Principles of Virology
Richard C. Condit
Viruses are unique in nature. They are the smallest of all self-replicating organisms, historically characterized by their ability to pass through filters that retain even the smallest bacteria. In their most basic form, viruses consist solely of a small segment of nucleic acid encased in a simple protein shell. Viruses have no metabolism of their own but rather are obliged to invade cells and parasitize subcellular machinery, subverting it to their own purposes. Many have argued that viruses are not even living,128 although to a seasoned virologist, they exhibit a life as robust as any other creature.
The apparent simplicity of viruses is deceptive. The truth is that as a group, viruses infect virtually every organism in nature, they display a dizzying diversity of structures and lifestyles, and they embody a profound complexity of function.
The study of viruses—virology—must accommodate both the uniqueness and the complexity of these organisms. The singular nature of viruses has spawned novel methods of classification and experimentation entirely peculiar to the discipline of virology. The complexity of viruses is constantly challenging scientists to adjust their thinking and their research to describe and understand some new twist in the central dogma revealed in a simple virus infection.
This chapter explores several concepts fundamental to virology as a whole, including virus taxonomy, virus cultivation and assay, and virus genetics. The chapter is not intended as a comprehensive or encyclopedic treatment of these topics, but rather as a relatively concise overview with sufficient documentation for more in-depth study. In addition to primary resources and practical experience, the presentation draws heavily on previous editions of Fields Virology35,36,37 for the taxonomy and genetics material, plus several excellent texts for material on virus cultivation and assay.20,34,41,59,70,76,81 It is hoped that this chapter will be of value to anyone learning virology at any stage: a novice trying to understand basic principles for the first time, an intermediate student of virology trying to understand the technical subtleties of virological protocols in the literature, or a bewildered scientist in the laboratory wondering why the host-range virus mutant received from a colleague does not seem to manifest the described host range.
Virus Taxonomy
A coherent and workable system of classification—a taxonomy—is a critical component of the discipline of virology. However, the unique nature of viruses has defied the strict application of many of the traditional tools of taxonomy used in other disciplines of biology. Thus, scientists who concern themselves with global taxonomy of organisms have traditionally either ignored viruses completely as nonliving entities or left them scattered throughout the major kingdoms, reasoning that viruses have more in common with their individual hosts than they do with each other.82,90 By contrast, for practical reasons at least, virologists agree that viruses should be considered together as a separate group of organisms regardless of host, be it plant, animal, fungus, protist, or bacterium, a philosophy borne out by the observation that in several cases viruses now classified in the same family—for example, family Reoviridae—infect hosts from different kingdoms. Interestingly, the discipline of virus taxonomy brings out the most erudite and thought-provoking, virtually philosophical discussions about the nature of viruses, probably because the decisions that must be made to distinguish one virus from another require the deepest thought about the nature of viruses and virus evolution. In the end, all of nature is a continuum, and the business of taxonomy has the unfortunate obligation of drawing boundaries within this continuum, an artificial and illogical task but necessary nevertheless. The execution of this obligation results today in a free-standing virus taxonomy, overseen by the International Committee on Taxonomy of Viruses (ICTV), with rules and tools unique to the discipline of virology. The process of virus taxonomy that has evolved
uses some of the hierarchical nomenclature of traditional taxonomy, identifying virus species and grouping these into genera, genera into families, and families into orders, but at the same time, to cope with both the uniqueness and diversity of viruses as a group, the classification process has been deliberately nonsystematic and thus is “based upon the opinionated usage of data”.92
uses some of the hierarchical nomenclature of traditional taxonomy, identifying virus species and grouping these into genera, genera into families, and families into orders, but at the same time, to cope with both the uniqueness and diversity of viruses as a group, the classification process has been deliberately nonsystematic and thus is “based upon the opinionated usage of data”.92
Most importantly, the virus taxonomy that has been developed works well. For the trained virologist, the mention of a virus family or genus name, such as “family Herpesviridae” or “genus Rotavirus” immediately conjures forth a set of characteristics that form the basis for further discussion or description. Virus taxonomy serves an important practical purpose as well, in that the identification of a limited number of biological characteristics, such as virion morphology, genome structure, or antigenic properties, quickly provides a focus for identification of an unknown agent for the clinician or epidemiologist and can significantly impact further investigation into treatment or prevention of a virus disease. Virus taxonomy is an evolving field, and what follows is a summary of the state of the art, including important historical landmarks that influenced the present system of virus taxonomy, a description of the system used for virus taxonomy and the means for implementation of that system, and a very brief overview of the taxonomy of viruses that infect humans and animals.
History and Rationale
Virology as a discipline is scarcely 100 years old, and thus the discipline of virus taxonomy is relatively young. In the early 1900s, viruses were initially classified as distinct from other organisms simply by virtue of their ability to pass through unglazed porcelain filters known to retain the smallest of bacteria. As increasing numbers of filterable agents became recognized, they were distinguished from each other by the only measurable properties available, namely the disease or symptoms caused in an infected organism. Therefore, animal viruses that caused liver pathology were grouped together as hepatitis viruses, and viruses that caused mottling in plants were grouped together as mosaic viruses. In the 1930s, an explosion of technology spawned a description of the physical properties of many viruses, providing numerous new characteristics for distinguishing viruses one from another. The technologies included procedures for purification of viruses, biochemical characterization of purified virions, serology, and perhaps most importantly, electron microscopy, in particular negative staining, which permitted detailed descriptions of virion morphology, even in relatively crude preparations of infected tissue. In the 1950s, these characterizations led to the distinction of three major animal virus groups, the myxoviruses, the herpesviruses, and the poxviruses. By the 1960s, because of the profusion of data describing numerous different viruses, it became clear that an organized effort was required to classify and name viruses, and thus the ICTV (originally the International Committee on Nomenclature of Viruses [ICNV]) was established in 1966. The ICTV functions today as a large, international group of virologists organized into appropriate study groups, whose charge it is to develop rules for the classification and naming of viruses and to coordinate the activities of study groups in the implementation of these rules.
