Predicted CYP2D6 metabolism phenotype
Implications for codeine metabolism
Therapeutic recommendations
Ultrarapid metabolizer (UM)
Increased formation of morphine following codeine administration, leading to higher risk of toxicity
Avoid codeine use due to potential for toxicity
Extensive metabolizer (EM)
Normal morphine formation
Use label-recommended age- or weight-specific dosing
Intermediate metabolizer (IM)
Reduced morphine formation
Use label-recommended age- or weight-specific dosing. If no response, consider alternative analgesics such as morphine or a non-opioid
Poor metabolizer (PM)
Greatly reduced morphine formation following codeine administration, leading to insufficient pain relief
Avoid codeine use due to lack of efficacy
Table 21.2
CPIC recommendations for CYP2D6-directed dosing of tricyclic antidepressantsa,b
Predicted CYP2D6 metabolism phenotype | Implications for tricyclic antidepressant metabolism | Therapeutic recommendations |
|---|---|---|
Ultrarapid metabolizer (UM) | Increased metabolism of tricyclics to less active compounds when compared to extensive metabolizers. Lower plasma concentrations will increase the probability of pharmacotherapy failure | Avoid tricyclic use due to potential lack of efficacy. Consider alternative drug not metabolized by CYP2D6 If a tricyclic is warranted, consider increasing the starting dose. Utilize therapeutic drug monitoring to guide dose adjustments |
Extensive metabolizer (EM) | Normal metabolism of tricyclics | Initiate therapy with recommended starting dose |
Intermediate metabolizer (IM) | Reduced metabolism of tricyclics to less active compounds when compared to extensive metabolizers. Higher plasma concentrations will increase the probability of side effects | Consider 25 % reduction of recommended starting dose. Utilize therapeutic drug monitoring to guide dose adjustments |
Poor metabolizer (PM) | Greatly reduced metabolism of tricyclics to less active compounds when compared to extensive metabolizers. Higher plasma concentrations will increase the probability of side effects | Avoid tricyclic use due to potential for side effects. Consider alternative drug not metabolized by CYP2D6 If a tricyclic is warranted, consider 50 % reduction of recommended starting dose. Utilize therapeutic drug monitoring to guide dose adjustments |
Available Assays
CYP2D6 genotyping typically is performed by platforms that simultaneously detect multiple variants across the gene (Fig. 21.1). Several commercial assays are currently available, including the AmpliChip® (Affymetrix/Roche [14]), Luminex [15], and AutoGenomics [16] assays, as well as other laboratory-developed tests (e.g., single-nucleotide extension assay) [17]. The AmpliChip® CYP450 Test (Roche Diagnostics, Indianapolis, IN) is an oligonucleotide microarray hybridization method that tests variants in both CYP2D6 and CYP2C19. The Tag-It™ Luminex (Luminex Molecular Diagnostics, Toronto, Canada) platform is a bead array with oligonucleotides bound to microspheres and genotyping by allele-specific primer extension. The AutoGenomics (Carlsbad, CA) platform is a film-based microarray tested on the INFINITI® PLUS Analyzer. These assays typically interrogate 15–20 important CYP2D6 variants including the deletion and duplication alleles.
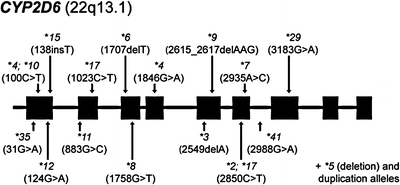

Figure 21.1
Gene diagram of CYP2D6 (and chromosome location) indicating common alleles and their molecular alteration (in parentheses), including the deletion (*5) and duplication alleles. Note that variants are referred to by their common names (GenBank Accession Number M33388)
Interpretation
CYP2D6 alleles are designated by the common star (*) allele nomenclature system, which often include multiple variants on the same haplotype. The CYP2D6*1 allele is the wild-type haplotype encoding normal enzyme activity; however, this is typically assigned in the absence of other detected variants. Consequently, when *1 is reported by targeted genotyping, a rare CYP2D6 star (*) allele not included in the genotyping panel would not be detected, which can only be identified by gene sequencing. Commonly interrogated CYP2D6 alleles are summarized in Table 21.3.
