Pharmacology of Bacterial Infections: DNA Replication, Transcription, and Translation
Alexander J. McAdam and Donald M. Coen
BIOCHEMISTRY OF BACTERIAL DNA REPLICATION, TRANSCRIPTION, AND TRANSLATION |
Inhibitors of Transcription: Fidaxomicin and Rifamycin Derivatives |
The central dogma processes—DNA replication, transcription, and translation—are generally similar in bacteria and humans. DNA is replicated and transcribed into RNA, and messenger RNA is translated into protein. However, there are important differences in the biochemistry of bacterial and human central dogma processes, and these differences can be exploited for the development and clinical use of antibiotics. Three such differences are targeted by the currently available antibacterial chemotherapeutic drugs: (1) topoisomerases, which regulate supercoiling of DNA and mediate segregation of replicated strands of DNA; (2) RNA polymerases, which transcribe DNA into RNA; and (3) ribosomes, which translate messenger RNA (mRNA) into protein. This chapter briefly reviews the biochemistry of central dogma processes in bacteria and discusses certain relevant differences between these processes in bacteria and humans. With this background, the chapter discusses the mechanisms by which pharmacologic agents interrupt bacterial DNA replication, transcription, and translation.
 It is the summer of 1976. Participants returning from an American Legion convention in Philadelphia are falling severely ill with a mysterious type of pneumonia. The outbreak centers on the Bellevue Stratford Hotel, where 150 hotel occupants and 32 passersby contract “Legionnaires’ disease.” Twenty-nine victims ultimately die. Conventional sputum stains, cultures, and even autopsy material show no consistent pathogens. The terror of an unknown epidemic disease sparks rumors and news reports of poison gases, tainted water supplies, terrorists, and deadly viruses.
It is the summer of 1976. Participants returning from an American Legion convention in Philadelphia are falling severely ill with a mysterious type of pneumonia. The outbreak centers on the Bellevue Stratford Hotel, where 150 hotel occupants and 32 passersby contract “Legionnaires’ disease.” Twenty-nine victims ultimately die. Conventional sputum stains, cultures, and even autopsy material show no consistent pathogens. The terror of an unknown epidemic disease sparks rumors and news reports of poison gases, tainted water supplies, terrorists, and deadly viruses.
Several months later, laboratory and field investigation teams from the Centers for Disease Control and Prevention (CDC) identify the causative aerobic Gram-negative bacterium and name it Legionella pneumophila. It is observed that patients treated with erythromycin, a macrolide antibiotic, and tetracycline have better outcomes than those treated with other agents. Today, newer macrolides, such as azithromycin, and fluoroquinolones are often used for treating Legionnaires’ disease—as well as many chlamydial, streptococcal, and staphylococcal infections.
Questions
1. Why are some antibiotics such as quinolones and aminoglycosides bactericidal, while other antibiotics such as tetracyclines and macrolides are bacteriostatic?
2. Which step in translation is blocked by tetracyclines and which by macrolides?
3. How do bacteria develop resistance to macrolide and fluoroquinolone antibiotics? Do they need to acquire exogenous DNA in order to become resistant to these antibiotics?
 BIOCHEMISTRY OF BACTERIAL DNA REPLICATION, TRANSCRIPTION, AND TRANSLATION
BIOCHEMISTRY OF BACTERIAL DNA REPLICATION, TRANSCRIPTION, AND TRANSLATION
The central dogma of molecular biology begins with the structure of DNA, which is the macromolecule that carries genetic information. To transmit all of the genetic information in a cell to two progeny cells, the parental DNA must be copied in its entirety (replicated), and the two resulting copies must be segregated—one copy going to each progeny cell. In order to express the genes that are present in the DNA, these specific portions of the DNA are copied (transcribed) into RNA. Some RNAs (mRNAs) are then read (translated) by the protein synthesis machinery in order to produce proteins. Certain other RNAs, such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), perform complex functions essential to protein synthesis. It is important to note that the following discussion of these bacterial processes is vastly simplified in order to emphasize the steps that are inhibited by antibiotics.