Early in its history, the ICTV wrestled with the fundamental problem of developing a taxonomic system for classification and naming of viruses that would accommodate the unique properties of viruses as a group and that could anticipate advancements in the identification and characterization of viruses. Perhaps the most critical issue was whether the classification of viruses should consider virus properties in a monothetical, hierarchical fashion or a polythetical, hierarchical fashion. A monothetic system of classification is defined as a system based on a single characteristic or a series of single characteristics. Polythetic is defined as sharing several common characteristics without any one of these characteristics being essential for membership in the group or class in question. Thus, a monothetical, hierarchical classification, modeled after the Linnaean system used for classification of plants and animals, would effectively rank individual virus properties, such as genome structure or virion symmetry, as being more or less important relative to each other and use these individual characteristics to sort viruses into subphyla, classes, orders, suborders, and families.79 Although the hierarchical ordering of viruses into groups and subgroups is desirable, a strictly monothetical approach to using virus properties in making assignments to groups was problematic because both the identification of individual properties to be used in the hierarchy and the assignment of a hierarchy to individual properties seemed too arbitrary. A polythetic approach to classification would group viruses by comparing simultaneously numerous properties of individual viruses without assigning a universal priority to any one property. Thus, using the polythetic approach, a given virus grouping is defined by a collection of properties rather than a single property, and virus groups in different branches of the taxonomy may be characterized by different collections of properties. One argument against the polythetic approach is that a truly systematic and comprehensive comparison of dozens of individual properties would be at least forbidding if not impossible. However, this problem could be avoided by the adoption of a nonsystematic approach, namely, using study groups of virologists within the ICTV to consider together numerous characteristics of a virus and make as rational an assignment to a group as possible. Therefore, the system that is currently being used is a nonsystematic, polythetical, hierarchical system. This system differs from any other taxonomic system in use for bacteria or other organisms; however, it is effective, useful, and has withstood the test of time.91 As our understanding of viruses increases, and as new techniques for characterization are developed, notably comparison of gene and genome sequences, the methods used for taxonomy will undoubtedly continue to evolve.
As a consequence of the polythetic approach to classification, the virus taxonomy that exists today has been filled initially from the middle of the hierarchy by assigning viruses to genera, and then elaborating the taxonomy upward by grouping genera into families and, to a limited extent, families into orders. By 1970, the ICTV had established two virus families each containing 2 genera, 24 floating genera, and 16 plant groups.133 A rigorous species definition,126 discussed later, was not approved by the ICTV until 1991 but has now been applied to the entire taxonomy and has become the primary level of classification for viruses. As of this writing, the currently accepted taxonomy recognizes 6 orders, 87 families, 19 subfamilies, 348 genera,
and 2,290 species. The complete virus taxonomy is far too extensive to relate here; however, examples of the results of the taxonomy are offered in Tables 2.1 and 2.2. Table 2.1 lists the distinguishing characteristics of the vertebrate animal virus families, whereas Table 2.2 provides an example of the entire taxonomic classification of one virus order, namely order Mononegavirales.
and 2,290 species. The complete virus taxonomy is far too extensive to relate here; however, examples of the results of the taxonomy are offered in Tables 2.1 and 2.2. Table 2.1 lists the distinguishing characteristics of the vertebrate animal virus families, whereas Table 2.2 provides an example of the entire taxonomic classification of one virus order, namely order Mononegavirales.
Table 2.1 Summary Characteristics of Vertebrate Virus Families | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The International Committee on Taxonomy of Viruses Universal System of Virus Taxonomy
Structure and Function
The ICTV is a committee of the Virology Division of the International Union of Microbiological Societies. The objectives of the ICTV are to develop an internationally agreed taxonomy
and nomenclature for viruses, to maintain an index of virus names, and to communicate the proceedings of the committee to the international community of virologists. The ICTV publishes an update of the taxonomy at approximately 3-year intervals.32,33,39,85,86,92,133 At the time of this writing, the ninth report is being completed. The official taxonomy is also available on line at the ICTV website: http://www.ictvonline.org.
and nomenclature for viruses, to maintain an index of virus names, and to communicate the proceedings of the committee to the international community of virologists. The ICTV publishes an update of the taxonomy at approximately 3-year intervals.32,33,39,85,86,92,133 At the time of this writing, the ninth report is being completed. The official taxonomy is also available on line at the ICTV website: http://www.ictvonline.org.
Table 2.2 Taxonomy of the Order Mononegavirales | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Virus Properties and Their Use in Taxonomy
As introduced previously, the taxonomic method adopted for use in virology is polythetic, meaning that any given virus group is described using a collection of individual properties. The description of a virus group is nonsystematic in that there exists no fixed list of properties that must be considered for all viruses and no strict formula for the ordered consideration of properties. Instead, a set of properties describing a given virus is simply compared with other viruses described in a similar fashion to formulate rational groupings. Characters such as virion morphology, genome organization, method of replication, and the number and size of structural and nonstructural viral proteins are used for distinguishing different virus families and genera. Characters such as genome sequence relatedness, natural host range, cell and tissue tropism, pathogenicity and cytopathology, mode of transmission, physicochemical properties of virions, and antigenic properties of viral proteins are used for distinguishing virus species within the same genus.127
The Hierarchy
The ICTV has adopted a universal classification scheme that employs the hierarchical taxonomic levels of order, family, subfamily, genus, and species. Because the polythetic approach to classification introduces viruses into the middle of the hierarchy, and because the ICTV has taken a relatively conservative approach to grouping taxa, levels higher than order are not currently used. Interestingly, groupings above the level of order may prove to be inappropriate: Higher taxons imply a common ancestry for viruses, whereas multiple independent lineages for viruses now seems the more likely evolutionary scenario.32 Taxonomic levels lower than species, such as clades, strains, and variants, are not officially considered by the ICTV but are left to specialty groups.