Table 21.3
Examples of commonly tested pharmacogenetic alleles and their effect on enzyme activity
Gene | Loss-of-function alleles | Reduced-function alleles | Increased-function alleles |
|---|---|---|---|
CYP2D6 | *3, *4, *5, *6, *7, *8, *12, *14 | *9, *10, *17, *29, *41 | Functional allele duplication |
CYP2C19 | *2, *3, *4, *5, *6, *7, *8 | *9, *10 | *17 |
CYP2C9 | *3, *6 | *2, *5, *8, *11 | – |
TPMT | *2, *3A, *3B, *3C, *4 | – | – |
Two functional CYP2D6 alleles or one functional and one reduced-function CYP2D6 allele predict normal or extensive metabolizers. Two reduced-function alleles or one reduced-function and one nonfunctional allele predict intermediate metabolizers. Two nonfunctional alleles predict a poor metabolizer phenotype. Duplicated functional CYP2D6 alleles predict an ultrarapid metabolizer phenotype [8]. In addition to duplicated functional alleles, duplicated nonfunctional or reduced-function alleles also have been described. As such, determining which CYP2D6 allele is duplicated is important for proper interpretation when a gene duplication is identified in addition to a heterozygous genotype [18].
CYP2D6 genotyping is complicated by the fact that several variants occur on a number of important star (*) alleles and commercial assays cannot determine the phase of identified genotypes [19]. For example, the nonfunctional *4 allele is defined by several variants, including 100C > T and 1846G > A. The 1846G > A variant is the defining functional mutation for *4, which disrupts exon splicing and results in a frameshift and loss of enzyme activity. However, the *10 reduced-function allele also has the 100C > T variant but without 1846G > A. Of note, 1846G > A can also exist on a haplotype without 100C > T (*4M), and this allele is very rare. When one copy of each variant is detected, the most probable genotype is heterozygous *4 (one copy of a nonfunctional allele), since 100C > T and 1846G > A are presumed to be on the same haplotype. As genotyping platforms generally do not predict CYP2D6 haplotypes, individual laboratories are responsible for interpreting results and reporting CYP2D6 genotypes. Without haplotyping by more involved molecular assays, it is possible that one copy of both 100C > T and 1846G > A could be misinterpreted as a *4M/*10 compound heterozygote (one nonfunctional allele and one decreased-function allele). Although this scenario would be rare in the general population, the *4/*10 genotype is more appropriately defined by two copies of 100C > T and one copy of 1846G > A.
Further interpretation challenges exist when a duplication is detected with a reduced-function or nonfunctional allele, as most platforms are not capable of identifying which allele is duplicated. Furthermore, the number of duplicated CYP2D6 copies on an allele is not typically determined. When in combination with a reduced-function allele, the number of copies may become clinically relevant in predicting the phenotype. Family studies (such as parental testing) may help determine the phase of identified variants in these scenarios, if warranted.
Laboratory Issues
The CYP2D6 gene is highly homologous to a related pseudogene (CYP2D7P1), which complicates the design of PCR primers used for genotyping. As such, primers are designed to avoid co-amplification of pseudogenes by employing a specific long-range PCR of the entire CYP2D6 gene. Given the technical challenges with long-range PCR, suboptimal or degraded DNA may not perform well for many commercial CYP2D6 assays.
When a CYP2D6 platform is unable to identify if a duplication is a functional or nonfunctional allele in a heterozygous sample, laboratories may report an “indeterminant” result. As noted above, family studies can facilitate identifying which allele is duplicated in these cases. Although laboratory guidelines for CYP2D6 genotyping in relation to tamoxifen therapy have recently been reported [19], no current professional guidelines detail which alleles should be included in clinical CYP2D6 assays. Therefore, different laboratories may include different CYP2D6 alleles in their testing panels, which can result in conflicting CYP2D6 genotypes and predicted phenotypes between laboratories.
CYP2C19
Cytochrome P450-2C19 (CYP2C19) is another important member of the cytochrome P450 superfamily that metabolizes commonly prescribed medications including anticonvulsants, antidepressants, antifungals, proton-pump inhibitors, antithrombotics, and chemotherapy, antimalarial, and antiulcer drugs [20]. Another drug metabolized by CYP2C19 is the antiplatelet agent clopidogrel, commonly prescribed for patients with acute coronary syndromes and those undergoing percutaneous coronary intervention (PCI). CYP2C19 converts clopidogrel in a two-step enzymatic reaction to an active metabolite, which binds irreversibly to platelet receptors and inhibits aggregation for the duration of the platelet life span. About 25 % of ACS/PCI patients have reduced platelet inhibition due in part to CYP2C19 loss-of-function alleles [21], which reduces the effectiveness of clopidogrel treatment.
Although over 30 CYP2C19 variant alleles have been identified, the effect of some rare alleles on enzyme function has not been established. Notable among the CYP2C19 alleles are *2 to *8, which impart loss of function, and *17, which has been reported as an increased-function allele (http://www.cypalleles.ki.se/cyp2c19.htm). In contrast to the role of the CYP2C19*2 to *8 alleles in reduced clopidogrel effectiveness, *17 may be associated with an enhanced response to clopidogrel and some antidepressants [22]. Variant CYP2C19 allele frequencies vary between racial and ethnic groups, with *2 being present in approximately 30 % of Asians and 15 % of Caucasians and African Americans, while *3 has a frequency of approximately 8 % in the Asian population, but is rare in other populations.