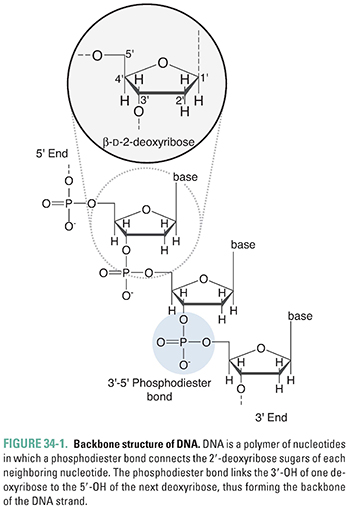
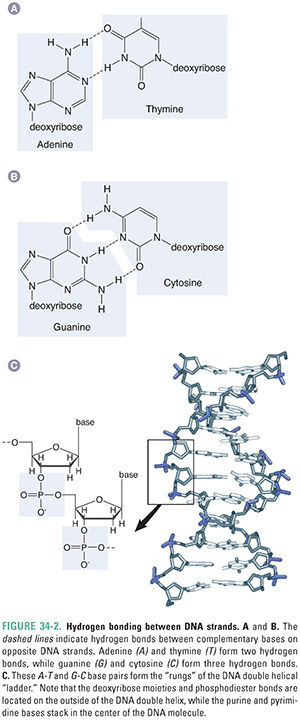
DNA is composed of two strands of polymerized deoxyribonucleotides that wind around one another in a “double helix” conformation. The 3′-hydroxyl group of each nucleotide’s deoxyribose ring is joined by a phosphate group to the 5′-hydroxyl group of the next nucleotide, thereby forming the phosphodiester backbone of each side of the double helical “ladder” (Figs. 34-1 and 34-2). The purines adenine (A) and guanine (G) and the pyrimidines thymine (T) and cytosine (C), which are covalently linked to the deoxyribose ring, associate with one another (A with T, G with C) via hydrogen bonds to form the “rungs” of the ladder (Fig. 34-2). It is the linear sequence of bases that encodes the genetic information of a cell. How the nucleotide precursors to these bases are synthesized is reviewed in Chapter 39, Pharmacology of Cancer: Genome Synthesis, Stability, and Maintenance. DNA structure is essentially the same between bacteria and eukaryotes. However, bacterial chromosomes are usually circular DNAs, while eukaryotic chromosomes, including our own, are linear molecules.
DNA Replication, Segregation, and Topoisomerases
The faithful replication and segregation of bacterial DNA to progeny cells involve numerous steps, many of which could make good targets for antibacterial drugs. To date, the enzymes in this process that have been most successfully targeted are topoisomerases. These enzymes perform several functions during DNA replication and segregation.
During DNA replication in both bacterial and eukaryotic cells, complementary strands of DNA are synthesized bidirectionally, forming two so-called replication forks. To initiate this process, the two DNA strands that compose the double helix must unwind and separate. In so doing, the DNA strands form excess “supercoils” in which the helical polymer overtwists as it rotates in the same direction as the turn of the helix. Supercoils increase tension in DNA strands and thereby interfere with further unwinding. In the absence of a process to relieve the stress created by the supercoils, the entire chromosome would have to rotate; this process would be complex and energy-consuming and could entangle the entire molecule.
Moreover, when DNA replication is completed, the two progeny DNA copies are wrapped around each other. In bacteria, because the chromosomes are circular, the intertwined progeny copies form interlocking rings (catenanes). These intertwined rings must be separated (resolved) before they can be segregated to the progeny cells.
Topoisomerases perform both of these functions—removing excess DNA supercoils during DNA replication and separating intertwined progeny DNA. Topoisomerases catalyze these activities by breaking, rotating, and resealing DNA strands. There are two types of topoisomerases. Type I topoisomerases form and reseal single-stranded breaks in DNA to decrease positive supercoiling (Fig. 34-3). Type II topoisomerases form and reseal double-stranded breaks (Fig. 34-4). Both types of topoisomerases can remove excess DNA supercoils during DNA replication. However, only type II topoisomerases can resolve intertwined copies of double-stranded DNA to permit segregation of the DNA to daughter cells. Type II enzymes are both more complex and more versatile than type I topoisomerases, and the type II enzyme is a more common molecular target for chemotherapeutic agents.
