Figure 54.1
The most common DNA evidence in US crime laboratories is a vaginal swab from a rape kit. This photomicrograph is a stained vaginal smear from a rape kit. The arrows indicate spermatozoa. In addition to the DNA from the male contributor, there is DNA from the female epithelial squamous and white blood cells, as well as that of the microbial flora
Sexual Assaults (Swabs)
In the USA, rape kits have dominated the evidential submissions to forensic DNA laboratories (Fig. 54.1). Often, the demand for DNA testing on rape kits outstrips the ability of crime laboratories’ testing capacity and large backlogs may exist despite substantial NIJ grant programs to reduce them [7]. Typical rape kits include vaginal, anal, and oral evidentiary swabs, buccal reference swabs, pubic combings, and exemplars of pubic and scalp hairs. If a condom was used by a rapist and later found, it may yield semen from the male perpetrator on the inside and vaginal epithelial cells from the female victim on the outside. Vaginal swab specimens are inherently mixed samples. Most commonly the DNA of the spermatozoa is partially purified by a differential extraction procedure in which the female fraction is released using a gentle lysis medium, after which male DNA is released from the sperm using a solution containing a strong reducing agent (dithiothreitol) to break the disulfide bonds in the capsules of the spermatozoan heads [8]. Laser capture of spermatozoa from microscopic slides has also been successfully used, while immunologic affinity methods have thus far been disappointing. Y-chromosome DNA markers (described below) are an alternative method of capturing male identity information.
Other Violent Crimes (Blood, Other Evidence)
Blood and similar specimens from homicides and other violent crimes are the next-most-common evidentiary materials submitted to forensic laboratories [9]. In an early study of biological evidence at crime scenes, blood was found to be present in 60 % of murders, assaults, and batteries [10]. The DNA can come from myriad items and materials. Saliva may be deposited on beverage containers, envelope seals, gum, cigarettes, or food (Fig. 54.2). Investigators have followed suspects to obtain “abandoned” specimens, such as facial tissues, cigarette butts, gum, or drinking glass. Cords used as a murder weapon for strangulation can yield both victim and perpetrator DNA (Fig. 54.3). Shed hairs, which contain little or no nuclear DNA (nDNA), still harbor mitochondrial DNA (mtDNA), which also can be used for identification purposes. Fingernail swabbing or scrapings occasionally yield foreign DNA if a victim struggled and scratched the perpetrator, and similarly bite marks can be swabbed for DNA. Reference samples may come from toothbrushes, razors, combs, clothing, and medical specimens. One of the authors (DF) has shown DNA to be useful to identify the bombmaker of deflagrated improvised explosive devices [11].



Figure 54.2
DNA testing identified a masked bandit when his peach strudel that was left at the scene of an armed robbery was used for DNA testing

Figure 54.3
This vacuum cleaner cord was used as a ligature for a strangulation murder. Swabbings of the cord along its length revealed the victim’s DNA in the center and a mixture of victim and accused DNA on outer areas of the cord
Property Crimes (Touch DNA)
Increasingly, jurisdictions are performing DNA testing in property crimes, including theft, burglary, robbery, and arson, among others. The vast majority of US crimes are property crimes: 9.3 million property crimes compared to 1.3 million violent crimes in 2009 [12]. The case closure (clearance) rate for property crimes is < 20 % [13] and DNA testing for such crimes has been found to be cost-effective [14]. Furthermore, it is generally thought that some individuals progress from nonviolent to violent crimes; often from petty theft to burglary to rape, and thus interdiction of a criminal career progression may break the cycle and prevent major crimes [15, 16]. In general, DNA testing for property crimes involves “touch DNA” from handled objects. The possibility of testing such trace or “low copy number” (LCN) DNA was introduced in 1997 when Dr. van Oorschot reported that minute quantities of DNA can be recovered from fingerprints [17]. Conventional laboratory testing will successfully type DNA from approximately 100 cells (0.5–1 ng at 6.5 pg/diploid cell), although many forensic laboratories may be successful down to as few as 15–20 cells (approximately 100 pg). LCN DNA is generally defined as <100 pg, but 35 pg is often considered an analytical threshold. LCN DNA testing for property crimes was pioneered by Drs. Peter Gill and Dave Werrett at the Forensic Science Service (now disbanded) in the UK [18] and later in the USA by Dr. Mechthild Prinz and Theresa Caragine in the New York City Office of the Chief Medical Examiner’s Department of Forensic Biology [19]. Such testing involves minimizing reaction volumes and increasing polymerase chain reaction (PCR) cycling (see below). However, only a portion of the specimens yield a useful profile (perhaps 10–20 %). LCN DNA testing is problematic due to detection of contamination from prior handling, in-laboratory contamination, and inconsistent results that stem from random sampling of one or both alleles when both exist at very small levels (so-called stochastic sampling effects). These difficulties are compounded by the destructive nature of DNA testing, which may negate the possibility of retesting. For these reasons, some have suggested that LCN analysis should only be used for investigative purposes, and not as probative evidence in court. No national standards are yet in place and the FBI has generally recommended against such testing [20]. Nonetheless, LCN testing is increasingly used. The object is swabbed and resultant DNA extracts may be amplified two or three times, wherein analysts hope to obtain pure (single) profile results that are assumed to be from the last person who pulled the trigger of a gun or handled a knife. Forensic laboratories performing this testing will generally simply disregard any results other than clear single profiles.