A virus species is defined as “a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche”.126 The formal definition of a polythetic class is “a class whose members always have several properties in common although no single common attribute is present in all of its members”.127 Thus, no single property can be used to define a given species, and application of this formal definition of a polythetic class to species accounts nicely for the inherent variability found among members of a species. The qualification of a replicating lineage implies that members of a species experience evolution over time with consequent variation, but that members share a common ancestor. The qualification of occupation of an ecological niche acknowledges that the biology of a virus, including such properties as host range, pathogenesis, transmission, and habitat, are fundamental components of the characterization of a virus. A type species has been identified for each genus. The type species is not necessarily the best characterized or most representative species in a genus; rather, it is usually the virus that initially necessitated the creation of the genus and therefore best defines or identifies the genus.
Taxonomic levels higher than species are formally defined by the ICTV only in a relative sense, namely a genus is a group of species sharing certain common characters, a subfamily is a group of genera sharing certain common characters, a family is a group of genera or subfamilies
sharing certain common characters, and an order is a group of families sharing certain common characters. As the virus taxonomy has evolved, these higher taxa have acquired some monothetic character. They remain polythetic in that they may be characterized by more than one virus property; however, they violate the formal definition of a polythetic class in that one or more defining properties may be required of all candidate viruses for membership in the taxon. Not all taxonomic levels need be used for a given grouping of viruses, thus whereas most species are grouped into genera and genera into families, not all families contain subfamilies, and only a few families have been grouped into orders. Consequently, the family is the highest consistently used taxonomic grouping, it therefore carries the most generalized description of a given virus group, and as a result has become the benchmark of the taxonomic system. Most families have distinct virion morphology, genome structure, and/or replication strategy (see Table 2.1).
sharing certain common characters, and an order is a group of families sharing certain common characters. As the virus taxonomy has evolved, these higher taxa have acquired some monothetic character. They remain polythetic in that they may be characterized by more than one virus property; however, they violate the formal definition of a polythetic class in that one or more defining properties may be required of all candidate viruses for membership in the taxon. Not all taxonomic levels need be used for a given grouping of viruses, thus whereas most species are grouped into genera and genera into families, not all families contain subfamilies, and only a few families have been grouped into orders. Consequently, the family is the highest consistently used taxonomic grouping, it therefore carries the most generalized description of a given virus group, and as a result has become the benchmark of the taxonomic system. Most families have distinct virion morphology, genome structure, and/or replication strategy (see Table 2.1).
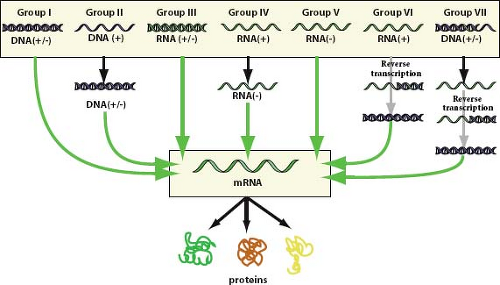 Figure 2.1. The Baltimore classification, a virus classification scheme based on the form of nucleic acid present in virion particles and the pathway for expression of the genetic material as messenger RNA.1 The original scheme contained groups I through VI and has been expanded to accommodate DNA-containing, reverse transcribing viruses. Viruses containing ambisense single-stranded RNA genomes are grouped under negative sense single-stranded RNA viruses. (Reprinted from Hulo C, de Castro E, Masson P, et al. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 2011;39 (Database issue):D576–D582; ViralZone, Swiss Institute of Bioinformatics, http://www.expasy.ch/viralzone/, with permission.) |
Nomenclature
The ICTV has adopted a formal nomenclature for viruses, specifying suffixes for the various taxa, and rules for written descriptions of viruses. Names for genera, subfamilies, families, and orders must all be single words, ending with the suffixes -virus, -virinae, -viridae, and -virales, respectively. Species names may contain more than one word and have no specific ending. In written usage, the formal virus taxonomic names are capitalized and written in italics, and preceded by the name of the taxon, which is neither capitalized nor italicized. For species names that contain more than one word, the first word plus any proper nouns are capitalized. As an example, the full formal written description of human respiratory syncytial virus is as follows: order Mononegavirales, family Paramyxoviridae, subfamily Pneumovirinae, genus Pneumovirus, species Human respiratory syncytial virus. The ICTV acknowledges that vernacular (informal) taxonomic names are widely used; however, they should not be italicized or capitalized. For example, the vernacular name “herpesvirus” refers to a member of the family Herpesviridae.
Informal Groupings and Alternate Classification Schemes
For convenience in presenting or tabulating the virus taxonomy, informal categorical groupings of taxa are often used. The criteria applied for such groupings typically include nature of the viral genome (DNA or RNA), strandedness of the viral genome (single stranded or double stranded), polarity of the genome (positive sense, negative sense, or ambisense), and reverse transcription. Separate categories accommodate subviral agents (including viroids, satellites, and prions) and unassigned viruses. The Baltimore classification system, named after its creator David Baltimore, is a widely used scheme based on the nature of the genome packaged in virions and the pathway of nucleic acid synthesis that each group takes to accomplish messenger RNA (mRNA) synthesis.1 This classification divides viruses into seven categories as depicted in Figure 2.1. Most usages of this system group ambisense virus families (family Arenaviridae
and family Bunyaviridae) along with negative sense, single-stranded RNA (ssRNA) viruses. The families of vertebrate viruses listed in Table 2.1 have been grouped according to the Baltimore classification, with ambisense viruses split into an eighth genome category.
and family Bunyaviridae) along with negative sense, single-stranded RNA (ssRNA) viruses. The families of vertebrate viruses listed in Table 2.1 have been grouped according to the Baltimore classification, with ambisense viruses split into an eighth genome category.
