, since 2012 MERS-CoV continues to cause outbreaks in the Middle East with secondary spread to Europe, Africa, Asia, and North America.
The prevalence of each viral pathogen can vary depending on environmental conditions (climate, season, geographic location). For example, in the New York City area, adenovirus (ADV) tends to circulate with the same prevalence year round (Fig. 52.1a), while RSV can be detected sporadically year round with the peak season in October through February (Fig. 52.1b) (unpublished data provided by author). The “classic” influenza season begins generally in late October and wanes during April. HMPV prevalence tends to rise as RSV season wanes in February and peaks in the spring, PIV-1 and PIV-4 are most prevalent in the summer through fall, while PIV-3 prevails in the spring and PIV-2 in the fall. Virus prevalence also can be patient population driven (e.g., pediatric, adult, geriatric, outpatient, or inpatient). For example, RSV and HMPV are primarily found in children < 5 years of age, but can cause severe disease in all age groups [64, 65, 94, 97]. In the immunosuppressed population, other less common respiratory viral pathogens, such as herpes simplex virus (HSV) and cytomegalovirus (CMV), must be considered as a potential cause of RTIs.
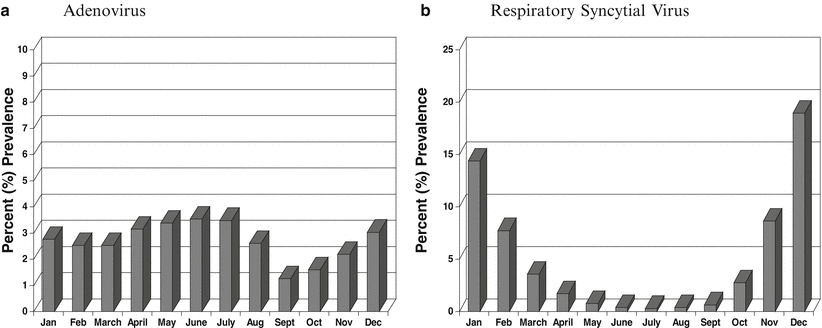

Figure 52.1
Mean prevalence of adenovirus (a) and respiratory syncytial virus (b) by month for the years 2000 through 2011. Adenovirus causes infections throughout the year, while respiratory syncytial virus infections are more seasonal
The major bacterial pathogens responsible for CAP (Table 57.1) include S. pneumoniae (20–60 %), Staphylococcus aureus (methicillin-susceptible [MSSA] and methicillin-resistant [MRSA]) (3–5 %), Haemophilus influenzae (3–10 %), variable gram-negative rods (3–10 %), and rarely Neisseria meningitidis (<1 %) depending on the patient and underlying diseases such as chronic obstructive pulmonary disease (COPD) or asthma [10, 146]. Moraxella cattharalis is generally associated with otitis media in children and exacerbation of asthma and COPD in adults. The classic atypical pathogens that cause CAP include C. pneumoniae, M. pneumoniae, and Legionella pneumophila, which also can be nosocomially acquired in institutional settings [10, 17, 46, 67, 222].
The majority of RTIs (up to 70 %) caused by C. pneumoniae are asymptomatic or have minimal symptoms. C. pneumoniae accounts for 6–20 % of CAP and 5 % of cases of sinusitis, pharyngitis, tracheitis, bronchiolitis, bronchitis, and exacerbations of chronic bronchitis and asthma in both immunocompetent and immunosuppressed persons [17, 42]. C. pneumoniae can be present with other bacterial pathogens in up to 30 % of adult cases of CAP. C. pneumoniae infection can present with varied clinical courses from mild, self-limiting disease to severe forms of pneumonia, particularly in patients with cardiopulmonary disease and in the elderly [17]. All age groups are affected; however, C. pneumoniae infections are rarely found in young children < 5 years of age. However, by age 20, 50 % of persons have detectable antibody levels, with the elderly demonstrating a seropositivity rate between 70 – 80 % [221].
M. pneumoniae is estimated to cause 6–20 % of the cases of CAP [222]. Although most infections are asymptomatic, mild, and often self-limiting, approximately 1–5 % of infections may require hospitalization and can lead to serious extrapulmonary complications. LRTIs are more common in school age children and adolescents, with the prevalence in pediatric LRTI ranging from 10–40 %. M. pneumoniae infection can cause outbreaks in the community and institutions such as schools, prisons, and hospitals.
Legionella spp. causes two distinct clinical entities. Pontiac Fever is a self-limiting flu-like illness and Legionaire’s disease is a severe multisystem disease involving pneumonia. Cases can be sporadic or part of outbreaks due to environmental exposure [51, 67]. L. pneumophila is responsible for 2–8 % of CAP and is responsible for 2–15 % of all CAP that require hospitalization. L. pneumophila (serogroup 1) is responsible for 90 % of the diagnosed disease, most probably because the major diagnostic tests are specific for this serogroup. Risk factors for contracting Legionellosis include smoking, immunosuppression, age ≥ 65, chronic lung disease such as emphysema, diabetes, kidney disease, cancer, or contact with environmental systems such as air conditioning cooling towers, evaporative condensers, whirlpools, and hot spring baths.