Other Forensic DNA Testing Applications
Forensic DNA identity testing also can be used in other forensic and non-forensic contexts. For example, urine samples from drug testing may be analyzed to confirm that the sample is truly from the person who allegedly generated it [21]. DNA testing is used for disaster victim identification [22]. In cases involving nonhuman DNA (discussed below), individual, group (clade), or species may be determined, linking items such as a plant leaf or animal hair to a criminal case, or proving illegal poaching activity [23]. “Microbial forensics” has been developed for source attribution of terrorist pathogens, such as the anthrax letter attacks [24, 25]. Genetic analyses can identify the type of body fluid or tissue (e.g., urine, semen, saliva) based on the RNAs expressed. DNA testing can be used for investigatory purposes by supplying information about the perpetrator using phenotypic markers (described below), as well as through partial (“low stringency”) matches that may detect relatives who represent investigatory leads (described below). In non-criminalistic applications, the same tests used in forensic identity testing can be used for determinations of parentage and sample switch disputes [26].
Genetic Systems and Methods for DNA Typing
Genetic Variation
In everyday life, we easily recognize individuals through obvious biological variation among individuals. Positive identification or individualization is a statement of uniqueness, which is theoretically impossible to prove. However, forensic identity testing harnesses the extraordinary statistical discriminatory power of genetic variation to support a policy-based, administrative, or judicial determination of identity [27]. Indeed, forensic DNA testing often is thought of as tantamount to positive identification. Genetic variation occurs in a continuum of biological classification, from kingdom to genus, clades, and individuals. Specifically, forensic DNA identity testing is based on the detection and comparison of polymorphisms (poly—many; morphs—types) in the DNA among individuals. Statistically, there is variation at approximately one in every thousand base pairs (bp) between every two unrelated humans. However, this variation is not random; many protein-coding regions are highly conserved, as mutations in genes succumb to natural selection. Most polymorphisms occur in the noncoding DNA, which predominates in the human genome (>98 %) and is more tolerant of mutation than the protein-coding DNA regions. Differences between individuals can be due to single nucleotide polymorphisms (SNPs) or variations in length of a specific region or locus in the genome; that is, length polymorphisms. Such polymorphisms result in different forms, or alleles, of genetic markers. All individuals have two copies of each autosomal chromosome: one inherited maternally and the other paternally. Routine forensic DNA testing, using short tandem repeat (STR) typing, (described below) involves length polymorphisms in repetitive DNA from noncoding regions of the chromosomes, although it employs only a small fraction of the differences in the human genome among individuals.