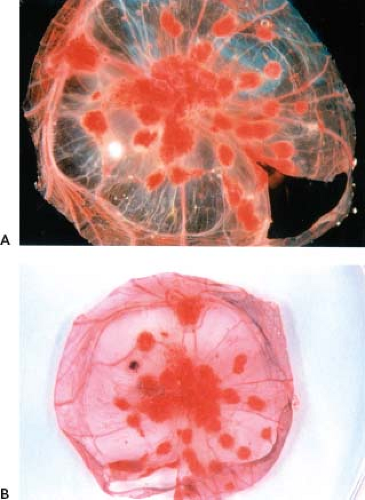 Figure 2.2. Cowpox-induced pock formation on the chorioallantoic membrane of chick embryos. The chorioallantoic membrane of intact chicken embryos, 11 days old, were inoculated with cowpox, and the eggs were incubated for an additional 3 days at 37.5°C. Chorioallantoic membranes were then dissected from the eggs and photographed. The membrane shown in A was untreated, whereas the membrane in B was stained with NBT, an indicator of activated heterophils.40 Wild-type cowpox forms red hemorrhagic pocks on the membrane (A and B). Spontaneous deletion mutants of cowpox virulence genes occur at a high frequency, resulting in infiltration of inflammatory cells into the pock. The infiltration of inflammatory cells causes the pocks to appear white in unstained membrane preparations or dark blue on NBT-stained membranes. The unstained membrane preparation (A) contains a single white pock, whereas the NBT-stained preparation (B) contains a single blue pock. NBT, nitroblue tetrazolium. (Courtesy of Dr. R. Moyer.) |
Universal Virus Database
To facilitate the management and distribution of virological data, the ICTV has established the universal virus database of the ICTV (ICTVdB). The ICTVdB is accessible on the Internet at http://www.ictvdb.org. Constructed from virus descriptions in the published reports of the ICTV, the database comprises searchable descriptions of all virus families, genera, and type species, including microscopic images of many viruses. The ICTVdB is a powerful resource for management of and access to virological data, and promises to considerably extend the reach and capability of the ICTV.
Virus Cultivation and Assay
Different branches of science are defined in large part by their techniques, and virology is no exception. Whereas the study of viruses uses some general methods that are common to other disciplines, the unique nature of viruses and virus infections requires a unique set of technical tools designed specifically for their investigation. Conversely, what we know and can know about viruses is delimited by the techniques used; therefore, a genuine understanding of virology requires a clear understanding of virological methods. What follows is a summary of the major techniques essential and unique to all of virology, presented as fundamental background for understanding the discipline.
Initial Detection and Isolation
The presence of a virus is evidenced initially by effects on a host organism or, in the case of a few animal viruses, by effects on cultured cells. Effects on animal hosts obviously include a broad spectrum of symptoms, including skin and mucous membrane lesions; digestive, respiratory, or neurological disorders; immune dysfunction; specific organ failure such as hepatitis or myocarditis; and death. Effects on cultured cells include a variety of morphological changes in infected cells, termed cytopathic effects and described in detail later in this chapter and in Chapter 15. Both adenovirus108 and the polyomavirus SV40121 were discovered as cell culture contaminants before they were detected in their natural hosts.
Viruses can be isolated from an infected host by harvesting excreted or secreted material, blood, or tissue and testing for induction of the original symptoms in the identical host, or induction of some abnormal pathology in a substitute host or in cell culture. Historically, dogs, cats, rabbits, rats, guinea pigs, hamsters, mice, and chickens have all been found to be useful in laboratory investigations,70 although most animal methods have now been replaced by cell culture methods.81 Once the presence of a virus has been established, it is often desirable to prepare a genetically pure clone, either by limiting serial dilution or by plaque purification.
Viruses that are cultivated in anything other than the natural host may adapt to the novel situation through acquisition of genetic alterations that provide a replication advantage in the new host. Such adaptive changes may be accompanied by a loss of fitness in the original host, most notably by a loss of virulence or pathogenicity. Whereas this adaptation and attenuation may present problems to the basic scientist interested in understanding the replication of the virus in its natural state, it also forms the basis of construction of attenuated viral vaccines.
Hosts for Virus Cultivation
Laboratory Animals and Embryonated Chicken Eggs
Prior to the advent of cell culture, animal viruses could be propagated only on whole animals or embryonated chicken eggs. Whole animals could include the natural host or laboratory animals such as rabbits, mice, rats, and hamsters. In the case of laboratory animals, newborn or suckling rodents often provide the best hosts. Today, laboratory animals are seldom used for routine cultivation of virus; however, they still play an essential role in studies of viral pathogenesis.
The use of embryonated chicken eggs was introduced to virology by Goodpasture et al44 in 1932 and developed subsequently by Beveridge and Burnet.4 The developing chick embryo, 10 to 14 days after fertilization, provides a variety of differentiated tissues, including the amnion, allantois, chorion, and yolk sac, which serve as substrates for growth of a wide variety of viruses, including orthomyxoviruses, paramyxoviruses, rhabdoviruses, togaviruses, herpesviruses, and poxviruses.70 Members of each of these virus families may replicate in several tissues of the developing egg, or replication may be confined to a single tissue. Several viruses from each of the previously mentioned groups cause discrete and characteristic foci when introduced onto the chorioallantoic membrane of embryonated eggs, thus providing a method for identification of virus types, or for quantifying virus stocks or assessing virus pathogenicity (Fig. 2.2). Although embryonated eggs have been almost wholly replaced by cell culture techniques, they are still the most convenient method for growing high titer stocks of some viruses and thus continue to be used both in research laboratories and for vaccine production.
Cell Culture
The growth and maintenance of animal cells in vitro, described generally (albeit incorrectly) as tissue culture, can be formally divided into three different techniques: organ culture, primary explant culture, and cell culture. In organ culture, the original three-dimensional architecture of a tissue is preserved under culture conditions that provide a gas–liquid interface. In primary explant culture, minced pieces of tissue placed in liquid medium in a culture vessel provide a source for outgrowth of individual cells. In cell culture, tissue is disaggregated into individual cells prior to culturing. Only cell culture will be discussed in detail here, because it is the most commonly used tissue culture technique in virology.