Coxiella burnetii, an obligate gram-negative intracellular bacterium has primary reservoirs in cattle, sheep, and goats. Transmission to humans occurs primarily through inhalation of aerosols from contaminated soil or animal waste. Most C. burnetti infections are manifested as Q Fever, a self-limited, influenza-like febrile illness (88–100 %) of abrupt onset, manifested by chills, headache, myalgia, fatigue, and sweats; ([150], CDC MMWR [30]). However, pneumonia is predominant in North America and usually mild in nature. Patients have dyspnea, pleuritic chest pain, and a dry, nonproductive cough. Rarely, C. burnetti infection occasionally can progress to acute respiratory distress syndrome (ARDS) ([150], CDC MMWR [30]).
B. pertussis is the cause of whooping cough [43, 119]. Pertussis-like illness can be attributed to Bordetella parapertussis, Bordetella holmseii, and rarely Bordetella bronchioseptica infections. Overall, Bordetella infections have increased dramatically over the last 10 years due to waning immunity, incomplete antibody response to vaccination with acellular vaccines, or lack of vaccination. Recent epidemics have occurred between 2010 and 2014. In 2010, more than 27,000 cases were reported, of which over 9,000 occurred in California. Nationwide by 2013 more than 28,000 cases were reported to the CDC (http://www.cdc.gov/pertussis/surv-reporting.html). The highest incidence was noted in infants < 1 year of age, but a significant amount of disease occurred among children aged 7–10 years. One large outbreak of pertussis-like illness in Ohio from 2010 to 2011 was attributed to both B. pertussis (68 % of the cases) and B. holmseii (29 % of the cases) [190]. Outbreaks of pertussis continue, highlighting the need for primary vaccination and the administration of “Tdap” booster immunizations.
HAP and VAP are more often associated with drug-resistant, multidrug-resistant, or pan-resistant bacteria, such as MRSA, extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriacae, or carbapenemase-producing Enterobacteriacae, Pseudomonas spp., Acinetobacter spp., and Stenotrophomonas maltophilia [4]. For patients in the intensive care unit (ICU) or immunocompromised patients, other pathogens such as Nocardia spp, Corynebacterium spp., Pneumocystis jiroveci, Fusarium spp., Aspergillus spp, Cryptococcus spp., and the zygomycetes need to be considered in the differential diagnosis.
Clinical Utility
Limitations of Conventional Diagnostic Procedures
Conventional virus detection methods include rapid antigen detection tests (RADTs), direct fluorescent antibody tests (DFAs), rapid cell culture, and traditional tube culture [125]. Although these methods are acceptable diagnostic tools in certain clinical settings, they are often inferior in the breadth of pathogens identified, assay sensitivity, and result turnaround time, when compared to NAATs [18, 28, 39, 73, 74, 79, 125, 157]. The sensitivity and specificity of all the diagnostic tests, but in particular the conventional tests, are highly dependent on the viral target, age of the patient, duration of symptoms prior to sample collection, sample collection methods, and the transport and storage conditions [1, 39, 48, 49, 85, 95, 118, 127, 131, 164].
RADTs are generally the simplest tests to perform, many are waived tests under the Clinical Laboratory Improvement Amendments (CLIA), and results are generally available within 15–30 min. Despite these benefits, RADTS have limited utility due to the narrow scope of pathogens detected (RSV, FluA, and FluB) [125], and modest to poor sensitivities [21, 39, 45, 61, 75, 79, 125, 199] which can range from 50–90 % for RSV [47] and 10–85 % for influenza viruses depending on the comparator method. Overall, the specificities of RADTs are good [21, 39, 45, 61, 75, 79, 125, 199]; however, the specificity for the detection of influenza A (H1N1) pdm09 was significantly lower than previously reported [197, 198]. RADTs generally perform better when testing pediatric samples since children shed higher titers of virus and for longer time periods than adults, especially the elderly [28, 85, 93].
DFAs detect a broader range of viruses (ADV, FluA, FluB, HMPV, PIV-1, PIV-2, PIV-3, and RSV) and can be performed in 30–60 min. The sensitivities of DFAs vary by virus, ranging from a high of approximately 60–85 % for RSV to a low of 50 % for ADV when compared to NAATs [116, 125]. DFAs are generally very specific, although specificity can be dependent on the level of technical expertise of the reader.
Rapid cell culture (Quidel/Diagnostic Hybrids, Athens, OH) can detect ADV, FluA, FluB, PIV-1, PIV-2, PIV-3, and RSV and has demonstrated sensitivities that range from a high of > 80 % for some FluA strains to a low of 50 % for RSV [125, 130, 152] and excellent specificities of greater than 95 % when compared to NAATs. Rapid cell culture is generally positive within 48 h for > 90 % of the seven viruses detected.
Depending on the cell lines used and antibodies available for confirmation, traditional tube culture can have a broader scope of pathogen detection when compared to RADTs, DFAs, and rapid cell culture [125]. Traditional culture will identify ADV, enterovirus (EV), HMPV, FluA, FluB, PIV-1, PIV-2, PIV-3, RSV, and HRV, plus additional viruses associated with lower RTIs in immunocompromised patients, including CMV, HSV-1, HSV-2, varicella zoster virus (VZV). Often laboratories do not specifically screen for HRV by culture, although HRV is the most common respiratory virus detected and has been shown to cause significant and serious disease in young, elderly, immunosuppressed patients, as well as patients with underlying chronic lung disease such as COPD and asthma [83]. In addition, many additional important viruses (229E-CoV, OC43-CoV, NL63-CoV, HKU-1-CoV, SARS-CoV, MERS-CoV), PIV-4, and potentially HBoV) that cause both URTIs and LRTIs are not routinely identified by traditional culture [83, 105, 200]. Finally, due to time-to-virus-detection by traditional culture (generally 3–7 days for most respiratory viruses and 3–4 weeks for slow-growing viruses such as CMV), results are usually not available within a time frame (48 h) that could affect patient management (i.e., initiate appropriate antiviral therapy and/or discontinue inappropriate antibiotic therapy). In summary, RADTs, DFAs, and rapid cell culture may provide results within a clinically relevant time frame but with limited pathogen scope and reduced sensitivity compared to NAATs. In addition, RADTs, DFAs, and rapid/traditional cell culture rarely detect more than one virus from a single sample.