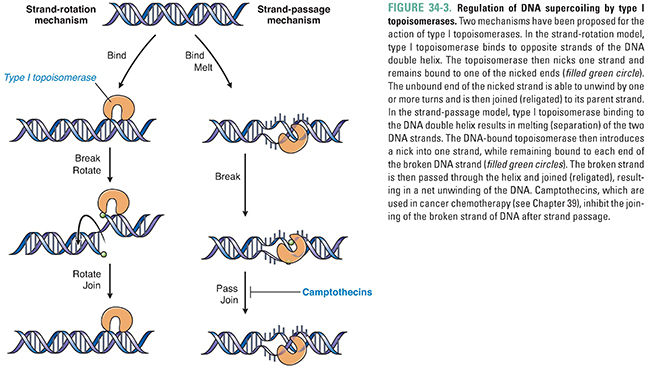
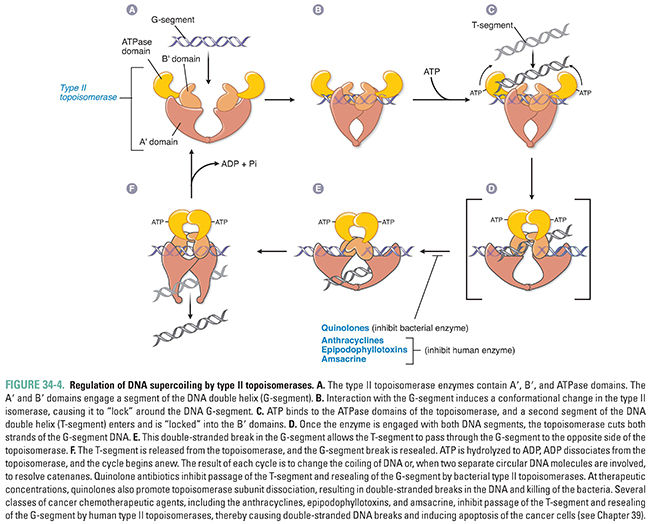
The mechanism of action of a type II topoisomerase proceeds in two steps. First, the enzyme binds a segment of DNA and forms covalent bonds with phosphates from each strand, thereby nicking both strands. Second, the enzyme causes a second stretch of DNA from the same molecule to pass through the break, relieving supercoiling (Fig. 34-4). This passage of double-stranded DNA through a double-stranded break permits separation of intertwined copies of DNA following replication and, thereby, segregation of DNA into progeny cells.
Two main type II topoisomerases are present in bacteria. The first to be identified, DNA gyrase, is a type II topoisomerase that is unusual in that it can introduce negative supercoils before the DNA strands separate and thereby neutralize positive supercoils that form during DNA unwinding. The second is topoisomerase IV. DNA gyrase is particularly crucial for segregation in some bacteria, while topoisomerase IV is the critical enzyme in other bacteria.
Because supercoiling is important for transcription as well as segregation, topoisomerases influence this central dogma process as well. Given their multiple functions, topoisomerases are usually engaged with DNA, and this is important for their roles as drug targets. These enzymes are important not only as antibacterial drug targets but also as targets for cancer chemotherapy (see Chapter 39).
Gene expression begins with transcription, which involves the synthesis of single-stranded RNA transcripts from a DNA template. Transcription is catalyzed by the enzyme RNA polymerase. In bacteria, five subunits (2 α, 1 β, 1 β′, and 1 σ) associate to form the holoenzyme. As discussed below, the σ subunit is instrumental for initiating transcription, while the rest of the RNA polymerase enzyme—also known as the core enzyme—contains the catalytic machinery for RNA synthesis.