Cultured cells currently provide the most widely used and most powerful hosts for cultivation and assay of viruses. Cell cultures are of three basic types—primary cell cultures, cell strains, and cell lines—that may be derived from many animal species and that differ substantially in their characteristics. Viruses often behave differently on different types of cultured cells; in addition, each of the culture types possess technical
advantages and disadvantages. For these reasons, an appreciation of the use of cultured cells in animal virology requires an understanding of several fundamentals of cell culture itself. A detailed description of the theory and practice of cell and tissue culture is provided by Freshney,41 and several additional texts provide excellent summaries of cell culture as it specifically applies to virology.20,34,59
advantages and disadvantages. For these reasons, an appreciation of the use of cultured cells in animal virology requires an understanding of several fundamentals of cell culture itself. A detailed description of the theory and practice of cell and tissue culture is provided by Freshney,41 and several additional texts provide excellent summaries of cell culture as it specifically applies to virology.20,34,59
Primary Cell Culture
A primary cell culture is defined as a culture of cells obtained from the original tissue that have been cultivated in vitro for the first time and that have not been subcultured. Primary cell cultures can be established from whole animal embryos or from selected tissues from embryos, newborn animals, or adult animals of almost any species. The most commonly used cell cultures in virology derive from primates, including humans and monkeys; rodents, including hamsters, rats, and mice; and birds, most notably chickens. Cells to be cultured are obtained by mincing tissue and dispersing individual cells by treatment with proteases and/or collagenase to disrupt cell–cell interactions and interactions of cells with the extracellular matrix. With the exception of cells from the hemopoietic system, normal vertebrate cells will grow and divide only when attached to a solid surface. Dispersed cells are therefore placed in a plastic flask or dish, the surface of which has been treated to promote cell attachment. The cells are incubated in a buffered nutrient medium in the presence of blood serum, which contains a complex mixture of hormones and factors required for the growth of normal cells. The blood serum may come from a variety of sources, although bovine serum is most commonly used. Under these conditions, cells will attach to the surface of the dish, and they will divide and migrate until the surface of the dish is covered with a single layer of cells, a monolayer, whereupon they will remain viable but cease to divide. If the cell monolayer is “wounded” by scraping cells from an isolated area, cells on the border of the wound will resume division and migration until the monolayer is reformed, whereupon cell division again ceases. These and other observations lead to the conclusion that the arrest of division observed when cells reach confluency results from cell–cell contact and therefore is called contact inhibition. Primary cultures may contain a mixture of cell types and retain the closest resemblance to the tissue of origin.
Subcultivation
Cells from a primary culture may be subcultured to obtain larger numbers of cells. Cells are removed from the culture dish and disaggregated by treating the primary cell monolayer with a chelating agent, usually EDTA, or a protease, usually trypsin, or both, giving rise to a single cell suspension. This suspension is then diluted to a fraction of the original monolayer cell density and placed in a culture dish with fresh growth medium, whereupon the cells attach to the surface of the dish and resume cell division until once again a monolayer is formed and cell division ceases. Cultures established in this fashion from primary cell cultures may be called secondary cultures. Subsequently, cells may be repeatedly subcultured in the same fashion. Each subculturing event is called a passage, and each passage may comprise several cell generations, depending on the dilution used during the passage. Most vertebrate cells divide at the rate of approximately one doubling every 24 hours at 37°C. Thus, a passage performed with an eightfold dilution will require three cell doublings over 3 days before the cells regain confluency.
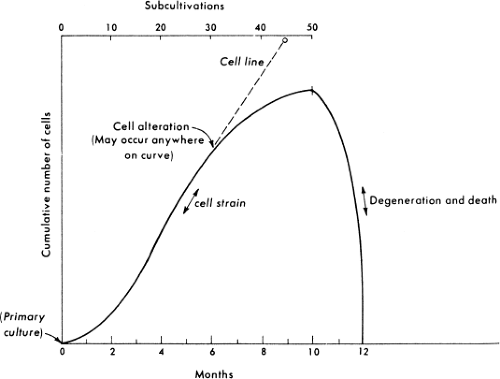
Cell Strains
Normal vertebrate cells cannot be passaged indefinitely in culture. Instead, after a limited number of cell generations, usually 20 to 100 depending on the age and species of the original animal, cultured normal cells cease to divide, then degenerate and die, a phenomenon called crisis or senescence51 (Fig. 2.3). Starting with the establishment of a secondary culture and until cells either senesce or become transformed as described later, the culture is termed a cell strain to distinguish it from a primary culture
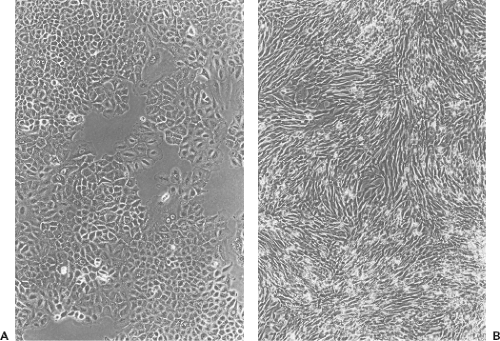
on the one hand, or a transformed, immortal cell line on the other hand. During culture, cells in a strain retain their original karyotype and are thus called euploid; however, culturing induces profound changes in the composition and characteristics of the cell strain, which are manifested early during the passage history and may continue during passage. Whereas primary cell cultures may contain a mixture of cell types that survive the original plating of cells, only a few cell types survive subculturing; thus, by the second or third passage, typically only one cell type remains in the cell strain. Cell strains are usually composed of one of two basic cell types—fibroblast-like or epithelial-like—characterized based on their morphology and growth characteristics (Fig. 2.4). Fibroblasts have an elongated, spindle shape, whereas epithelial cells have a polygonal shape. Although after only a few passages only one cell type may remain in a cell strain, continued passage may select for faster-growing variants, such that the
characteristics of a cell strain may change with increasing passage number. Despite the fact that normal cell strains experience senescence in culture, they may be maintained for many years by expanding the culture to a large number of cells early during the passage history and storing numerous small samples of low passage cells by freezing. Therefore, as a given strain approaches high passage number and senescence, low passage cells of the same strain may be thawed and cultured.