Clinical Utility
CYP2C19 genotyping can identify individuals who should avoid medications or may require modified doses of medications metabolized by CYP2C19. For ACS/PCI patients being treated with clopidogrel, CYP2C19*2 and *3 carriers have reduced platelet inhibition compared to extensive metabolizers due to reduced metabolic activation of the prodrug. Consequently, an alternative antiplatelet agent has been recommended for ACS/PCI patients who are CYP2C19 intermediate and poor metabolizers [23]. For tamoxifen, CYP2C19*17 carriers have been shown to produce higher concentrations of the active endoxifen metabolite and may have a more favorable outcome than *1, *2, and *3 carriers [24]. CPIC guidelines are available for the use of CYP2C19 genotyping results for patients treated with tricyclic antidepressants [13, 25] (Table 21.4 ) and clopidogrel [23, 25] (Table 21.5 ).
Table 21.4
CPIC recommendations for CYP2C19-directed dosing of tricyclic antidepressantsa,b
Predicted CYP2C19 metabolism phenotype | Implications for tricyclic antidepressant metabolism | Therapeutic recommendations |
|---|---|---|
Ultrarapid metabolizer (UM) | Increased metabolism of amitriptyline when compared to extensive metabolizers | Consider alternative drug not metabolized by CYP2C19. If a tricyclic is warranted, utilize therapeutic drug monitoring to guide dose adjustments |
Extensive metabolizer (EM) | Normal metabolism of amitriptyline | Initiate therapy with recommended starting dose |
Intermediate metabolizer (IM) | Reduced metabolism of amitriptyline when compared to extensive metabolizers | Initiate therapy with recommended starting dose |
Poor metabolizer (PM) | Greatly reduced metabolism of amitriptyline when compared to extensive metabolizers. Higher plasma concentrations of amitriptyline will increase the probability of side effects | Consider 50 % reduction of recommended starting dose. Utilize therapeutic drug monitoring to guide dose adjustments |
Table 21.5
CPIC recommendations for CYP2C19-directed antiplatelet therapy when considering clopidogrel for ACS patients undergoing PCIa
Predicted CYP2D6 metabolism phenotype | Implications for clopidogrel | Therapeutic recommendations |
|---|---|---|
Ultrarapid metabolizer (UM) | Increased platelet inhibition; decreased residual platelet aggregationb | Clopidogrel label-recommended dosage and administration |
Extensive metabolizer (EM) | Normal platelet inhibition; normal residual platelet aggregation | Clopidogrel label-recommended dosage and administration |
Intermediate metabolizer (IM) | Reduced platelet inhibition; increased residual platelet aggregation; increased risk for adverse cardiovascular events | Alternative antiplatelet therapy (if no contraindication); e.g., prasugrel, ticagrelor |
Poor metabolizer (PM) | Significantly reduced platelet inhibition; increased residual platelet aggregation; increased risk for adverse cardiovascular events | Alternative antiplatelet therapy (if no contraindication); e.g., prasugrel, ticagrelor |
Available Assays
A number of multiplexed molecular assays are available to identify selected variant CYP2C19 alleles. For example, CYP2C19 is combined with CYP2D6 in the AmpliChip® assay (Roche) described above. Additional CYP2C19 assays are commercially available from Luminex Molecular Diagnostics (xTAG® CYP2C19 Kit), AutoGenomics (Infiniti® CYP2C19 Assay), Nanosphere (Verigene® CYP2C19 Test), GenMark Diagnostics (eSensor® 2C19 Test), Sequenom (iPLEX® ADME CYP2C19 Panel), and Spartan Bioscience (Spartan RX CYP2C19 Test). All these assays include the common nonfunctional alleles (*2 and *3, with or without *4 to *8), and some also include *17 and other variants (Fig. 21.2; Table 21.3 ).
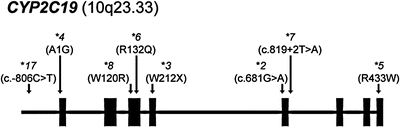

Figure 21.2
Gene diagram of CYP2C19 (and chromosome location) indicating common alleles and their molecular alteration (in parentheses)
Interpretation
Based on identified CYP2C19 genotype, individuals typically are categorized as extensive (*1/*1), intermediate (e.g., *1/*2, *1/*3), or poor (e.g., *2/*2, *2/*3) metabolizers [20]. The frequencies of CYP2C19 poor metabolizers are 2–5 % of Caucasians and African Americans and approximately 15 % of Asians [23, 26]. Additionally, individuals who have at least one copy of the CYP2C19*17 allele often are categorized as ultrarapid metabolizers (e.g., *17/*17). However, given that *17 is unable to completely compensate for the *2 loss-of-function allele [27], *2/*17 compound heterozygotes can be classified as intermediate metabolizers, although some laboratories may report the predicted phenotype for this genotype as “unknown” or “indeterminate.”
