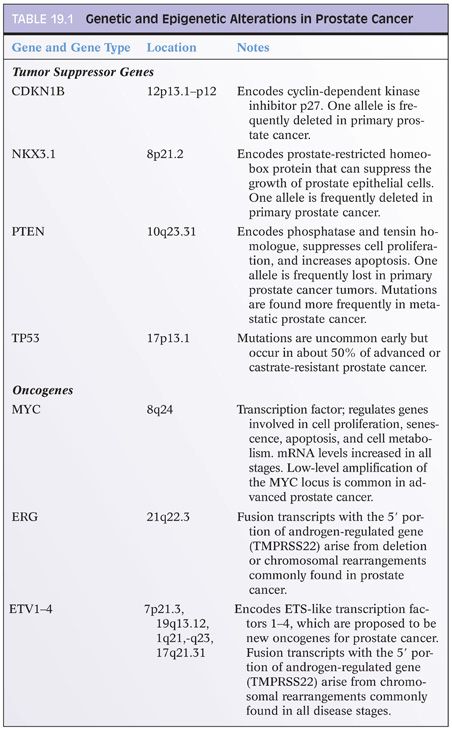
Epigenetic Changes in Prostate Cancer
Changes in DNA methylation marks, accompanied by epigenetic gene silencing, appear to be the earliest somatic genome changes in prostate cancer.21 New generation of assay strategies for detection of specific DNA sequences carrying 5-meC offers promising opportunities for potential clinical tests for prostate cancer screening, detection, diagnosis, staging, and risk stratification. Hypermethylation of glutathione S-transferase-π (GSTP1) transcriptional regulatory sequences has been consistently detected in more than 90% of prostate cancers. GSTP1 encodes an enzyme responsible for detoxifying electrophiles and oxidants, thus shielding cell from genome damage. Loss of GSTP1 expression appears to be an early event in the initiation of prostatic carcinogenesis, as evidenced by the presence of GSTP1 methylation in 5% to 10% of proliferative inflammatory atrophy (PIA) lesions thought by some to be the earliest prostate cancer precursors, and in more than 70% of high-grade prostatic intraepithelial neoplasia (PIN) lesions.49,50 In addition to GSTP1, more than 40 other genes have been shown to be altered by epigenetic hypermethylation.51 Yegnasubramanian et al.52,53 found hypermethylation of GSTP1, APC, RASSF1a, COX2, and MDR1 to be detected both in localized and in metastatic prostate cancer, whereas hypermethylation of other genes such as ERα, hMLH1, and p14/INK4a were more likely to be found in latter stages of prostate cancer progression, suggesting “two waves” of epigenetic alterations in prostate cancer.
ERG-ETS Gene Fusions
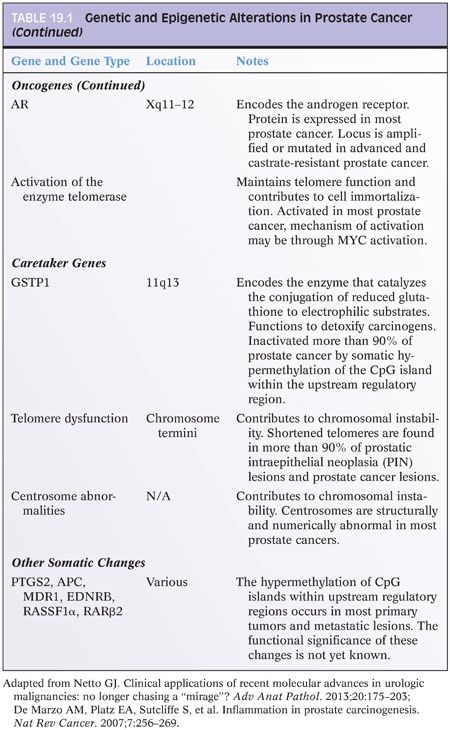
In 2005, Tomlins et al.54,55 identified a recurrent chromosomal rearrangement in over one-half of their analyzed prostate cancer cases. The recurrent chromosomal rearrangements led to a fusion of the androgen-responsive promoter elements of the TMPRSS2 gene (21q22) to one of three members of the ETS transcription factors family members ERG, ETV1, and ETV4 located at chromosomes 21q22, 7p21, and 17q21, respectively. Although the prognostic role of assessing TMPRSS2-ETS rearrangements in prostate cancer tissue samples has been called into question by recent well-designed large cohort studies including ours,56,57 the discovery had great implications in terms of furthering our understanding of the development and pathogenesis of prostate cancer and providing a new marker for molecular diagnosis in prostate cancer.58–67 The potential diagnostic and prognostic role of detecting TMPRSS2-ERG fusion in postprostate massage urine samples requires further investigation.68–70 Figure 19.2 depicts a commonly used FISH split-apart–based approach for the evaluation of ERG gene fusion.