Restriction Fragment Length Polymorphisms
Historical Context
In the mid-1980s, most DNA-based forensic analysis involved restriction fragment length polymorphism (RFLP) testing, first described by Dr. Edwin Southern in 1975 [28]. Such testing merely gave a binary result and was too little information for too much work. Drs. Wyman and White detailed a polymorphic RFLP marker in 1980, in which variation between human individuals was observed [29]. However, the beginning of the forensic DNA typing revolution began with the 1985 publication of a landmark article by Dr. Alec Jeffreys of Leicester, England, in which he coined the term “DNA fingerprint” and suggested the potential application of DNA fingerprinting in forensic investigations [30, 31]. His technique involved use of “minisatellites,” which was a multilocus probe RFLP system that yielded a bar code pattern that seemed to be different for every person (Fig. 54.4). Jeffreys conducted the first DNA identity tests in 1986 in a disputed immigration case and a double rape-homicide, which resulted in the 1987 exoneration of Richard Bucklin and then the 1988 conviction of Colin Pitchfork [32, 33]. In the USA, single-locus probe RFLP analysis was pioneered by Dr. Arthur Eisenberg (then at Lifecodes Corporation), that was more robust and permitted statistical evaluation (Fig. 54.5). In 1986–1987, commercial laboratories, particularly involving Dr. Edward Blake of the Serologic Research Institute, Drs. Michael Baird and Arthur Eisenberg of Lifecodes Corporation, and Dr. Robin Cotton of Cellmark Diagnostics, undertook forensic DNA testing in the USA, and in 1987 Tommy Lee Andrews became the initial American to be convicted of a crime (rape) using DNA data [34]. The FBI, led by Dr. Bruce Budowle, began performing DNA typing casework in December 1988. A few months later, in March 1989, Virginia became the first state crime laboratory with an operational DNA unit, directed by Dr. Paul Ferrara. RFLP testing was the mainstay of most criminalistic DNA typing for a decade. At the same time, Dr. Henry Erlich and coworkers of Cetus Corporation, developed a faster PCR-based (see below) HLA DQ-alpha dot-blot system (and later the Polymarker system, Fig. 54.6), but it did not have sufficient discriminatory power for widespread adoption by the forensic community. Nevertheless, the first use of DNA tests in litigation in the USA was in 1986, in the case of Commonwealth v. Pestinikas, using HLA DQ-alpha to show that organs had not been switched in an autopsy [35]. In the early 1990s, PCR-based STR systems (described below) were developed and eventually became the standard forensic DNA test worldwide. STR methods replaced RFLP systems due to robustness, sensitivity, statistically discrete systems, ease of automation, and economy. Other systems, such as Y-chromosome markers, mtDNA sequencing, and phenotypic markers also are sometimes used (described below) (see Table 54.1).
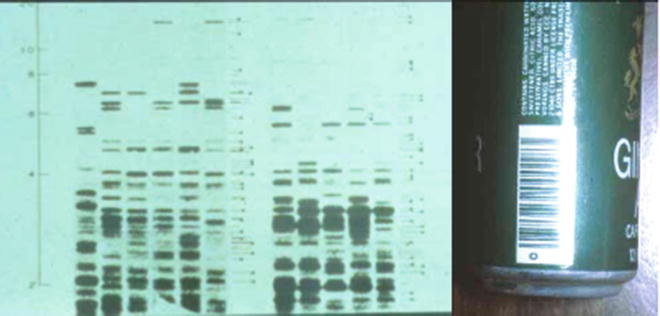
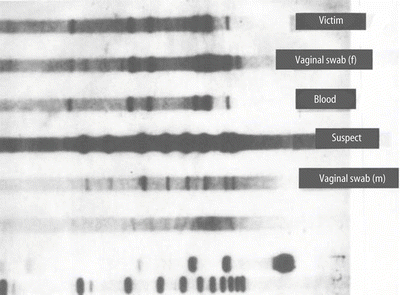
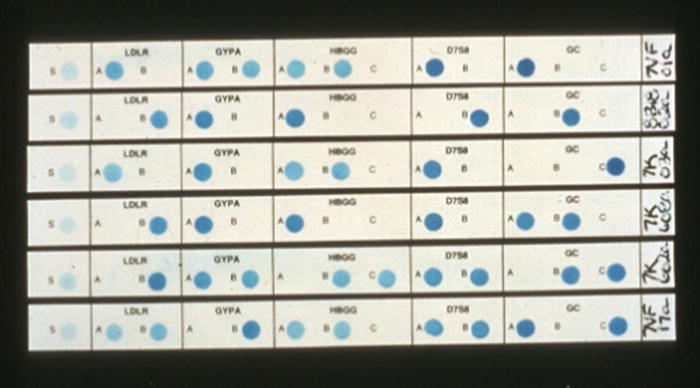

Figure 54.4
In 1985, Alec Jeffreys first described a DNA fingerprint. He used a multilocus minisatellite probe that resulted in a band pattern similar to a bar code, such as the one shown on the can to the right. The various lanes of the autoradiograph are from different individuals, demonstrating that each shows a unique pattern of bands. This multilocus probe method of DNA typing is no longer used in forensic identification

Figure 54.5
RFLP autoradiograph with five analytical lanes and three control lanes. The DNA profile of the reference sample from a female rape victim matches the DNA profile of blood found at the scene and that of the female fraction of a vaginal swab. The DNA profile of the suspect reference specimen matches the male fraction of a vaginal swab but does not match the DNA profile of the female victim