The standard methods for the detection of bacterial pathogens causing pneumonia are gram stain in combination with microbiological culture of lower respiratory tract specimens and blood. Often culture of respiratory specimens is not ordered on hospitalized patients and rarely performed in the outpatient setting. A gram stain result can be available within a few hours but results do not always correlate with culture [10]. Culture and antibiotic susceptibility results are usually available in 2–5 days after sample collection, and detection rates for pathogens are relatively low. A meta-analysis that evaluated 122 reports on CAP for the time period of 1966–1995 showed that a bacterial pathogen was only identified in 18 % of the samples tested [68]. A urinary antigen test for S. pneumoniae offers a substantial improvement over culture, with a sensitivity of 82 % and a specificity of 97 % in bacteremic adults [209, 210]. However, in non-bacteremic adults and in children, both the sensitivity and specificity are lower [210]. False-positive S. pneumoniae antigen tests have been reported relating to antibiotic interference. Legionella spp. are identified by growth on buffered charcoal yeast extract agar [158]. The sensitivity of Legionella culture can vary significantly from <10–80 % and DFA sensitivity from 25–70 % [158]. A urinary antigen test for L. pneumophila improves detection but is suboptimal since detection is limited to serogroup 1. For L. pneumophila serogroup 1 the sensitivity of the test varies from 70 – 100 %. Most laboratories do not culture for Mycoplasma or Chlamydophila [140].
Traditionally, B. pertussis was identified using culture on Bordet–Gengou media and/or DFA. However, the sensitivity of culture ranges from 12 – 60 %, the sensitivity of DFA ranges from 11 – 68 % [119]. Additionally, culture can take many days. Therefore, NAATS have become the gold standard for the rapid and sensitive (70–99 %) identification of pertussis [119].
Serologic antibody testing is available for some of the respiratory pathogens and can provide supplemental information. However, due to a delay in the development of detectable IgM or IgG antibodies for certain pathogens (e.g., L. pneumophila, C. pneumoniae, B. pertussis), usefulness for diagnosis in a clinically relevant time frame is very limited [17, 151, 158, 221]. Shortcomings of serological testing include the timing of the serum samples, difficulty in obtaining appropriately paired serum samples, and the high background of IgG antibody prevalence in some adult populations [140]. Serologic diagnosis can be misinterpreted due to prior immunization or infections, such as in the case of influenza, and require demonstration of a significant rise in antibody titers from initial to convalescent samples. Finally, persons with immune suppression may not develop antibodies or they may be of an insufficient level for detection, limiting the functionality of serology for monitoring vaccine response and for epidemiology studies to determine prevalence rates.
Application of Molecular Assays for the Detection of Respiratory Pathogens
Clinical Utility of NAAT for Respiratory Pathogens
Prior to the 2009 FluA(H1N1) pandemic, the infectious causes of CAP were mostly inferred based on clinical presentation which can be highly inaccurate since some bacteria, atypical pathogens, and many of the respiratory viruses cause illnesses with similar clinical symptoms [177, 194]. One study demonstrated that physicians recognized influenza in only 28 % of hospitalized children and 17 % of non-hospitalized children with laboratory-confirmed influenza when the diagnosis was based only on clinical symptoms [177]. Most diagnostic testing was limited to RADTs for FluA, FluB, and RSV as few hospitals offered comprehensive DFAs, viral culture, or laboratory-developed NAATs.
The superior ability of NAATs to rapidly and accurately detect both known and novel pathogens was best exemplified during the chaos of the 2009 FluA H1N1 pandemic [18, 39, 79, 197, 198]. Fortunately, at the start of the pandemic two NAATs were FDA-cleared for the detection of influenza viruses, the Prodesse PROFLU+ (Hologic, San Diego, CA) for the detection and differentiation of FluA and FluB ([124]; and one highly multiplexed NAAT: the Luminex xTAG RVP Respiratory Virus Panel (Luminex Molecular Diagnostics, Toronto, Canada) [113, 114, 143]. The xTAG assay enabled laboratories to detect FluA(H1N1)pdm09 and differentiate the seasonal FluA H1N1 (FluA-H1) and seasonal FluA H3N2 (FluA-H3), but also identify many other circulating viruses [79, 78]. As shown in Fig. 52.2, the number of samples positive for a respiratory virus increased dramatically with the use of the highly multiplexed xTAG RVP assay (64 %) compared to traditional test methods, including RADTS (17 %), DFA (18 %), and rapid cell culture (31 %) [79]. Interestingly, mixed viral infections containing up to four viral pathogens were identified in hospitalized patients [79]. Similarly, other studies have shown that when broad test panels are used more than one virus will be identified in 3–30 % of respiratory samples [8, 166]. Although the significance of mixed viral infections needs to be more clearly defined, the clinical impact needs to be considered as potentially severe in patients with comorbidities, immunosuppression, or other critical illnesses. During respiratory virus seasons with high influenza rates, patients with the same pathogen often are placed in hospital rooms together (cohorting) due to limited private rooms [19, 39, 40, 62, 126, 155, 165, 217, 219]. The consequences of a second viral infection in an already seriously ill hospitalized patient could be substantial, indicating that comprehensive test panels are essential in this setting.