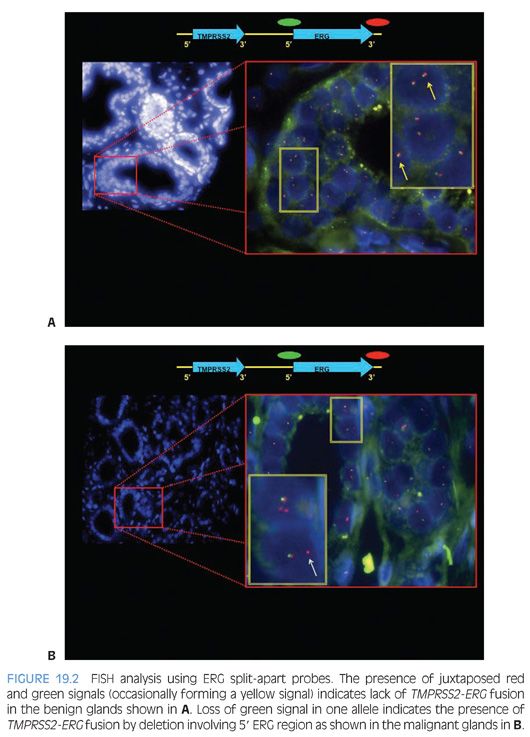
Recently, commercial anti-ERG monoclonal antibodies became available that makes it possible to use IHC for evaluating ERG protein expression as a surrogate approach to detecting TMPRSS2-ERG fusion by FISH. We, and others, have demonstrated a strong correlation between ERG overexpression by IHC and ERG fusion status with over 86% sensitivity and specificity rates. ERG IHC may offer an accurate, simpler, and less costly alternative for evaluation of ERG fusion status in prostate cancer on needle biopsy and radical prostatectomy samples (Fig. 19.3).71,72
PI3k/mTOR Pathway
The PI3K/mammalian target of rapamycin (mTOR) pathway plays an important role in cell growth, proliferation, and oncogenesis in prostate cancer.73–79 PTEN is a negative regulator of this pathway. Several recent well-designed retrospective studies have revealed that loss of PTEN tumor suppressor gene activity and the ensuing mTOR pathway activation is associated with poor prognosis in prostate cancer. In a recent large nested case–control, tissue microarray–based study from our institution, we were able to show loss of immunoexpression of PTEN to be a predictor of biochemical recurrence following radical prostatectomy independent of Gleason grade, cancer stage, and other clinicopathologic parameters.80 In a second study from our group by Lotan et al.,81 the prognostic role of PTEN alteration was further linked to adverse pathologic features and decreased time to metastatic disease in a surgical cohort of high-risk prostate cancer patients. The correlation of PTEN immunostains with genomic loss of PTEN gene was also established in a later study. The mTOR pathway is also a potential target for prostate cancer treatment and several rapamycin analogs are currently being tested as potential therapeutic agents for prostate cancer.78,82 We previously reported the results of a pilot study evaluating the pharmacodynamic efficacy of neoadjuvant rapamycin therapy in prostate cancer.82 Using IHC analysis, we found a significant decrease in Phos-S6 protein, the main downstream effector of mTOR pathway, in patients receiving neoadjuvant mTOR inhibitor agent.82
Other Tumor Suppressor Genes and Oncogenes
Among tumor suppressor genes, the role of p53 expression in predicting prognosis in prostate carcinoma has been extensively studied. Brewster et al.83 found p53 expression and Gleason score in needle biopsy to be independent predictors of biochemical relapse after radical prostatectomy. Another study found p53 status on prostatectomy but not needle biopsies to be predictive raising the issue of sampling.84 Many studies evaluating prostatectomy specimens found p53 to be of prognostic significance independent of grade, stage, and margin status.29,36,85–90 As discussed in the following text, more recent genome-wide studies seems to support the prognostic role of p53 alterations.91 The majority of studies of another tumor suppressor gene p27, a cell cycle inhibitor, have also supported a correlation with progression after prostatectomy. Although less robust evidence exists for the prognostic role of p21,92 a downstream mediator of p53, and transcription factors such as NKX3.1,22,93 preponderance of evidence supports a prognostic role for Bcl225,83,85,87,89 and myc oncogenes94,95 as potential adjuncts to histologic prognostic parameters.