Figure 54.6
Polymarker strips from different individuals using five genetic systems detected by PCR amplification and reverse dot-blot hybridization probes. GC group-specific component, GYPA glycophorin A, HBGG hemoglobin gamma-globin chain, LDLR low-density lipoprotein receptor,
Table 54.1
Summary of DNA typing system usage in crime laboratories
Typing method | PCR-based | Late 1980s | 1990s | 2000s | Utility |
|---|---|---|---|---|---|
RFLP | No | Dominates | Dominates | Abandoned | Routine casework |
Dot blots | Yes | Used | Used | Abandoned | Routine casework |
STRs | Yes | In research | Used | Dominates | Routine casework |
mtDNA | Yes | In research | Used | Used | Hairs, degraded samples |
Y-STRs | Yes | In research | Used | Vaginal swabs in rape cases | |
SNPs | Yes | In research | Very degraded samples |
Early Cases Using DNA Testing
Queen v Pitchfork
The first criminal investigation using DNA typing was in a double rape-homicide (of Linda Mann in 1983 and of Dawn Ashworth in 1986) on a deserted footpath in the English countryside, known as the “Black Pad Murders.” Richard Buckland, a person of low intelligence and sexual fetishes, became the focus of early suspicion and was charged but then exonerated by the new Jeffreys DNA tests. Males in the community between 13 and 30 years of age were asked to volunteer blood samples for DNA testing. There were no matches despite 4,500 “bloodings.” However, police discovered that a man named Ian Kelly had substituted his blood for Colin Pitchfork’s sample. Pitchfork was subsequently DNA matched and then convicted of both homicides.
Pennsylvania v Pestinikas
The first use of DNA typing in the USA was in a 1986 nursing home negligent homicide case. Forensic Science Associates performed DNA tests to prove that organs in the autopsy had not been switched as was alleged by one expert. The DNA in this case had become highly degraded, averaging fragments of approximately 100 bp.
Florida v Andrews
The first US criminal conviction based on DNA typing was of a serial rapist, Tommy Lee Andrews (1987). A series of breaking and entering women’s homes and rapes began in 1986 in Orlando, Florida. A stakeout resulted in an arrest, and Lifecodes Corporation matched the suspect’s DNA to vaginal swabs of two of the rape victims.
PCR Amplification as Sample Preparation
Today, all major methods for routine forensic DNA testing begin with amplification of the DNA target by PCR. Dr. Kary Mullis shared the 1993 Nobel Prize in Chemistry for PCR development in 1983. Forensically valuable human leukocyte antigen (HLA) polymorphisms were among the earliest targets to be amplified by PCR in the laboratory [36]. PCR amplification is relatively easy to perform, inexpensive, quick, and amenable to automation. It also permits chemical labeling of the amplified fragments, as well as simultaneous amplification of several loci in a single reaction (multiplex). PCR amplification allows the routine testing of nanogram quantities of DNA, and can be optimized for testing of even picogram quantities, enabling the use of new classes of evidentiary specimens. However, such sensitivity requires extreme care to prevent contamination, including laboratory facilities with separate pre- and post-amplification areas, unidirectional handling of evidence intake through final analysis, limited laboratory access by untrained personnel, and knowledge of each analyst’s DNA profile to identify any contamination. Lastly, PCR can be successful on evidentiary material in which the DNA has become degraded and only a few fragments with the intact target sequence remain. Although amplification methods other than PCR exist, the conservative forensic community will not likely be quick to adopt an alternative to PCR unless there is a very good reason.