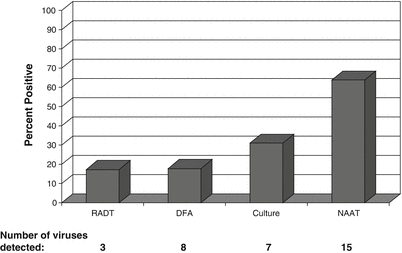

Figure 52.2
Comparison of viral test methods for identification of respiratory pathogens. Percentage of respiratory samples (n = 35,456) positive by each test methods: RADT (rapid antigen detection test) detects three viruses [influenza A (FluA), influenza B (FluB), and respiratory syncytial virus (RSV)]; DFA (direct fluorescent antibody assay) detects eight viruses [adenovirus (ADV), FluA, FluB, human metapneumovirus (HMPV), parainfluenza viruses (PIV) 1, 2, and 3, and RSV]; Culture (R-Mix rapid cell culture) detects seven viruses (ADV, FluA, FluB, PIV-1, PIV-2, PIV-3, and RSV); NAAT (nucleic acid amplification testing) detects 15 viruses (ADV, coronaviruses (CoV: OC43, NL63, HKU1, 229E), enterovirus/rhinovirus group (EV/HRV), FluA (A/H1, A/H3), FluB, HMPV, PIV (1–4), and RSV
Consequently, there has been a major shift in testing practices as numerous FDA-cleared single-and multi-analyte molecular tests for the detection of respiratory pathogens have become available (Table 52.2). Viral respiratory pathogens are particularly suited for detection using NAATs since the number of targets is relatively limited and the detection of a respiratory virus is generally considered diagnostic, although asymptomatic carriage of certain respiratory viruses has been reported in several studies [2, 14, 104, 181]. For the majority of viral and atypical bacterial respiratory pathogens, NAATs offer enhanced sensitivity over culture, RADTs and DFAs (see Tables 52.2 and 52.3 for references), and the specificity varies with the target and assay design but is generally very high. NAATs also are suited for detection of respiratory pathogens that are not routinely or easily cultured (e.g., C. pneumoniae, M. pneumoniae, HBoV, HMPV, and PIV-4), for pathogens dangerous to culture (e.g., SARS-CoV, MERS-CoV), and for pathogens where the time-to-detection by traditional testing is often too delayed to impact patient care (e.g., influenza and CMV by cell culture).
Table 52.2
FDA cleared tests for the detection of bacterial and viral respiratory pathogensa,b
Manufacturer/testc | Amplification and detection platform(s)d | Extraction Platforms | Targetse | Specimen types approvedf | Method(s)g | References |
|---|---|---|---|---|---|---|
Alere i NAT Flu A/B | Alere i Instrument | Included | FluA, FluB | NS | Isothermal amplification, Fluorescence detection | |
Argene/bioMerieux Argene R-Gene Adenovirus Assay | Cepheid SmartCycler | bioMerieux NucliSENS easyMAG | ADV | NPS | Real-Time PCR, Fluorescence detection | [148] |
BioFire/bioMerieux Film Array Respiratory Virus Panel | BioFire Film Array | Included | ADV, CoV (OC43, NL63, 229E, HKU-1), HMPV, FluA (H1, H3, 2009-H1N1), HRV/EV, PIV 1,2,3,4, M. pneumoniae, C. pneumoniae, B. pertussis | NPS | Real-Time RT-PCR/PCR, Fluorescence detection | |
CDC Influenza Divisiong CDC Human Influenza Virus Real-Time RT-PCR Diagnostic Panels 1. Influenza A/B typing Kit 2. Influenza A subtyping Kit 3. Influenza A /H5 (Asian lineage) Kit 4. Influenza B lineage genotyping assay | ABI 7500 Fast Dx | Qiagen QIAamp, Qiagen QIAcube, Roche Magna Pure compact, Roche Magna Pure LC, bioMerieux NucliSENS easyMAG | 1. FluA, FluB 2. A/H1, A/H3, A/2009 H1 3. A/H5N1 (Asian lineage) 4. B/Victoria, B/Yamagata lineages | Varies by test including: NPS, NS, NA, NW, NPS/TS, BAL, TA, BW, VC | Real-Time RT-PCR, Fluorescence detection | NA |
Cepheid 1. XpertFlu Assay 2. Xpert Flu/RSV XC Assay | Cepheid GeneXpert | Included | 1. FluA (A/2009 H1), FluB 2. FluA, FluB, RSV | 1. NPS, NA, NW NPS, NW, NA (in VTM) | Real-Time RT-PCR, Fluorescence detection | |
Focus Diagnostics 1. Simplexa Influenza A H1N1 2. Simplexa FluA/B and RSV 3. Simplexa Flu A/B Direct | 3 M Integrated Cycler | 1 and 2: Qiagen QIAamp Viral RNA 3: Included | 1. FluA (A/2009 H1) 2. FluA, FluB, RSV 3. FluA, FluB, RSV | 1. NPS, NA, NPA 2. NPS, NA, NPA 3. NPS | Real-Time RT-PCR, Fluorescence detection | |