on the one hand, or a transformed, immortal cell line on the other hand. During culture, cells in a strain retain their original karyotype and are thus called euploid; however, culturing induces profound changes in the composition and characteristics of the cell strain, which are manifested early during the passage history and may continue during passage. Whereas primary cell cultures may contain a mixture of cell types that survive the original plating of cells, only a few cell types survive subculturing; thus, by the second or third passage, typically only one cell type remains in the cell strain. Cell strains are usually composed of one of two basic cell types—fibroblast-like or epithelial-like—characterized based on their morphology and growth characteristics (Fig. 2.4). Fibroblasts have an elongated, spindle shape, whereas epithelial cells have a polygonal shape. Although after only a few passages only one cell type may remain in a cell strain, continued passage may select for faster-growing variants, such that the
characteristics of a cell strain may change with increasing passage number. Despite the fact that normal cell strains experience senescence in culture, they may be maintained for many years by expanding the culture to a large number of cells early during the passage history and storing numerous small samples of low passage cells by freezing. Therefore, as a given strain approaches high passage number and senescence, low passage cells of the same strain may be thawed and cultured.
Cell Lines
At any time during the culture of a cell strain, cells in the culture may become transformed such that they are no longer subject to crisis and senescence but can be passaged indefinitely. Transformation is a complex phenomenon, discussed in more detail later and in Chapter 7; however, in the context of cell culture, the most important characteristic of transformation is that the transformed cells become immortalized. Immortal cell cultures are called cell lines, or sometimes continuous cell lines, to distinguish them from primary cultures and cell strains. Immortalization can occur spontaneously during passage of a cell strain, or it can be induced by treatment with chemical mutagens, infection with tumorigenic viruses, or transfection with oncogenes. In addition, cells cultured from tumor tissue frequently readily establish immortal cell lines in culture. Spontaneous immortalization does not occur in cultured cells from all animal species. Thus, immortalization occurs frequently during culture of rodent cells (e.g., in mouse and hamster cell strains), and it has been observed in monkey kidney cells, although it occurs rarely, if at all, during the culture of chicken or human cells. Immortalization is typically accompanied by genetic changes such that cells become aneuploid, containing abnormalities in the number and structure of chromosomes relative to the parent species, and not all cells in a culture of a continuous cell line necessarily display the same karyotype. Like cell strains, cell lines are usually composed of cells that are either fibroblast-like or epithelial-like in morphology.
As with the propagation of cell strains, continued culture of a cell line may result in selection of specific variants that outgrow other cells in the culture over time, and thus with passage the character of a cell line may change substantially, and cell lines of the same origin cultured in different laboratories over a period of years may have significantly different characteristics. It is prudent, therefore, to freeze stocks of cell lines having specific desirable properties so that these cells can be recovered if the properties disappear during culture. Likewise, it makes sense to obtain a cell line showing certain desired characteristics directly from the laboratory that described those characteristics, because cells from alternate sources may differ in character.
Transformation
Transformed cells are distinguished from normal cells by myriad properties that can be grouped into three fundamental types of changes: immortalization, aberrant growth control, and malignancy. Immortalization refers simply to the ability to be cultured indefinitely, as described previously. Aberrant growth control comprises a number of properties, several of which have relevance to experimental virology, including loss of contact inhibition, anchorage independence, and tumorigenicity. Loss of contact inhibition means that cells no longer cease to grow as soon as a monolayer is formed, and cells will now grow on top of one another. Anchorage independence means that the cells no longer need to attach to a solid surface to grow. Anchorage independence is often assayed as the ability to form colonies suspended in a semisolid medium such as agar, and a practical consequence of anchorage independence is the ability to grow in liquid suspension. Tumorigenicity refers to the ability of cells to form a tumor in an experimental animal, and malignancy refers to the ability to form an invasive tumor in vivo. While malignancy is obviously of vital importance as a phenomenon in its own right, it has limited application in virology except within the specific discipline of tumor virology (Chapter 7). Importantly, the many properties of transformed cells are not necessarily interdependent, and no one property is an absolute prerequisite for another. Thus, transformation is thought to be a multistep genetic phenomenon, and varying degrees of transformation are measurable. Tumorigenicity is often regarded as the most stringent assay for a fully transformed cell and is most closely correlated with anchorage independence.
The fact that the various characteristics of transformed cells are not interdependent has important consequences for experimental virology, especially in the assay of tumor viruses. Specifically, a transformed cell line that is immortalized but still contact inhibited may be used in a viral transformation assay that measures the further transformation to loss of contact inhibition. When cells in a monolayer are transformed by a tumor virus and lose contact inhibition, they grow on top of a confluent monolayer, forming a focus, literally a pile of cells, which is readily distinguishable from the rest of the monolayer. This property forms the basis for quantitative biological assay of tumor viruses,129 described in more detail later.
Advantages and Disadvantages of Different Cultured Cell Types
The various types of cultured cells described previously have specific application to different problems encountered in experimental virology. For most applications, an adherent cell line provides the most useful host cell. Cell lines are relatively easy to maintain because they can be passaged indefinitely, and adherence is a prerequisite for a plaque assay, described later. A distinct technical advantage of adherent cells is that the culture medium can easily be changed for the purposes of infection or metabolic labeling by simply aspirating and replacing fluid from a monolayer, a process that requires repeated centrifugations with suspension cells. By contrast, relative to adherent cell lines, suspension cell lines are easier to sample than adherent cells, and they produce large numbers of cells from a relatively small volume of medium in a single culture vessel, which has significant advantages for some high-volume applications in virology. Unfortunately, not all viruses will grow on a cell line, and often under these circumstances, a primary cell culture will suffice. This may reflect a requirement for a particular cell type found only under conditions of primary cell culture, or it may reflect a requirement for a state of metabolism or differentiation closely resembling the in vivo situation, which is more likely to exist in a primary culture than it is in a cell line.