Short Tandem Repeats
STRs are repeat length polymorphisms that have become the mainstay of current forensic identity profiling around the world. Core repeat units in STR systems are tetranucleotide or pentanucleotide elements (i.e., have four or five nucleotides in each core repeat, respectively), with resulting amplicon sizes of approximately 100–450 bp (see Fig. 54.7). STR analysis is robust, amenable to automation, highly sensitive, relatively insensitive to degraded DNA, and yields discrete alleles. Multiplexed amplification of multiple STR loci achieves extraordinary discriminatory powers (typically >10−12) (see Fig. 54.8). As a result, PCR-based STR testing has become dominant in forensic DNA laboratories (Table 54.1).
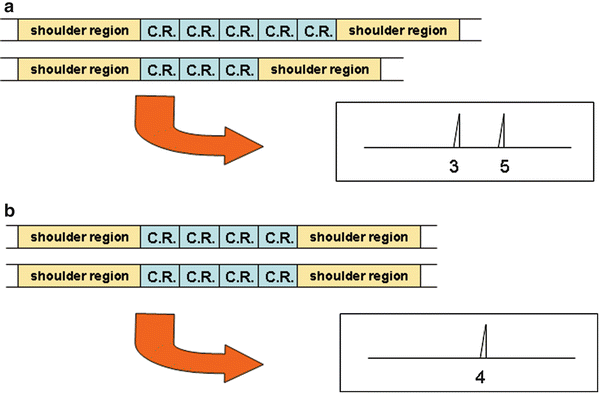
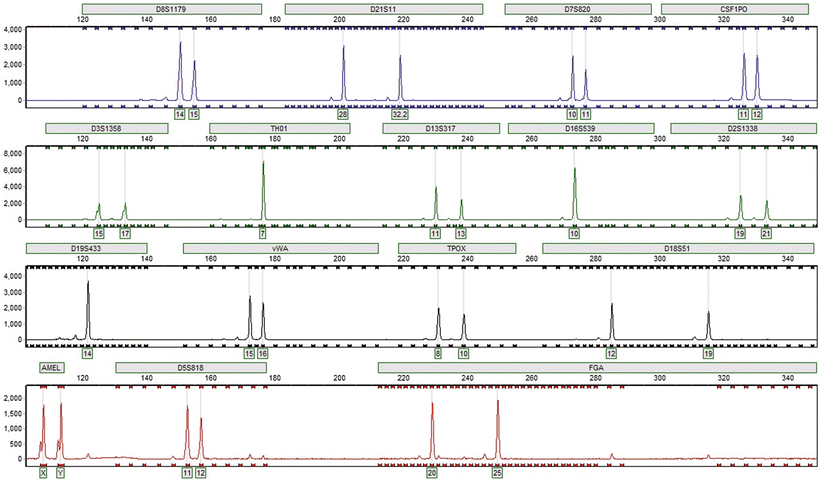

Figure 54.7
Diagram of short tandem repeat DNA segments composed of varying numbers of core repeats (C.R.) and accompanying electropherograms showing the corresponding allele peaks: (a) heterozygous pattern with alleles of 3 and 5 repeats, (b) homozygous pattern with allele of 4 repeats. The shoulder region is the flanking constant region to which PCR primers hybridize

Figure 54.8
Electropherogram of multiplexed fluorescently labeled PCR amplicons of STR loci demonstrating the allelic determinations (boxes). The X-axis reflects time and the Y-axis reflects fluorescence intensity. Four fluorophore colors permit separate analysis of genetic loci with overlapping sizes. A fifth dye channel is used for a size standard that is not shown. This person is a 15,16 genotype at the vWA locus, a 7,7 genotype (7 phenotype) at the TH01 locus, has a 32.2 variant allele in the D21S11 locus, and is a male according to the amelogenin locus
In the late 1980s, Dr. C. Thomas Caskey working with Holly Hammond, then at Baylor College of Medicine in Houston, Texas, was funded by NIJ to develop STR systems for forensic applications [37]. Subsequently, in 1991, STRs were first used in casework by one of the authors (VW) at the US Armed Forces DNA Identification Laboratory (AFDIL), through a subcontract with Cellmark Diagnostics, to identify service members who died in the first Persian Gulf War. However, it was Drs. Peter Gill and David Werrett at UK’s Forensic Science Service who, in the mid-1990s, began applying STR analysis (using in-house systems) to routine criminal casework [38, 39].