The process of transcription occurs in three stages: initiation, elongation, and termination (Fig. 34-5). During initiation, the RNA polymerase holoenzyme binds to and then separates the strands of a short segment of double-helical DNA after its σ subunit recognizes an upstream site. Once the double helix is unwound to form a single-stranded template, RNA polymerase initiates RNA synthesis at a start site on the DNA. Initiation entails conformational changes in the enzyme so that it opens and closes around DNA and it makes appropriate contacts with unwound DNA and the nascent RNA. During elongation, RNA polymerase synthesizes a complementary RNA strand by joining together ribonucleoside triphosphates via phosphodiester bonds. In the process, the σ subunit dissociates from the holoenzyme. RNA synthesis proceeds in the 5′→3′ direction, with the nascent RNA strand emerging from an exit channel of the enzyme, until a termination sequence is reached.
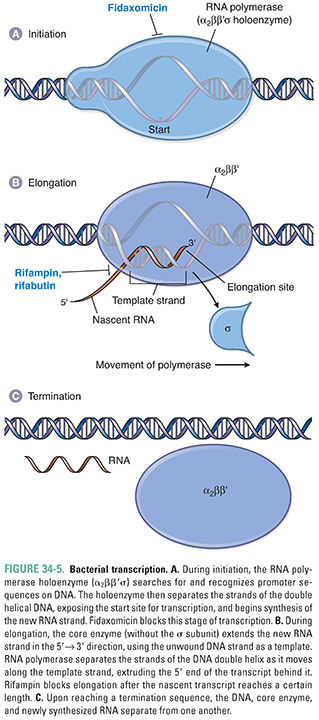
The RNA polymerase enzyme differs between bacteria and humans and thus can serve as a selective target for antibacterial drug action. In bacteria, one RNA polymerase synthesizes all of the RNA in the cell (except for the short RNA primers needed for DNA replication, which are made by primase). In contrast, eukaryotes express three different nuclear RNA polymerases, and each enzyme is considerably more complex in its subunit structure than the bacterial counterpart.
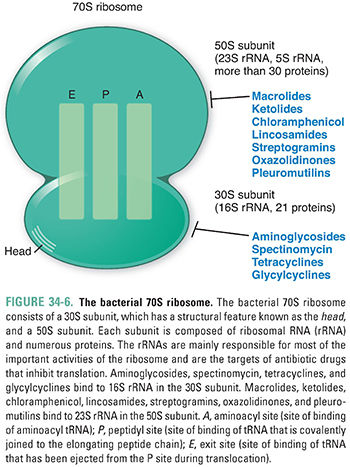
Once the mRNA transcripts are synthesized from a DNA template, these transcripts are translated by the bacterial translational machinery. Although the overall process of translation is similar between bacteria and higher organisms, there are a number of pharmacologically exploitable differences in the details of the mechanisms. In particular, the composition of the rRNA molecules differs between bacterial and human ribosomes. Thus, bacterial ribosomes can also serve as selective targets for antibiotics.
The ribosome of a representative bacterium, Escherichia coli, has a sedimentation coefficient of 70S and is composed of a 30S subunit and a 50S subunit. The 30S subunit contains a single 16S rRNA molecule and 21 different proteins, while the 50S subunit contains two rRNA molecules—23S rRNA and 5S rRNA—and more than 30 different proteins. Importantly, it is the rRNAs rather than the protein components of the ribosome that are mainly responsible for the ribosome’s key activities, namely, decoding the mRNA, linking together amino acids, and translocating the translation machine. The 70S ribosome contains three sites that bind tRNAs during translation: the A or “aminoacyl” site (also known as the “acceptor” site), which binds incoming tRNA molecules carrying the various amino acids; the P or “peptidyl” site, which contains the growing peptide chain; and the E or “exit” site, which binds the tRNAs that have been used during translation before they are ejected from the ribosome (Fig. 34-6).
Translation, like transcription, can be divided into three stages (Fig. 34-7). During initiation, the components of the translation system assemble together, abetted by proteins called initiation factors. First, the mRNA joins with the 30S subunit of the bacterial ribosome and with a specific tRNA molecule linked to formylated methionine (fMet), the first amino acid encoded by every bacterial mRNA. The tRNA-formylated methionine molecule (fMet-tRNAf) binds to its initiation codon (AUG) on the mRNA. Next, the 50S subunit joins with the 30S subunit to form the complete 70S ribosome. The fMet-tRNAf now occupies the P site of the 70S ribosome.