Lastly, some viruses do not grow in cell culture at all. In such cases, investigators are reliant either on the old expedients of natural hosts, laboratory animals, or embryonated eggs, or on some more modern advances in tissue culture and recombinant DNA technology. The papillomaviruses, which cause warts, provide an enlightening example of this situation (Chapter 54). Although the viral nature of papillomatosis was
demonstrated more than 90 years ago, progress on the study of papillomaviruses was seriously hampered in the virology heyday of the mid 20th century because the viruses grow well only on the natural host; they do not grow in culture. The inability to grow in culture is now reasonably well understood, and results from a tight coupling of the regulation of viral gene expression with the differentiation state of the target epithelial cell, which in turn is tightly coupled to the three-dimensional architecture of the epidermis, which is lost in culture. Specialized tissue culture techniques have now been developed that result in the faithful reconstruction of an epidermis by seeding primary keratinocytes on a “feeder” layer composed of an appropriate cell line and incubating these cells on a “raft” or grid at a liquid–air interface. On these raft cultures, the entire replication cycle of a papillomavirus can be reproduced in vitro, albeit with difficulty.7 In the meantime, it is significant that a large fraction of the genetics and biology of papillomaviruses was determined primarily through the use of recombinant DNA technology, without ever growing virus in culture. Thus, the genetic structure of both the model bovine papillomavirus and many human papillomaviruses has been determined by cloning genomic DNA from natural infections, and regulation and function of many genes can be gleaned from sequence alone, from in vitro assays on individual gene products expressed in vitro, and from cell transformation assays that use all or parts of a papillomavirus genome. In summary, the inability to grow a virus in culture, although it increases the challenge, no longer presents an insurmountable impediment to understanding a virus.
demonstrated more than 90 years ago, progress on the study of papillomaviruses was seriously hampered in the virology heyday of the mid 20th century because the viruses grow well only on the natural host; they do not grow in culture. The inability to grow in culture is now reasonably well understood, and results from a tight coupling of the regulation of viral gene expression with the differentiation state of the target epithelial cell, which in turn is tightly coupled to the three-dimensional architecture of the epidermis, which is lost in culture. Specialized tissue culture techniques have now been developed that result in the faithful reconstruction of an epidermis by seeding primary keratinocytes on a “feeder” layer composed of an appropriate cell line and incubating these cells on a “raft” or grid at a liquid–air interface. On these raft cultures, the entire replication cycle of a papillomavirus can be reproduced in vitro, albeit with difficulty.7 In the meantime, it is significant that a large fraction of the genetics and biology of papillomaviruses was determined primarily through the use of recombinant DNA technology, without ever growing virus in culture. Thus, the genetic structure of both the model bovine papillomavirus and many human papillomaviruses has been determined by cloning genomic DNA from natural infections, and regulation and function of many genes can be gleaned from sequence alone, from in vitro assays on individual gene products expressed in vitro, and from cell transformation assays that use all or parts of a papillomavirus genome. In summary, the inability to grow a virus in culture, although it increases the challenge, no longer presents an insurmountable impediment to understanding a virus.
Recognition of Viral Growth in Culture
Two principal methods exist for the recognition of a virus infection in culture: cytopathic effect and hemadsorption. Cytopathic effect comprises two different phenomena: (a) morphological changes induced in individual cells or groups of cells by virus infection that are easily recognizable under a light microscope, and (b) inclusion bodies, which are more subtle alterations to the intracellular architecture of individual cells. Hemadsorption refers to indirect measurement of viral protein synthesis in infected cells, detected by adsorption of erythrocytes to the surface of infected cells. Cytopathic effect is the simplest and most widely used criterion for infection; however, not all viruses cause a cytopathic effect, and in these cases, other methods must suffice.
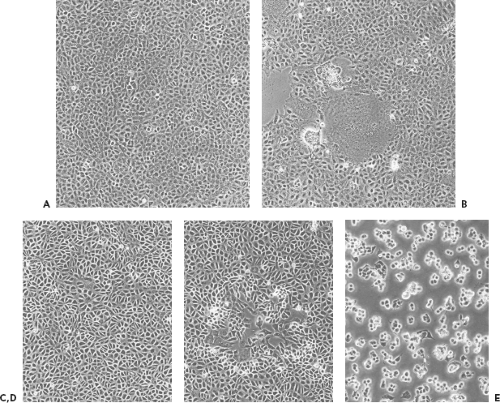
Morphological changes induced by virus infection comprise a number of cell phenomena, including rounding, shrinkage, increased refractility, fusion, aggregation, loss of adherence or lysis. Morphological changes caused by a given virus may include several of these phenomena in various combinations, and the character of the cytopathic effect may change reproducibly during the course of infection. Morphological changes caused by a given virus are very reproducible and can be so precisely characteristic of the virus type that significant clues to the identity of a virus can be gleaned from the cytopathic effect alone (Chapter 15). Figure 2.5 depicts different cytopathic effects caused by two viruses—measles and vaccinia. Most important to the trained virologist, a simple microscopic examination of a cell culture can reveal whether an infection is present, what fraction of cells are infected, and how advanced the infection is. In addition, because cytopathology results directly from the action of virus gene products, virus mutants can be obtained that are altered in cytopathology, yielding either a conveniently marked virus or a tool to study cytopathology per se.
The term inclusion bodies refers generally to the observation of intracellular structures specific to an infected cell and discernible by light microscopy. The effects are highly specific for a particular virus type so that, as with morphological alterations, the presence of a specific type of inclusion body can be diagnostic of a specific virus infection. Electron microscopy, combined with a more detailed understanding of the biology of many viruses, reveals that inclusion bodies usually represent focal points of virus replication and assembly, which differ in appearance depending on the virus. For example, Negri bodies formed during a rabies virus infection represent collections of virus nucleocapsids84 (Chapter 31).