GenMark eSensor Respiratory Viral Panel | Thermocycler GenMark eSensor XT-8 | bioMerieux NucliSENS easyMAG | FluA, (A/H1, A/H3, A/2009 H1), FluB, RSV (A, B), ADV (B/E, C), PIV (1–3), HMPV, HRV | NPS | RT-PCR/PCR, Electrochemical detection of bound signal probes | |
Hologic/Gen-Probe/Prodesse 1. ProFlu+ 2. ProFAST+ 3. ProAdeno+ 4. ProParaFlu+ 5. ProhMPV+ | Cepheid Smartcycler II | bioMerieux NucliSENS easyMAG, Roche Magna Pure LC, Roche Magna Pure Total NA | 1. FluA, FluB, RSV 2. A/H1, A/H3, A/2009 H1 3. ADV 4. PIV 1,2,3 5. HMPV | All NPS | Real-Time RT-PCR/PCR, Fluorescence detection | |
IntelligentMDx IMDx FluA/B and RSV | Abbott m2000rt | Abbott m2000sp | FluA (A/H1, A/H3, A/2009 H1), FluB, RSV | NPS | Real-Time RT-PCR, Fluorescence detection | NA |
Iquum/Roche Liat Influenza A and B | Iquum/Roche Liat Analyzer | Included | FluA, FluB | NPS | Real-Time RT-PCR, Fluorescence detection | NA |
Luminex 1. xTag Respiratory Virus Panel 2. xTag RVP Fast | Thermocyclers Luminex Lx100/200 | bioMerieux NucliSENS easyMAG, bioMerieux NucliSENS miniMAG, Roche MagnaPure | 1. ADV, hMPV, HRV, FluA (A/H1, A/H3), FluB, PIV 1,2,3, RSV (A, B) 2. AdV, HMPV, HRV, FluA (A/H1, A/H3), FluB, RSV (A, B) | 1. NPS 2. NPS | RT-PCR/PCR Primer extension xTAG Bead Array Fluorescent detection | |
Meridian Bioscience 1. illumigene Pertussis 2. illumigene Mycoplasma | Meridian Illumipro-10 | 1. Heat 2. Qiagen QIAmp DSP DNA miniKit | 1. B. pertussis 2. M. pneumoniae | 1. NPS 2. NPS, TS | Isothermal amplification with turbimetric detection | [184] |
Nanosphere Verigene Respiratory Virus Nucleic Acid Test Plus | Nanosphere Verigene Processor SP Verigene Reader | Included | FluA (A/H1, A/H3, A/2009 H1), FluB, RSV (A, B) | NPS | RT-PCR Gold nanoparticle detection | |
Qiagen Artus Influenza A/B Rotor-gene RT-PCR Kit | Qiagen Roto-Gene Q MDx | Qiagen QIAsymphony RGQ | FluA, FluB | NPS | Real-Time RT-PCR, Fluorescent detection | [73] |
Quidel 1. Lyra Influenza A + B Assay 2. Lyra RSV + HMPV Assay 3. Lyra Parainfluenza Virus Assay 4. Lyra Adenovirus Assay 5. Amplivue Bordetella Assay | 1. and 2. Cepheid SmartCycler II, ABI 7500 Fast Dx, Life Technologies QuantStudio 3. and 4. ABI 7500 Fast Dx 5. Thermocycler and Amplivue Cassette | 1–4: bioMerieux NucliSENS easyMAG Heat treatment | 1. FluA, FluB 2. HMPV, RSV 3. PIV-1,2,3 4. ADV 5. B. pertussis | 1. and 2. NPS, NS, NA, NW 3 and 4. NPS, NS 5. NPS | 1–4. Real-Time RT-PCR/PCR, Fluorescent detection 5. Helicase dependent amplification, Lateral flow | NA |
US Army JBAIDS 1. Influenza A/H5g 2. Influenza A&B Detection Kitg 3. Influenza A Subtyping Kitg | Idaho Technologies JBAIDS | Included | 1. H5N1 (Asian lineage) 2. FluA and FluB 3. A/H1, A/H3, A/2009 H1 | 1. NPS, TS 2. NPS, NPW 3. NPS, NPW | Real-Time RT-PCR, Fluorescent detection | NA |
Table 52.3
Research use only, investigational use only, or CE marked tests for the detection of respiratory pathogensa
Manufacturer/testb | Amplification and detection platform(s) | Extraction platforms | Targetsc | Specimen types | Method(s)d | References |
|---|---|---|---|---|---|---|
Abbott Ibis PLEX-ID/Flu assay PLEX-ID Respiratory Virus Assay | Abbott Ibis T5000 platform | Thermo King-Fisher | Pan-influenza (PB1) Five pan-FluA (NP, M1, PA, PB2, NS1) ADV (A-F), CoV HMPV, FluA, FluB, PIV (1–3), RSV | Respiratory samples (not specified) NPA | Broad range RT-PCR, Electrospray Ionization Mass Spectrometry, (RT-PCR/ESI-MS) | |
Autogenomics 1. Infiniti RVP Plus 2. Infiniti Flu A-sH1N1 | Thermocycler Autogenomics Infiniti Analyzer | Not specified | 1. ADV (A, B, C, E) FluA (A/2009-H1N1), FluB, RSV, PIV (1–4), HRV (A, B), EV (A, B, C, D), CoV (HKU1, OC43, NL63, 229E), HMPV (A, B) 2. FluA, A/2009 H1 | Respiratory samples (not specified) | RT-PCR BioFilmChip Microarray | NA |