It is crucially important to recognize that potential variability in performance characteristics exists even with the new molecular markers. Sources of variability include differences in molecular methodologies, tissue fixation and processing, inter- and intraobserver variability (in IHC-based biomarkers), and differences in cutoff points.3 Furthermore, illustration of statistical significance for a particular biomarker does not alone assure its utility in a given patient. Therefore, a promising prognostic or therapeutic target biomarker should endure a rigorous “evidence-based” analysis and be validated in large size, prospective clinical trials before transition into standard practice.96
Integrated Genomics
In a sentinel gene expression profiling study using cDNA microarrays containing 26,000 genes, Lapointe et al.9 identified three subclasses of prostate tumors based on distinct patterns of gene expression. High-grade and advanced stage tumors as well as tumors associated with recurrence were disproportionately represented among two of the three subtypes, one of which also included most lymph node metastases. Furthermore, two surrogate genes were differentially expressed among tumor subgroups by IHC. These included (a) MUC1, a gene highly expressed in the subgroups with “aggressive” clinicopathologic features and (b) AZGP1, a gene highly expressed in the favorable subgroup. The surrogate genes were strong predictors of tumor recurrence independent of tumor grade, stage, and preoperative PSA levels. Such a study suggests that prostate tumors can be usefully classified according to their gene expression patterns, and these tumor subtypes may provide a basis for improved prognostication and treatment stratification. Lapointe et al.97 complemented their aforementioned gene expression findings by looking for associated copy number alterations using array-based comparative genomic hybridization (array CGH). They were able to identify recurrent copy number genetic aberrations that corresponds to three prognostically distinct groups of prostate cancer: (a) deletions at 5q21 and 6q15 deletion group associated with favorable outcome group, (b) an 8p21 (NKX3-1) and 21q22 (resulting in TMPRSS2-ERG fusion) deletion group, and (c) 8q24 (MYC) and 16p13 gains and loss at 10q23 (PTEN) and 16q23 groups correlating with metastatic disease and aggressive outcome.
In a recent genome-wide analysis of prostate cancer, Taylor et al.98 elegantly illustrated how detailed annotation of prostate cancer genomes can impact our understanding of the disease and its treatment strategy. Assessing DNA copy number, mRNA expression, and focused exon resequencing in 218 prostate cancer tumors, the authors identified the role of nuclear receptor coactivator NCOA2 as a novel oncogene in 11% of prostate cancer cases. TMPRSS2-ERG fusion was associated with novel prostate-specific deletion at chromosome 3p14 that may implicate FOXP1, RYBP, and SHQ1 as potential cooperative tumor suppressors. Most intriguing was their ability to define clusters of low-risk and high-risk disease beyond that achieved by Gleason score using DNA copy number data. Six clusters of prostate cancer tumors are identified by unsupervised hierarchical clustering with distinct risk for biochemical recurrence.
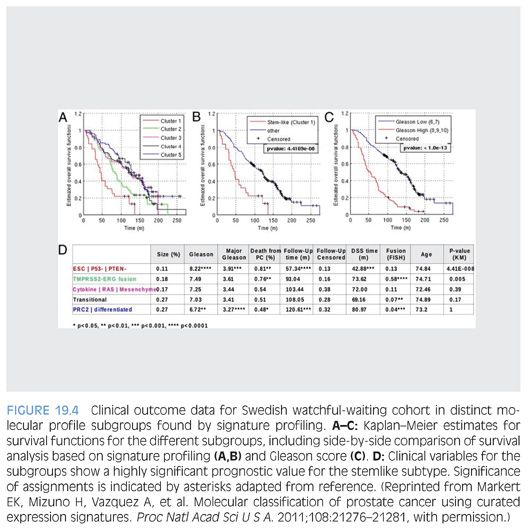
Markert et al.91 also illustrated the potential use of molecular signatures as a prognosticator in prostate cancer. The authors assessed microarray dataset characterizing 281 prostate cancer patients from a Swedish watchful-waiting cohort. mRNA microarray signature profiles for gene signatures reflecting embryonic stem cell (ESC), induced pluripotent stem cell (iPSC), and polycomb repressive complex-2 phenotypes (PRC2), in addition to inactivation of the tumor suppressors p53 and PTEN loss and the TMPRSS2-ERG fusion, were assessed. Unsupervised clustering identified prostate cancer subset with “stemlike signatures” combined with p53 and PTEN inactivation to be associated with very poor survival outcome. Prostate cancer tumors characterized by TMPRSS2-ERG fusion had intermediate survival outcome, whereas remaining groups demonstrated more favorable outcome (Fig. 19.4). The exciting findings were further validated in an independent clinical cohort at Memorial Sloan-Kettering Cancer Center. This classification was independent of Gleason score and therefore can provide additional added value in prognostication in patients with lower Gleason grade prostate cancer.