Hemadsorption refers to the ability of red blood cells to attach specifically to virus-infected cells.111 Many viruses synthesize cell attachment proteins, which carry out their function wholly or in part by binding substituents such as sialic acid that are abundant on a wide variety of cell types, including erythrocytes. Often, these viral proteins are expressed on the surface of the infected cell—for example, in preparation for maturation of an enveloped virus through a budding process. Thus, a cluster of infected cells may be easily detectable to the naked eye as areas that stain red after exposure to an appropriate preparation of red blood cells. Hemadsorption can be a particularly useful assay for detecting infections by viruses that cause little or no cytopathic effect.
Virus Cultivation
From the discussion presented previously, it may be obvious that ultimately the exact method chosen for growing virus on any particular occasion will depend on a variety of factors, including (a) the goals of the experiment, namely whether large amounts of one virus variant or small amounts of several variants are to be grown; (b) limitations in the in vitro host range of the virus, namely whether it will grow on embryonated eggs, primary cell cultures, continuous adherent cell lines, or suspension cell lines; and (c) the relative technical ease of alternative possible procedures. Furthermore, the precise method for harvesting a virus culture will depend on the biology of the virus—for example, whether it buds from the infected cell, lyses the infected cell, or leaves the cell intact and stays tightly cell associated. As a simple example, consider cultivation of a budding, cytopathic virus on an adherent cell line. Confluent monolayers of an appropriate cell line are exposed to virus diluted to infect a fraction of the cells, and progress of the infection is monitored by observing the development of the cytopathic effect until the infection is judged complete based on experience with the relationship between cytopathic effect and maximum virus yield. A crude preparation of virus can be harvested simply by collecting the culture fluid; it may not even be necessary to remove cells or cell debris. Most viruses can be stored frozen indefinitely either as crude or purified, concentrated preparations.
Quantitative Assay of Viruses
Two major types of quantitative assays for viruses exist: physical and biological. Physical assays, such as hemagglutination, electron microscopic particle counts, optical density measurements, or immunological methods, quantify only the presence of virus particles whether or not the particles are infectious. Biological assays, such as the plaque assay or various endpoint
methods that have in common the assay of infectivity in cultured cells or in vivo, measure only the presence of infectivity and may not count all particles present in a preparation, even many that are in fact infectious. Thus, a clear understanding of the nature and efficiency of both physical and biological quantitative virus assays is required to make effective use of the data obtained from any assay.
methods that have in common the assay of infectivity in cultured cells or in vivo, measure only the presence of infectivity and may not count all particles present in a preparation, even many that are in fact infectious. Thus, a clear understanding of the nature and efficiency of both physical and biological quantitative virus assays is required to make effective use of the data obtained from any assay.
Biological Assays
The Plaque Assay
The plaque assay is the most elegant, the most quantitative, and the most useful biological assay for viruses. Developed originally for the study of bacteriophage by d’Herelle18 in the early 1900s, the plaque assay was adapted to animal viruses by Dulbecco and Vogt28 in 1953, an advance that revolutionized animal virology by introducing a methodology that was relatively simple and precisely quantitative, which enabled the cloning of individual genetic variants of a virus, and which permitted a qualitative assay for individual virus variants that differ in growth properties or cytopathology.
The plaque assay is based simply on the ability of a single infectious virus particle to give rise to a macroscopic area of cytopathology on an otherwise normal monolayer of cultured cells. Specifically, if a single cell in a monolayer is infected with a single virus particle, new virus resulting from the initial infection can infect surrounding cells, which in turn produce virus that infects additional surrounding cells. Over a period of days (the exact length of time depending on the particular virus), the initial infection thus gives rise through multiple rounds of infection to an area of infection, called a plaque. Photomicrographs of plaques are shown in Figure 2.5, and stained monolayers containing plaques are shown in Figure 2.6.
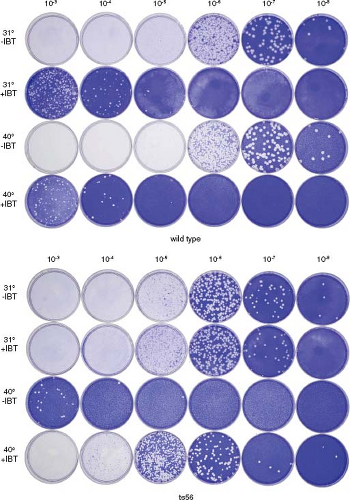 Figure 2.6. Plaque assay. Monolayers of the African green monkey kidney cell line BSC40 were infected with 0.5-mL portions of 10-fold serial dilutions of wild-type vaccinia virus or the temperature-sensitive vaccinia mutant, ts56, as indicated. Infected monolayers were overlayed with semisolid medium and incubated at 31°C or 40°C, the permissive and nonpermissive temperatures for ts56, in the presence of 45 μM isatin-β-thiosemicarbazone (IBT) or in the absence of drug as indicated, for 1 week. Overlays were removed, and monolayers were stained with crystal violet. Wild-type vaccinia virus forms plaques at both 31°C and 40°C; however, plaque formation is inhibited by IBT. Spontaneous IBT-resistant mutants in the wild-type virus stock are revealed as plaques forming at 10−3, 10−4, and 10−5 dilutions in the presence of IBT. ts56 carries a single-base missense mutation in the vaccinia gene G2R.87 G2R is an essential gene that when completely inactivated renders virus dependent on IBT; hence, ts56 is not only temperature sensitive, forming plaques at 31°C but not at 40°C in the absence of IBT, but it is also IBT dependent at 40°C, forming plaques in the presence but not the absence of IBT. ts56 is slightly defective at 31°C; it forms smaller than wild-type plaques and is IBT resistant, forming plaques both in the presence and absence of drug, a phenotype intermediate between the wild-type IBT-sensitive phenotype and the null G2R mutant IBT-dependent phenotype. Wild-type, temperature-insensitive revertants present in the ts56 stock are revealed as plaques growing on the 10−3 plate at 40°C. Based on this assay, the titer of the wild-type stock is 2.0 × 109 pfu/mL, and the titer of the ts56 stock is 6.0 × 108 pfu/mL. IBT, isatin-β-thiosemicarbazone.
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

|