bioMerieux 1. NucliSENS Influenza A + B 2. NucliSENS RSV A + B 3. NucliSENS HMPV 4. NucliSENS Mycoplasma 5. NucliSENS Chlamydophila | bioMerieux NucliSENS easyQ | bioMerieux NucliSENS easyMAG | 1. FluA and FluB 2. RSV (A, B) 3. HMPV 4. M. pneumoniae, C. pneumoniae | NPA, NPS, NS, NA, BAL | NASBA, Fluorescence detection | |
Curetis Pneumonia Panel Bacterial | Curetis Unyvero System | Included | A. baumanii, E. coli Enterobacter spp., M. morganii K. oxytoca, K. pneumoniae H. influenzae, Proteus spp. M. catarrhalis, S. aureus P. aeruginosa, S. marcescens S. maltophilia, S. pneumoniae C. pneumoniae, L. pneumophila P. jiroveci, Plus 18 antibiotic resistance markers | BAL, TA, BW, BB, PB | Multiplex End Point PCR | |
Hologic/Gen-Probe ProPneumo-1 | Smartcycler II Qiagen Roto-Gene Applied Biosystems GeneAmp PCR 7500 | bioMerieux NucliSENS easyMag | C. pneumoniae M. pneumoniae | NPS, NPW, BAL, sputum | Real-Time RT-PCR, Fluorescence detection | [96] |
Icubate 1.Respiratory Panel V (viral) 2.Flu Typing 3. Respiratory Panel B (Bacterial) | Icubate Processor Incubator | Included | 1. FluA (A/2009 H1), FluB, PIV (1–4), RSV (A,B), HMPV (A,B), HRV, ADV (3/7, 4), Cox A, B, Echo, HRV, Corona (OC43, NL63, 229E, HKU-1), HBoV 2. FluA (A/H1, A/H3, A/2009 H1, A/H5 (avian), N1 (shared), N2 (seasonal), FluB H. influenzae (non-typeable) H. influenzae (a, b, c, d) H. influenzae (e, f), S. aureus N. meningitidis, S. pneumoniae C. pneumoniae, L. pneumophila M. pneumoniae | Respiratory samples (not specified) | ARM-PCR technology (Amplicon Rescued Multiplex PCR), End point detection | NA |
Luminex FluA/B, RSV Assay | Aries | included | FluA/B and RSV | NPS | Multi-code Real-Time RT-PCR, Fluorescence detection | NA |
Pathofinder 1. Respifinder 15 2. Respifinder 19 3. Respifinder Smart 22 | Thermocycler ABI310, ABI3100, ABI3130, ABI3730, ABI3500 Beckman CEQ Roche Lightcycler 480 Corbett RotorGgene 3000/6000 Qiagen RotorGene Q | Not specified | 1. FluA (H5N1), FluB, PIV (1–4), RSV (A,B), hMPV, HRV, ADV, CoV (OC43, NL63, 229E) 2. 15 plus: B. pertussis, M. pneumoniae, C. pneumoniae, L. pneumophila 3. FluA (H1N1-2009), FluB, PIV (1–4), RSV (A,B), hMPV, EV/HRV, ADV, CoV (OC43, NL63, 229E, HKU-1), HBoV B. pertussis, C. pneumoniae, L. pneumophila, M. pneumoniae | NPW, NPA, NPW, BAL, Sputum | RT-PCR/PCR, Capillary electrophoresis, Multiplex PCR, Melt curve analysis | |
Qiagen Resplex II | GeneAmp PCR system 9700 LiquiChip 200 Workstation | QIAamp viral RNA QIAamp MiniElute QIASymphony QIAxtractor | FluA, FluB, PIV (1–4), RSV (A,B), HMPV, HRV, Cox/Echo Pan-ADV (B,E), CoV (OC43, NL63, 229E, HKU-1), HBoV | Respiratory Samples (not specified) | RT-PCR/PCR, Bead Array, Hybridization detection | |
Seegene 1. Seeplex Respiratory Assay Group of 12: 2. Seeplex Respiratory Assay Group of 15: 3. Seeplex Typing Influenza A Virus | Variety including: Qiagen Rotor-gene Applied Biosystems GeneAmp PCR system 9700 | Variety including: NucliSENS easyMAG Qiagen BioRobot MDx iNtRON Biotechnology Viral Gene-spin Kit | 1. Set A: ADV, CoV (229E/NL63), PIV (1–3) 1. Set B: FluA/B, RSV (A,B), HRV A, CoV (OC43) 2. Set A: ADV (A/B/C/D/E), PIV (1–3), CoV (229E/NL63) 2. Set B: CoV (OC43), HRV (A/B/C), FluA, RSV (A, B) 2. Set C: HBoV (1/2/3/4), FluB, HMPV, EV, PIV 4 3. FluA (generic), A/H1, A/H3, A/2009-H1 | Respiratory Samples (not specified) | Real-Time RT-PCR, Fluorescence detection |
The expanded scope of pathogen detection from a previous low of three viral pathogens detected by RADTs to 17 viral and three bacterial pathogens detected by NAATs greatly enhances the clinical laboratory’s diagnostic capabilities. The data derived from studies utilizing comprehensive viral and bacterial NAATs has and will continue to provide invaluable insights into the clinical manifestations of viral and bacterial infections, significance of mixed viral infections, and surprisingly the realization that viruses can colonize a host without overt disease.