Recognizing the importance of cross-jurisdictional matches, the FBI convened a panel of forensic scientists in 1998 to select a panel of STR loci for use in their National DNA Index System (NDIS). Thirteen loci, all containing tetranucleotide repeats, were chosen: D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, CSF1PO, FGA, THO1, TPOX, and vWA (Table 54.2) [40, 41]. These 13 core loci have become standard for forensic casework in much of the world and are referred to as the “CODIS” loci, after the Combined DNA Index System (CODIS) software into which DNA profiles are entered [42]. Databases are maintained of the STR alleles of convicted felons, casework profiles, and missing persons, although exactly which profiles can or must be uploaded varies based on state requirements. The commonality of genetic systems (i.e., STR loci) used in forensic casework enables computer searches for matches across jurisdictions.
Table 54.2
Nationally indexed “13 CODIS STR Core Loci”*
Locus | Location | GenBank# | Alleles | Repeats | Motif | H | Mut % |
|---|---|---|---|---|---|---|---|
D3S1358 | 3p21.31 | AC099539 | 25 | 8–21 | Compound TCTG/TCTA | 0.795 | 0.12 |
D5S818 | 5q23.2 | AC008512 | 15 | 7–18 | Simple AGAT | 0.682 | 0.11 |
D7S820 | 7q21.11 | AC004848 | 30 | 5–16 | Simple GATA | 0.806 | 0.1 |
D8S1179 | 8q24.13 | AF216671 | 15 | 7–20 | Compound TCTA/TCTG | 0.78 | 0.14 |
D13S317 | 13q31.1 | AL353628 | 17 | 5–16 | Simple TATC | 0.771 | 0.14 |
D16S539 | 16q24.1 | AC024591 | 19 | 5–16 | Simple GATA | 0.767 | 0.11 |
D18S51 | 18q21.33 | AP001534 | 51 | 7–40 | Simple AGAA | 0.876 | 0.22 |
D21S11 | 21q21.1 | AP000433 | 89 | 12–41.2 | Complex TCTA/TCTG | 0.853 | 0.19 |
CSF1PO | 5q33.1 | X14720 | 20 | 5–16 | Simple TAGA | 0.734 | 0.16 |
FGA | 4q31.3 | M64982 | 80 | 12.2–51.2 | Compound CTTT/TTCC | 0.86 | 0.28 |
THO1 | 11p15.5 | D00269 | 20 | 3–14 | Simple TCAT | 0.783 | 0.01 |
TPOX | 2p25.3 | M68651 | 15 | 4–16 | Simple GAAT | 0.621 | 0.01 |
vWA | 12p13.31 | M25858 | 29 | 10–25 | Compound TCTG/TCTA | 0.811 | 0.17 |
All CODIS STR loci are tetranucleotide repeats. In general, smaller fragments are preferred for amplification of potentially degraded samples. Additionally, preferential amplification, where smaller target DNA fragments are amplified preferentially over larger targets, becomes an issue with large core repeats as the size discrepancy between overall allele lengths increases substantially. However, “stutter” can become problematic if the core repeat size (for example, dinucleotide and trinucleotide repeats) is too small. Stutter peaks are produced when the DNA polymerase slips during amplification, resulting in PCR products that have fewer or more repeat units than the starting template, with the major stutter product generally one repeat unit less than the template. Dinucleotide and trinucleotide repeats have substantial stutter, while pentanucleotide or larger repeats have almost none.
Commercial kits for amplification of the CODIS core STR loci are available in various combinations of multiplex primer sets from two companies: Promega Corporation and Applied Biosystems Inc. (ABI, a subsidiary of Life Technologies, Cartsbad, CA). ABI sells the Identifiler series, which includes all 13 core loci as well as amelogenin (below), D2S1338, and D19S433 in a single reaction. Promega offers the PowerPlex series, such as PowerPlex 16 that includes the 13 core loci, plus amelogenin and two pentanucleotide repeat loci, Penta D and Penta E. These two companies regularly produce new products that add loci to the multiplex, increasing discriminatory power. Mini-STRs, such as Applied Biosystem’s Minifiler, are traditional STRs with primers designed to reduce the flanking regions and thus the amplicon size, so that typing results can be obtained from more substantially degraded specimens.
