Term
Definition
Significance
Complete or full donor chimerism
Only donor detected
Suggests complete hematopoietic engraftment, but low levels of recipient cells may not be detected depending on the method used to assess
Mixed chimerism
Both donor and recipient detected
May indicate increased risk of relapse; dynamic change most important (increasing, decreasing, or stable); for RI-HSCT, mixed chimerism is expected initially
Subset chimerism
Mixed chimerism in a cellular subpopulation, e.g., T cells
Patterns of T cell and natural killer (NK) cell chimerism may be predictive of GVHD or graft loss in RI-HSCT recipients
Split chimerism
Recipient detectable only in some cell lineages
May indicate relapse or risk of relapse; important to correlate with clinical picture and original disease cell lineage
Microchimerism
<1 % recipient detected
Associated with solid organ transplantation; unclear significance in setting of allogeneic HSCT
Hematopoietic Stem Cell Transplantation
Each year, more than 28,000 patients undergo allogeneic HSCT worldwide. Allogeneic HSCT is accomplished by the infusion of either bone marrow or granulocyte colony stimulating factor (G-CSF)-mobilized peripheral blood stem cells from a related or unrelated donor. The stem cell infusion is preceded by the administration of a conditioning regimen (chemotherapy and/or radiotherapy), designed to eradicate residual malignant cells and induce sufficient immunosuppression to facilitate engraftment. Allogeneic HSCT permits the administration of myeloablative doses of chemotherapy and radiation, and offers important antitumor immunity through donor-derived T and NK cells. The genetic disparity between the donor and the recipient results in a therapeutic antitumor effect, often called the graft-vs-tumor (GVT) response, but also results in immune-mediated damage to normal recipient tissues, causing a clinical syndrome called GVHD. Reduced intensity HSCT (RI-HSCT) involves the administration of non-myeloablative doses of chemotherapy and radiation in order to reduce the toxicity associated with aggressive conditioning. The lower toxicity of RI-HSCT allows its use in patients deemed ineligible for myeloablative transplants due to age, comorbidities, or prior therapies. RI-HSCT eradicates malignant cells through a powerful GVT effect and not through chemotherapy or radiation.
Clinical Utility of Chimerism Analysis
Chimerism is a dynamic process following allogeneic HSCT, and its kinetics depend on the intensity of the conditioning regimen, sensitivity of different cell types to chemotherapy and radiation, the recipient’s prior therapies, composition of the graft, and other factors. Therefore, identification of longitudinal changes (increasing or decreasing mixed chimerism over time) is most useful clinically. The interpretation of chimerism testing depends on clinical context, type of transplant, and time after transplant. For example, the presence of “10 % residual recipient cells” may have completely different implications on day 21 vs day 100 after HSCT, or if the patient had a myeloablative or a reduced-intensity conditioning regimen prior to transplant. Similarly, the presence of “25 % donor cells” in a patient with aplastic appearing bone marrow is interpreted differently from a patient who has evidence for relapsed leukemia occupying the marrow. Here we describe the common clinical situations and various clinical outcomes that can be evaluated with chimerism testing.
Patterns of Engraftment
Recovery of normal hematopoiesis after HSCT occurs gradually after the stem cell infusion. In typical hematologic recovery, granulocytes recover first (within 10–24 days) and platelets and red blood cells ensue. Complete blood counts fail to reveal the origin of recovering hematopoiesis, which can be donor-or recipient-derived, and is very often mixed in the early phases after HSCT. Chimerism analysis is usually done in the first 100 days after the stem cell infusion to verify and document adequate engraftment of donor cells [1]. In myeloablative HSCT, if careful sequential studies are done, an early phase of mixed chimerism is usually observed, rapidly followed by conversion to complete donor chimerism (Fig. 56.1a). In cases of graft failure, transient mixed chimerism transitions back to predominantly recipient chimerism, followed by disappearance of donor cells (Fig. 56.1b). Therefore, chimerism studies are useful in the evaluation of poor hematological recovery, which may be the result of engraftment failure, infection, GVHD, or ABO incompatibility. In RI-HSCT, mixed chimerism may persist for long periods of time without indicating engraftment failure.