Impact on Antimicrobial Selection and Stewardship
Aside from the use of RADTs during influenza season, RTI diagnostic testing including viral DFAs and culture, bacterial culture, NAATs, serologic testing for the atypical pathogens, and urinary antigen testing for S. pneumoniae and L. pneumophila are still underutilized in the outpatient setting [10, 189]. Therefore, treatment of patients with CAP is generally empiric and based on guidelines by the American Thoracic Society and IDSA [146] rather than on a confirmed laboratory diagnosis. Antibiotic selection must cover both the most prevalent bacterial pathogens and the atypical pathogens.
Despite the high prevalence of viral infections, approximately 22.6 million (55 %) out of 41 million antibiotic prescriptions were prescribed for viral lower and upper RTIs, despite the fact that a bacterial etiology was highly unlikely [82]. Conversely the identification of the viral pathogen can lead to the administration of an appropriate antiviral. For example, during the early weeks of the 2009 influenza A H1N1 pandemic, four different influenza viruses were circulating with varying antiviral susceptibility patterns. According to surveillance data provided by the Center for Disease Control and Prevention (CDC, Atlanta, GA), FluA-H1 strains demonstrated >99 % resistance to the first line therapeutic oseltamivir, a neuraminidase inhibitor, and susceptibility to adamantine, FluA-H3 was oseltamivir-susceptible and >99 % adamantine-resistant, FluA(H1N1)pdm09 was oseltamivir-susceptible and >99 % adamantine-resistant; FluB was oseltamivir-susceptible and adamantine-resistant. Therefore, the identification of what specific strain of influenza virus was causing the infection of specific patients was essential to ensure proper drug selection, especially in high-risk or critically ill patients. Additionally, the more appropriate use of influenza antivirals can be achieved when an accurate, rapid diagnosis is made. Recent studies demonstrated that rapid testing permitted the timely administration of oseltamivir [229] and allowed for a more rapid discontinuation of treatment in persons without documented influenza [191]. Finally, new viral therapeutic agents for respiratory viruses other than influenza are in development and/or in clinical trials [171, 233]. Proper administration of these new agents will depend on the laboratory providing accurate tests that detect a broad range of viral pathogens [189].
For HAP and VAP, initial empiric therapy choices may be standardized and initiated based on patient clinical status, underlying disease, and/or risk for infection with a multidrug-resistant pathogen. Initial therapy often is inadequate, thereby extending the course of the disease and increasing morbidity, mortality, and hospital length of stay. The mortality rate in ICU pneumonia cases ranges from 20–30 % when initial therapy was adequate, to 50–80 % when initial therapy was inappropriate and changed after culture results were obtained [112]. Furthermore, inadequate and/or unnecessary broad spectrum antibiotic therapy can enhance the spread of drug-resistant pathogens within institutional settings and increase the risk of hospital-acquired infections such as Clostridium difficile, MRSA, and vancomycin-resistant enterococci.
Comprehensive NAATs provide key information not only for antiviral therapy selection but can also aid in restricting antibiotic use to those circumstances where antibiotic therapy is appropriate and in promoting switches to targeted specific therapies, thus reducing the use of broad spectrum antibiotics when not indicated [170, 142, 189]. This practice is in keeping with the goals of antibiotic stewardship, especially considering the steady and critical rise of antibiotic resistance and limited or no options available for the treatment of multidrug or pan-resistant bacterial infections.
Prevention of Nosocomial Infections
The burden of nosocomial infections can be significant, incurring additional costs for supplemental diagnostic tests, extended hospitalization, and increased morbidity and mortality [19, 40, 62, 126, 155, 217, 219]. Therefore, rapid diagnostic tests are needed to identify infected patients upon admission, thereby preventing nosocomial transmission by facilitating isolation and appropriate cohorting decisions [165, 206]. Studies have documented significant nosocomial transmission of ADV, influenza, RSV, HMPV, PIV, and HRV in hospital units, chronic care facilities, and pediatric units [19, 40, 62, 126, 155, 217, 219, 232]. During the height of RSV season, when prevalence can be >50 %, high numbers of hospital admissions often require the cohorting of RSV-positive children due to a lack of private rooms. However, limiting diagnostic testing to RSV alone in a cohorting scenario could put other seriously ill children at risk for acquisition of a second viral infection with other pathogens such as HMPV or HRV [80]. The rapid identification of health care facility-acquired Legionella infection is essential so that the environment source can be identified and eradicated, thus preventing further transmission. In addition, PCR techniques are used to proactively routinely screen potential environmental sources.
Epidemiologic Surveillance and Outbreak Investigation
The identification of a wide range of viral pathogens is essential for epidemiologic surveillance and establishes both seasonal and population patterns which can serve as excellent indicators for predicting immunization scheduling (e.g., influenza), or for administering preventive measures (e.g., RSV immune globulin). Previously, such surveillance was done by state Departments of Health or on a national level by the CDC. Comprehensive NAATs now enable local laboratories to monitor in real time viral prevalence and provide to their clinicians and health care facilities regular updates on circulating viruses. Testing decisions should use local data and additional resources and epidemiology information provided on the CDC web site (http://www.cdc.gov/flu/).