Laboratory Issues
Although most commercial assays include the common CYP2C19*2 and *3 loss-of-function alleles, some assays also include the *4 to *8 alleles. Moreover, some assays include the *17 increased-function allele. Like other CYP450 genotyping assays, the wild-type *1 allele is assigned in the absence of other detected alleles. Consequently, different laboratories may test different allele panels which can result in conflicting CYP2C19 genotypes and predicted phenotypes depending on which alleles were interrogated.
CYP2C9 and VKORC1 (Warfarin Sensitivity)
Warfarin is a commonly prescribed vitamin K antagonist for the prevention of thromboembolism among patients with atrial fibrillation, deep vein thrombosis, and other indications. However, the drug has a very narrow therapeutic index and a large interindividual variability in response, in part due to inherited genetic variability within genes involved in warfarin pharmacokinetics and pharmacodynamics. For example, cytochrome P450-2C9 (CYP2C9) is the principal enzyme involved in S-warfarin inactivation and elimination. Like other cytochrome P450 genes, CYP2C9 is highly polymorphic with several known variant alleles encoding reduced enzyme activity (http://www.cypalleles.ki.se/cyp2c9.htm). Importantly, in vitro studies have shown that the common *2 and *3 alleles are functionally defective, exhibiting only approximately 70 % and 5 % of wild-type activity toward S-warfarin, respectively [28]. Consequently, these CYP2C9 alleles result in reduced warfarin inactivation, higher warfarin blood levels with standard warfarin doses, and increased bleeding risks [29, 30]. The frequencies of *2 and *3 are approximately 15 % and 6 %, respectively, among Caucasians and 2–4 % among Asians and African Americans [31]. Of note, other variant alleles are more prevalent in individuals of African descent (e.g., *5, *6, and *8) [31], which may provide additional utility for genetically guided warfarin dosing in these populations.
Warfarin acts as a vitamin K antagonist by inhibiting the regeneration of reduced vitamin K, an essential cofactor for the clotting cascade. The target enzyme for warfarin is VKORC1, which catalyzes the rate-limiting step in the vitamin K cycle [32]. Importantly, common VKORC1 haplotypes that result in reduced gene expression have been reproducibly implicated as the major genetic determinant of warfarin dose variability [33–38]. The most commonly interrogated VKORC1 allele associated with warfarin sensitivity is the c.-1639G > A promoter polymorphism (Fig. 21.3) [38]. Like CYP2C9, the frequencies of VKORC1 alleles vary between racial and ethnic groups, with c.-1639G > A allele frequencies of approximately 10 %, 45 %, and 70–90 %, among African-American, Caucasian, and Asian individuals, respectively [31]. Of note, rare VKORC1 coding region mutations also have been identified that are strongly correlated with warfarin resistance.
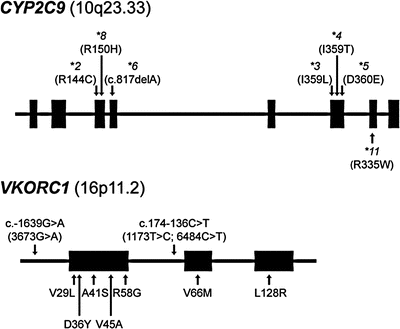

Figure 21.3
Gene diagrams of CYP2C9 and VKORC1 (and chromosome locations) indicating common alleles and their molecular alteration (in parentheses). Note the rare “warfarin-resistant” VKORC1 mutations listed below the VKORC1 gene, which are not commonly included in clinical CYP2C9 and VKORC1 genetic tests
Taken together, common CYP2C9 and VKORC1 variant alleles account for approximately 35 % of interindividual dose variability [36, 37], which can be used in conjunction with known clinical variables (e.g., age, race, body weight, gender) to predict individual therapeutic warfarin doses. These data prompted the US Food and Drug Administration (FDA) to modify the warfarin label noting the importance of pharmacogenetic testing with dosing recommendations based on CYP2C9 and VKORC1 genotypes (Table 21.6 ).
Table 21.6
Range of expected therapeutic warfarin doses based on CYP2C9 and VKORC1 genotypesa
CYP2C9 | ||||||
|---|---|---|---|---|---|---|
VKORC1 (c.-1639G > A) | *1 / *1 | *1 / *2 | *1 / *3 | *2 / *2 | *2 / *3 | *3 / *3 |
G/G | 5–7 mg | 5–7 mg | 3–4 mg | 3–4 mg | 3–4 mg | 0.5–2 mg |
G/A | 5–7 mg | 3–4 mg | 3–4 mg
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

| |||