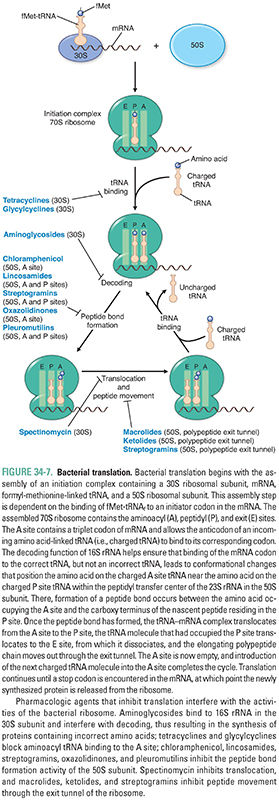
Elongation, abetted by elongation factors, involves the addition of amino acids to the carboxyl end of the growing polypeptide chain, as the ribosome moves from the 5′-end to the 3′-end of the mRNA that is being translated. tRNA molecules carrying specific amino acids (aminoacyl tRNAs) enter the ribosomal A site, and their three base anticodons base-pair to their complementary codons on the mRNA. Utilization of the correct tRNA requires not only anticodon–codon recognition between tRNA and mRNA, respectively, but also decoding functions provided largely by the 16S rRNA in the decoding center of the 30S ribosomal subunit. Binding of the correct tRNA to its codon in the A site leads to several conformational changes—in 16S rRNA within the decoding center, in a region of 23S rRNA of the 50S subunit that protrudes into the decoding center, in the A site tRNA, and in protein elongation factors. These movements culminate in the positioning of the tRNA-bound amino acid in the A site so that it is close to the fMet (on the P site tRNA) in a region of the 50S subunit known as the peptidyl transferase center. This center, which is formed largely by the 23S rRNA, catalyzes the formation of a peptide bond between fMet and the next amino acid. Once the peptide bond is formed linking fMet to the next amino acid, which, in turn, is linked to the tRNA in the A site, the tRNA in the A site is said to have “accepted” the fMet.
After the peptide bond has been formed, the ribosome undergoes more conformational changes, including rotation (“ratcheting”) of the 30S subunit relative to the 50S subunit and a swiveling motion of the “head” of the 30S subunit, so that both subunits advance three nucleotides toward the 3′-end of the mRNA. In this multistep process, the tRNAf that was originally linked to the fMet is ejected from the P site and binds to the E site, the tRNA that is now linked to two amino acids shifts from the A site to the unoccupied P site, and the A site becomes available. This process is known as translocation. As these translocation events occur, the nascent polypeptide starts to move through the exit tunnel of the ribosome. In this manner, polypeptide chain elongation results from multiple cycles of aminoacyl tRNA binding to the A site, peptide bond formation, and translocation.
During termination, proteins called release factors recognize the termination codon in the A site and activate discharge of the newly synthesized protein and dissociation of the ribosome–mRNA complex, with the 70S ribosome separating into its 50S and 30S subunits.
Three general points are worth noting about bacterial translation. First, the two ribosomal subunits demonstrate segregated functions: the 30S subunit is largely responsible for faithful decoding of the mRNA message, while the 50S subunit catalyzes peptide bond formation. Translocation, however, involves both subunits. Second, these functions are performed mainly by the RNA components of the ribosome. Third, inhibitors of protein synthesis affect the process of translation at different steps and, indeed, have greatly helped dissect these steps.