A study by S. Wong et al. demonstrated that the use of a multiplex NAAT (17 viruses) identified a virus in 59 % of the outbreaks and 29 % of the outbreak specimens that were negative using DFAs and a limited number of individual NAATs (ten viruses) [226]. Overall, the detection rate increased from 72–91 % for outbreaks and from 47–56 % for outbreak specimens. Comprehensive testing can identify reemerging or new emerging viral pathogens, as was the case during the influenza A H1N1 2009 pandemic.
Additionally, NAATs have been a key component in understanding both the epidemiology and features of large outbreaks of pertussis-like illness [190]. For example the large percentage of cases (29 %) of B. holmseii identified in an outbreak in Ohio in 2010–2011 was in contrast to some previous reports where the frequency of detection was very low [190]. The USA has seen peaks of pertussis activity every 3–5 years, with increasing cases since 1980. Accurate detection of pertussis is essential to better control the spread of disease from a public health standpoint through antimicrobial prophylaxis of asymptomatic household contacts, children less than 1 year of age, pregnant women in their third trimester, persons with preexisting conditions that are at risk for the development of severe respiratory failure, and all contacts in high-risk settings (http://www.cdc.gov/pertussis/outbreaks/pep.html).
Available Assays
Sample Types, Transport, and Storage
The recovery of respiratory pathogens is highly dependent on using the appropriate sample type and collection methods [139]. Laboratories using commercial FDA-cleared assays should refer to the manufacturer’s package information to determine what sample types have been validated for use with the specific test (Table 52.2). The use of alternate samples types is permitted after the laboratory has performed their own validation studies that establish acceptable performance characteristics when testing the alternate sample type.
Applicable upper respiratory tract specimens for viral, C. pneumoniae, and M. pneumoniae testing include nasopharyngeal (NP) washes, NP aspirates, NP swabs, and mid-turbinate swabs placed in viral transport media [1, 48, 49, 95, 118, 131, 164, 223]. Oropharyngeal swab specimens are less sensitive (54 %) than either NP swabs (73 %) or NP wash specimens (85 %) due to the substantially lower levels of virus present in the oropharynx than the nasopharynx [131]. However, the combined use of nasal-oropharyngeal swabs can enhance the recovery of both avian influenza and SARS-CoV [49]. NP flocked synthetic swabs should be used in lieu of traditional synthetic NP swabs since flocked NP swabs yield a greater recovery of viral pathogens, with sensitivity comparable to NP wash specimens [1, 48, 95]. The rates of positivity for B. pertussis by real-time PCR were shown to be comparable when specimens were collected with either NP rayon swabs on aluminum shafts in Amies gel with charcoal or NP flocked swabs in universal transport media [6].
Lower respiratory tract (LRT) samples appropriate for viral pathogens, C. pneumoniae, M. pneumoniae, L. pneumophila, and bacterial pathogens include induced sputum, bronchial alveolar lavages, bronchial washings, protected brushes, and Combicath specimens. Studies that examined the detection of FluA(H1N1)pdm09 found that in seriously ill patients requiring intensive care, upper respiratory tract samples can be negative while LRT samples are positive [120, 123, 156, 230].
Additional factors that influence pathogen recovery include the time of sample collection after the onset of clinical symptoms and the age of the patient (children tend to shed higher titers of virus and for longer periods of time than adults). Therefore, for optimal detection, samples should be collected within 3 days for adults and within 5 days for children after the onset of symptoms [39, 85, 95, 120, 123]. Samples should be transported to the laboratory as soon as possible, preferentially on wet ice or refrigerated (2–8 °C) if testing will be performed within 48 h. If testing is delayed, the samples should be stored at −80 °C. Multiple freeze-thaws should be avoided as this process can decrease pathogen titers.
Nucleic Acid Extraction
Target lysis in a stabilizing matrix to prevent target degradation by deoxyribonucleases (DNases) and/or ribonucleases (RNases), followed by isolation and purification of the nucleic acids (NAs) are essential and critical steps of every NAAT. This is particularly important for respiratory samples that can be highly viscous and contain inhibitory substances and enzymes that destroy the target NAs. Some sample types may require a pretreatment with proteinase K or a similar enzyme. Highly efficient commercial NA extraction systems ensure sufficient NA recovery and the removal of inhibitory substances that could result in inefficient or no amplification of the target NAs. Presently, the majority of NA extraction systems use a chaotropic agent to lyse viral particles or bacterial cells, silica particles or a membrane to capture the released NAs, and a series of wash steps to remove inhibitory substances. NAs are generally eluted in either RNase/DNase free water or a stabilizing buffer such as EDTA-Tris. Although NA extraction can be performed manually using for example spin columns, the majority of laboratories currently use automated platforms that can extract as few as one sample at a time or more than 96 samples in microwell plate formats. Many NAATs have been FDA-cleared with specific extraction platforms or methods and laboratories should be aware that the substitution of another extraction platform or method constitutes a major change in the assay protocol. From a regulatory perspective, if the extraction procedure is modified or changed then the entire test is now considered an LDT. Laboratories must be compliant with all applicable state and federal CLIA standards, and may choose to comply with the College of American Pathologists regulations to meet state or federal regulatory requirements.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