EMERGING MOLECULAR AND GENOMIC COMMERCIAL PROGNOSTIC ASSAYS
Genomic studies suggest that prostate cancers develop via a limited number of alternative preferred genetic pathways. The resultant molecular genetic subtypes provide a new framework for investigating prostate cancer biology and explain in part the clinical heterogeneity of the disease. Transitioning the aforementioned findings to the clinical arena will no doubt be accelerated by the increasing implementation of genomic and next-generation sequencing (NGS) technologies in clinical molecular diagnostic labs. Only few examples of such burgeoning genomic and molecular tests are discussed in the following text without implying any endorsement for their routine use. Although the role of some of these assays has been supported by initial studies, additional prospective validation studies are ongoing to justify their future implementation as standard of care. GenomeDx Decipher test99,100 is one example of such genomic classifiers. Ross et al.100 have recently illustrated that compared to clinicopathologic variables, Decipher genomic classifier applied to paraffin-embedded radical prostatectomy samples better predicted metastatic progression among an 85 prostate cancer patients cohort of men with biochemical recurrence from our institution. Although confirmatory studies are needed, such results suggest that use of such genomic classifiers may allow for better selection of men requiring earlier initiation of treatment at the time of biochemical recurrence.
The Oncotype DX Prostate Cancer Assay is a multigene reverse transcriptase polymerase chain reaction (RT-PCR) expression assay that was developed for use with fixed paraffin-embedded diagnostic prostate needle biopsies. The assay evaluates the expression of 12 cancer genes representing distinct biologic pathways with a known role in prostate tumorigenesis. These include the androgen pathway (AZGP1, KLK2, SRD5A2, and FAM13C), cellular organization (FLNC, GSN, TPM2, and GSTM2), proliferation (TPX2), and stromal response (BGN, COL1A1, and SFRP4). The expression of five reference genes is also assessed to control for sources of preanalytical and analytical variability as well as allow for variable RNA inputs. The calculated genomic prostate score (GPS) has been shown to predict adverse prostate cancer pathology beyond conventional clinical or pathologic factors in a recently completed clinical validation study.101,102
Metamark Genetics Inc, has developed an automated, quantitative, protein-based multiplex imaging platform designated ProMark. Metamark’s automated proteomics imaging platform is applied to standard formalin-fixed, paraffin-embedded tissue biopsy sections. The tissue sections are subjected to multiplex immunofluorescent staining with monoclonal antibodies as well as DAPI using a proprietary assay format that enables the quantitative biomarker measurements in the tumor epithelium regions only. Awaiting prospective validation studies, the assay could be of value in predicting indolent disease (organ confined, Gleason grade 3 = 3 or 3 + 4) in patients with positive biopsies that would hence be potential active surveillance candidates. Prospective studies are needed.
EMERGING EARLY DETECTION MARKERS AND TARGETS OF THERAPY
Markers of prostate cancer detection that can be applied to blood, urine, or prostatic secretion fluid (ejaculate or prostate massage fluids) have been the focus of active recent research. Markers that have been investigated in the urine or prostatic secretions include gene promoter hypermethylation profile assays56,103–105 and differential display code 3 (DD3), also known as PCA3.
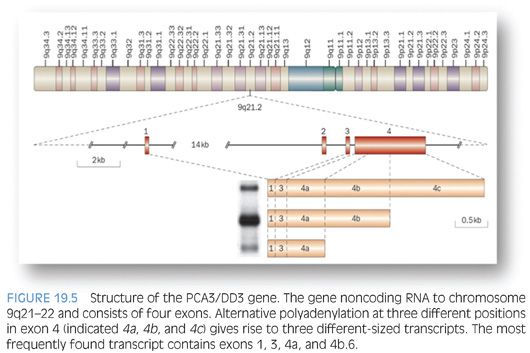
DD3 is a noncoding RNA that was initially identified by Bussemakers et al.106 as one of the most specific markers of prostate cancer. PCA3 gene is located on chromosome 9q21.2 (Fig. 19.5). Quantitative real-time RT-PCR assay detecting PCA3 can be applied to blood, urine, or prostatic fluid.107 Evaluation of PCA3 in urine samples, obtained following an “attentive” prostate massage, using transcription-mediated amplification (TMA) technology has shown to be superior to serum PSA in predicting biopsy outcome with sensitivity and specificity approximating 70% and 80%, respectively, and a negative predictive value of 90%108–111; it is currently offered by commercial laboratories as a Food and Drug Administration (FDA)–approved assay in the United States. Encouraging data from the Reduction by Dutasteride of Prostate Cancer Events (REDUCE) trial support a role for evaluation of PCA3 in postattentive prostate massage urine sample in predicting positive prostate needle biopsy in immediately subsequent as well as future biopsies following initial negative biopsy. PCA3 may also have a role in predicting the risk for higher Gleason score and larger tumor volume on radical retropubic prostatectomy. If confirmed, the latter could be of great value in treatment options algorithm and delineation of candidates for active surveillance.112–115 Multiplex urine assays to include PCA3, TMPRSS2-ERG, SPINK1, and GOLPH2 are also under evaluation with recent data suggesting an improved performance of such assays compared to PCA3 alone.116
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