 PHARMACOLOGIC CLASSES AND AGENTS
PHARMACOLOGIC CLASSES AND AGENTS
Three general categories of drugs target bacterial DNA replication, transcription, and translation: (1) drugs that target type II topoisomerases, (2) drugs that target RNA polymerase, and (3) drugs that target ribosomes. Quinolone antibiotics are broad-spectrum agents that not only inhibit certain topoisomerases but also convert these enzymes into DNA-damaging agents. Fidaxomicin and rifamycin derivatives bind to and inhibit bacterial RNA polymerase. Fidaxomicin is used to treat colitis caused by Clostridium difficile, and one rifamycin derivative, rifampin, is a mainstay in the therapy of tuberculosis. Multiple classes of drugs bind bacterial ribosomes to inhibit protein synthesis. Specifically, aminoglycosides, spectinomycin, tetracyclines, and glycylcyclines bind the 30S ribosomal subunit, while macrolides, ketolides, chloramphenicol, lincosamides, streptogramins, oxazolidinones, and pleuromutilins target the 50S ribosomal subunit. These inhibitors of protein synthesis generally act on both Gram-positive and Gram-negative organisms and are therefore in wide clinical use (see Chapter 35, Pharmacology of Bacterial and Mycobacterial Infections: Cell Wall Synthesis, for a discussion of Gram-positive and Gram-negative bacteria).
Elucidation of the mechanisms of action of the agents described below has depended crucially on the field of bacterial genetics. In particular, the molecular targets of antibiotics have been identified by (1) isolating bacteria that are resistant to the particular antibiotic (e.g., rifampin), (2) showing that the target molecule (e.g., RNA polymerase) exhibits biochemical resistance to the antibiotic, and (3) demonstrating that the drug-resistance mutation lies within the gene encoding the target (e.g., the gene encoding the β subunit of RNA polymerase). More recent work, using nuclear magnetic resonance spectroscopy and x-ray crystallography, has further elucidated the structures of the targets as well as the molecular nature of the various drug–target interactions. Indeed, the 2009 Nobel Prize in Chemistry was awarded for crystallographic analyses of ribosomes and their binding to certain antibiotics.
Inhibitors of Topoisomerases: Quinolones
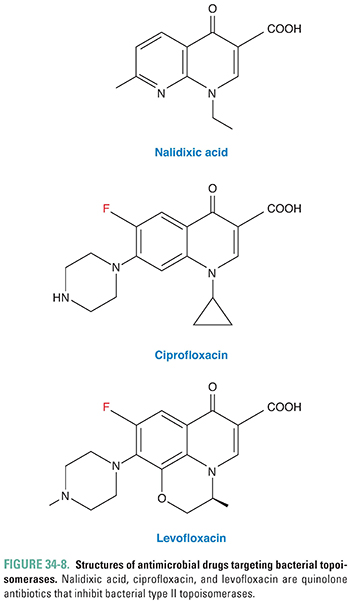
Quinolones are a major class of bactericidal antibiotics that act by inhibiting bacterial type II topoisomerases. One of the earliest quinolones to enter clinical use was nalidixic acid (Fig. 34-8), and the mechanism of action of the quinolones was elucidated largely by studying this drug. Fluoroquinolones (all ending in “-floxacin”) have a fluorine atom at position 6 (Fig. 34-8), which increases the potency and spectrum of bacteria killed by these agents compared to nalidixic acid. The older fluoroquinolones, ciprofloxacin, norfloxacin, and ofloxacin, are used to treat urinary tract infections and gastrointestinal infections caused by Gram-negative bacteria, including E. coli, Klebsiella pneumoniae, Campylobacter jejuni, and Enterobacter, Salmonella, and Shigella species. Newer fluoroquinolones, including gemifloxacin, moxifloxacin, and levofloxacin, retain activity against Gram-negative bacteria and also have activity against Streptococcus pneumoniae and bacteria that cause atypical pneumonia (Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella pneumophila), so these drugs are commonly used to treat bacterial pneumonia. Levofloxacin and moxifloxacin have activity against Mycobacterium tuberculosis and are sometimes used in combination with other antimycobacterial drugs to treat tuberculosis. Fluoroquinolone resistance is common in staphylococci, so other antibiotics are usually used to treat infections with these organisms. Bacteria typically evolve resistance to the quinolones through chromosomal mutations in the genes that encode type II topoisomerases or through alterations in the expression of membrane porins and efflux pumps that determine the concentration of drug inside the bacteria. Adverse effects are infrequent but can include nausea, vomiting, and diarrhea and, rarely, tendinitis, tendon rupture, and peripheral neuropathy.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree