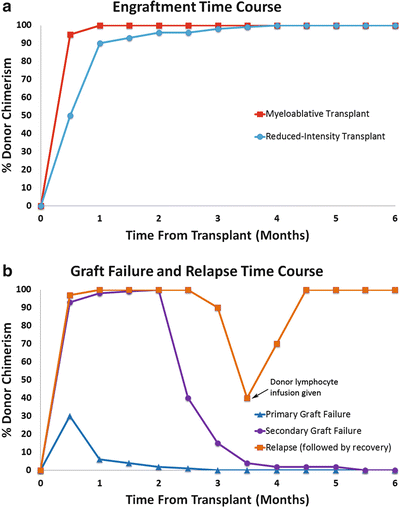

Figure 56.1
Comparisons of typical time courses of engraftment (a) and graft failure (b) after bone marrow transplant. The example of relapse (b) shows recovery of donor engraftment after infusion of donor lymphocytes
The kinetics of the transition from mixed to complete chimerism reflect the complexity of immune interactions between donor and recipient cells as well as differences in the HSCT process for different clinical situations. The major determinants of engraftment timing and levels of mixed chimerism include the type of conditioning regimen used (myeloablative vs reduced-intensity), graft type (bone marrow vs peripheral blood stem-cells vs umbilical cord blood [UCB]), graft source (related vs unrelated donor), human leukocyte antigen (HLA) compatibility, numbers of CD34+ and CD3+ cells in the graft, prior chemotherapy, degree of lymphodepletion prior to transplant, type of disease (acute myelogenous leukemia [AML], lymphoma, chronic myelogenous leukemia [CML], etc.), and the use of T cell depletion [2–11]. Acute GVHD is associated with accelerated engraftment, but the causal relationship is unclear. Faster engraftment may trigger GVHD by not allowing sufficient time for the immune system to develop tolerance [2, 3, 12, 13].
In RI-HSCT, higher levels of donor chimerism, and sometimes even early full chimerism, are more common in patients who had intensive chemotherapy prior to transplant, likely secondary to lymphodepletion, which leads to homeostatic expansion of donor cells and therefore faster engraftment [5, 14]. Strategies have been developed to intensify pre-transplant lymphodepletion in order to accelerate engraftment [11]. Chimerism analysis enables close monitoring in these cases.
In UCB transplants, slow engraftment and engraftment failure are common due to low numbers of CD34+ cells and HLA incompatibility. Chimerism is tested in these cases to monitor the engraftment process. Chimerism measurements as early as 14 days post-transplant can be helpful in predicting the risk for graft failure [15]. In order to overcome the barriers of slow engraftment and high incidence of graft failure after UCB transplants, a commonly used approach is the concurrent transplantation of two different UCB units, which accelerates engraftment and reduces the risk for graft failure [16, 17]. After double cord transplant, chimerism analysis often detects DNA from both UCB units at the same time, with occasional residual recipient cells, especially after reduced-intensity conditioning. The usual course is that both units contribute to early hematopoiesis, but by 21–100 days post-transplant, one of the units predominates, and eventually the minor unit can no longer be detected [17, 18]. The factors determining which unit “wins” include the CD3+ and CD34+ cell doses, the order of infusion, and the degree of HLA match [16, 19, 20]. Interestingly, cases have been reported where reversal of dominance occurs a long time after the transplant, even after documentation of full chimerism with one of the donors [20, 21]. Evaluation of chimerism in the setting of two donors is more complicated than with a single donor.
Evaluation of Graft Failure
Graft failure presents as either failure to achieve adequate engraftment and recovery of blood counts (primary graft failure) or loss of normal blood counts after initial engraftment (secondary graft failure) (Fig. 56.1b). In the evaluation of graft failure, it is important to differentiate between graft rejection, defined as the absence of donor-derived hematopoiesis (usually < 5 % donor chimerism) in a patient with a hypocellular marrow, and poor graft function, where donor cells are present but production of blood elements is reduced. Poor graft function may result from a low dose of CD34+ cells in the graft, presence of recipient-derived anti-HLA antibodies against the donor cells, use of bone marrow or UCB as opposed to peripheral blood stem cells, infection, and factors in the bone marrow environment [22, 23]. A poorly functioning graft may respond to infusion of additional stem cells. Graft rejection is more likely to be the result of immunologic rejection of the graft, and can be treated with modulation of immunosuppressive therapies, donor lymphocyte infusions, or a second HSCT [24]. In all cases of graft failure, an evaluation of a bone marrow biopsy is mandatory to exclude a relapsed malignancy. The combination of bone marrow histology and chimerism levels in bone marrow and blood are useful to help interpret the clinical picture.
Mixed chimerism does not always predict graft failure, and is very common after RI-HSCT. However, it is known that predominant donor chimerism precedes hematologic recovery. A persistently low level of donor chimerism in whole blood, T cells, or NK cells after transplant is associated with subsequent graft failure [1, 5, 25, 26]. Serial monitoring every 1–4 weeks can identify a downward trend which predicts graft failure. Serial monitoring is also useful to measure responses to treatments of graft failure, such as withdrawal of immunosuppression, infusion of additional stem cells or additional lymphocytes, or a second transplant from a different donor.
Association with GVHD
In some studies, early conversion to full donor chimerism was associated with increased incidence of GVHD, while mixed chimerism seemed to be protective against acute and/or chronic GVHD [25, 27–29]. Mechanistically, co-existence of recipient and donor cells may facilitate peripheral tolerance induction, but mixed chimerism is not fully protective against GVHD, and conversion to full donor chimerism is not required for initiation of GVHD [5, 30–32]. Studies of T cell subset chimerism are potentially more informative than whole blood, and show that high donor T cell chimerism is associated with GVHD [5, 9].
Chimerism and Relapse in Myeloablative Transplants
The utility of chimerism testing for prediction of disease relapse depends on several factors. Attempts to associate mixed chimerism with relapse and survival outcomes resulted in conflicting results, possibly related to differences in diseases and conditioning regimens studied, and the use of T cell depletion strategies in some studies. In myeloablative HSCT for CML, the presence of mixed chimerism or mixed T-cell chimerism within 90 days after transplant correlates with disease relapse and a poor outcome, but this seems to depend on the conditioning regimen [33], and has questionable value in diseases other than CML [7]. The timing of testing is critical; studies in CML have shown that not only mixed chimerism but also residual copies of BCR–ABL1 transcript are not predictive of relapse in the first 90 days after HSCT [34]. Therefore, in most clinical scenarios, early testing of mixed chimerism (day 30) should be reserved for evaluation of possible graft failure and not disease relapse [1].
Beyond the first 90 days post-transplant, many studies have demonstrated an association between persistence of recipient cells with relapse and worse survival [30, 35, 36]. More importantly, studies that looked at the dynamic changes in chimerism levels showed that a persistent or rapid trend of dropping chimerism levels often heralds relapse [31, 37–40], whereas stable mixed chimerism is not clearly associated with poor outcomes [27, 40–42]. One of the major limitations of these reports is that they included primarily patients with chronic phase CML, which rarely requires HSCT today [33, 36, 43–45]. In acute leukemias, trends seem to be more important than single measurements, and frequent monitoring is more likely to predict relapse successfully than less frequent testing since the kinetics of chimerism prior to relapse can show an abrupt change [45–47].
Chimerism in RI-HSCT
In RI-HSCT, early evaluation of chimerism may have a more important role than for myeloablative HSCT. The low intensity conditioning regimen, combined with post-transplant immunosuppression, allows the establishment of mixed chimerism in most patients [48, 49]. With time, mixed chimerism may transition to complete chimerism, or remain stable and serve as a platform for subsequent immunotherapies such as donor lymphocyte infusions (DLI) [50]. After RI-HSCT, the initial recovery of neutrophil counts, traditionally named “engraftment,” represents a mixed recovery of autologous hematopoiesis and engrafted donor cells, which quickly transition into complete myeloid chimerism (Fig. 56.1a) [5, 51]. Other lineages also recover gradually, but often remain in a mixed chimeric state for prolonged periods of time. Multiple factors determine the kinetics and the degree of donor chimerism achieved after RI-HSCT, and higher levels are achieved with a more intensive conditioning regimen, use of chemotherapy prior to transplant, use of G-CSF mobilized peripheral blood stem cells as opposed to bone marrow, and pre-transplant lymphopenia [5, 52]. The critical role of lymphodepletion prior to transplant in inducing full donor chimerism has led to the development of chemotherapy regimens that efficiently deplete recipient lymphocytes prior to RI-HSCT for lymphoma [11].
Is the degree of donor chimerism associated with outcomes of RI-HSCT? Early studies showed that full T cell chimerism precedes full myeloid chimerism and suggested that full chimerism precedes GVT and GVH responses [51, 53]. Other studies have found that low levels of T and NK cell chimerism predict graft rejection and lack of antitumor activity; high levels of T cell chimerism are associated with GVHD, while high levels of NK cell chimerism predict improved progression free survival [5, 26]. Some of these correlations might be specific to certain diseases and certain conditioning regimens [4, 6]. Recently, large retrospective studies have demonstrated the prognostic value of early (day 30 and day 100) measurement of donor chimerism in predicting relapse and survival after related or unrelated donor transplants, regardless of disease [52, 54]. Similarly, full donor T-cell chimerism on day 100 after double UCB transplants with reduced intensity conditioning seems to predict a lower incidence of relapse [55]. These studies strengthen the rationale for chimerism testing in the setting of RI-HSCT.