The 13 CODIS core STR loci are being expanded to a likely set of 24 loci. The impetus for this expansion is to reduce the likelihood of adventitious matches in large databases, to increase the compatibility with international databases, and to increase the discrimination power for missing person cases [43]. The putative additional loci include loci from the European Standard Set (ESS). ABI has responded with the Global Filer kit and Promega with the Powerplex Fusion kit. These new kits have been engineered to be more sensitive as well as capable of more rapid amplification (see discussion below).
Amelogenin and Other Sex Markers
An amelogenin assay is included in current commercial STR amplification kits as a sex marker [44]. The amelogenin gene is present as homologs on both the X and Y chromosomes, but there are a number of sequence differences in the noncoding regions. The locus used in forensic testing involves a 6 bp deletion on the X chromosome; thus, the X marker is shorter than the Y marker and males manifest two peaks whereas females manifest a single peak of twice the intensity (Fig. 54.9). The amelogenin sex marker system is robust and reliable, although in very rare instances, sex-typing discrepancies have been noted [45]. Other sex markers have been described, including ones that exist at higher copy number and are thus more sensitive than the single copy amelogenin.
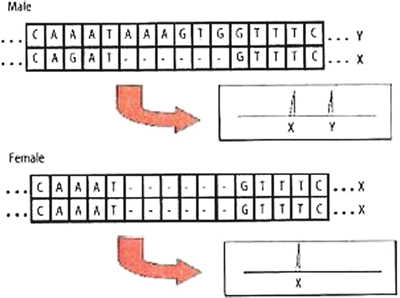

Figure 54.9
The amelogenin locus is 6 bp longer on the Y chromosome than on the X chromosome. Thus, a male will have two peaks and a female will have only one peak in the electropherogram
Male-Specific DNA Typing with Y-Chromosome Markers
Y-chromosome markers are not sex markers, but rather are male-specific identity systems that permit typing of the male DNA in mixed male/female specimens (e.g., vaginal swabs following rape or fingernail scrapings after assault). Y-chromosome markers are useful due to their strict paternal inheritance and can be helpful in lineage studies. The absence of recombination means that the exact same Y chromosome DNA alleles are present in distant paternal relatives of an individual. For example, Y-chromosome markers were used in determining the paternity of US President Thomas Jefferson among his distant descendants [46]. Y-chromosome markers are inherited together, and are reported together as a haplotype. Since they are not genetically independent, the population frequencies of each allele cannot be multiplied together; instead the counting method is used, where the number of times the haplotype is seen in a population database generates the frequency statistic. This means that the discriminatory power is much less than autosomal STRs and that a large number of loci is necessary to achieve substantial discrimination. Current commercial Y-chromosome markers are STRs that can be analyzed on the same equipment as standard STRs. More information on Y-STR haplotypes is available from various websites [47].
Single Nucleotide Polymorphisms
By far the most common polymorphisms in the human genome are SNPs [48–50]. It is estimated that there are approximately 15 million SNP sites out of the more than six billion bps of the diploid human genome. Very small DNA fragments can be interrogated for SNP alleles; thus, SNP genotyping can be applied for forensic identification despite extreme DNA degradation. The identification of human remains recovered from the World Trade Center disaster is one scenario in which SNPs showed an advantage over other DNA typing (Orchid-GeneScreen, now LabCorp, Burlington, NC), since the DNA was severely degraded in a large percentage of the specimens. Most SNPs are biallelic; that is, there are only two alleles, despite the fact that there are four possible nucleotide bases. Most SNPs are base substitutions; however, a smaller number are insertions or deletions (“indels”). Therefore, a large set of SNPs must be used to obtain decisive discriminatory values. In forensics, SNP analysis is not currently commercially available, but it is particularly amenable to automation and analysis with chip technologies. Indels are compatible with more standard fragment length technologies. SNPs may prove to be particularly valuable with Y chromosome or phenotypic markers (described below).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