Failure to achieve full chimerism after both myeloablative and RI-HSCT can be overcome by DLI, which often successfully induces transformation from mixed to full chimerism and enhances the GVT response [56, 57]. Monitoring whole blood or T cell chimerism after HSCT has been used to guide and monitor preemptive DLI strategies in patients with mixed chimerism, considered to be at high risk for rejection and disease relapse [35, 46, 51, 53, 58–62]. Randomized studies have not been conducted to provide solid evidence for the benefit of this approach, but it is clear that DLI can induce favorable antitumor responses in several diseases [37, 58, 62]. Cytokine therapy also has been used to enhance engraftment and increase donor chimerism levels [63].
Chimerism vs Minimal Residual Disease
Minimal residual disease (MRD) detection can be performed by amplification of disease-specific markers such as BCR–ABL1 transcripts. The analytic sensitivity of MRD analysis (generally between one cell in 1,000 and one cell in 1,000,000) is greater than that of standard methods for chimerism analysis (generally 1–5 %). In addition, MRD detection with a disease-specific marker detects only the malignant cells, while chimerism analysis detects all recipient cells without distinguishing between malignant and normal host cells. Chimerism testing is useful because it offers a universal way to detect residual recipient cells without the need to have a known disease-specific cytogenetic or molecular marker. A way to augment the sensitivity of chimerism analysis for early detection of relapse is by enriching the sample for cells that carry a phenotypic marker similar to the patient’s malignancy, i.e., enriching for CD19+ cells in a patient who underwent HSCT for B-lymphoblastic leukemia [64, 65]. Cells also can be enriched for either an abnormal phenotype or CD34 positivity (a surface marker of immaturity, which is carried by most acute leukemias) [66, 67]. Potentially the best way to predict disease relapse is to combine chimerism testing with other MRD assays, if available, to increase the predictive value of both tests [37, 68, 69].
Guidelines for Chimerism Testing in HSCT
The American Society for Blood and Marrow Transplantation published a set of guidelines for the testing of chimerism after allogeneic HSCT (Table 56.2) [53]. These guidelines support the use of STR or variable number tandem repeat (VNTR) analysis of peripheral blood samples as the standard method to analyze chimerism and specify that lineage specific chimerism is useful following non-myeloablative and reduced-intensity conditioning.
Table 56.2
Comparison of chimerism analysis in myeloablative and RI-HSCT based on recommendations of the National Marrow Donor Program and the International Bone Marrow Transplant Registry 2001 Workshop
Myeloablative HSCT | RI-HSCT (Non-myeloablative) | |
|---|---|---|
Recommended sample | Peripheral blood | Peripheral blood and cell lineage subpopulations |
Frequency of analysis | 3, 6, and 12 months after HSCT; after complete chimerism, based on change in clinical condition | Every 2–4 weeks |
Significance | Mixed chimerism, particularly increasing mixed chimerism, may help predict relapse; significance depends on disease and cell lineage of chimerism | Early patterns of chimerism may predict either GVHD or graft loss |
Method | Tandem repeat loci sensitive for detection of recipient allele(s) | Tandem repeat loci sensitive for detection of both recipient and donor allele(s) |
Other Applications of Chimerism Analysis
HSCT for Nonmalignant Conditions
RI-HSCT is commonly used to treat nonmalignant conditions, where mixed chimerism is sufficient to reverse a clinical phenotype such as congenital immunodeficiency, enzyme deficiency, or hemoglobinopathy. In these situations chimerism studies are helpful in predicting the clinical benefit. For example, in HSCT for congenital immunodeficiency, the increase in T cell chimerism correlates with immune reconstitution [70], and in HSCT for hemoglobinopathies, establishment of 25 % donor chimerism is sufficient to reverse the clinical phenotype of sickle cell anemia and thalassemia, and usually correlates with transfusion independence [71, 72]. Factors that are associated with stable mixed chimerism include younger donor age, sibling donor, and the type of conditioning regimen used. Acute GVHD occurs more often in patients who achieve full donor chimerism [73].
Donor-Derived Clonal Disorders
Donor-derived malignancies are rare after HSCT and solid organ transplants [74]. These malignancies can easily be confused with relapsed disease in a patient after HSCT, and therefore chimerism testing is important to identify the origin of the malignant clone. When the donor is still alive, this may require advising the donor about appropriate screening procedures. Similarly, chimerism analysis allows determination of the origin of proliferating lymphocytes in post-transplant lymphoproliferative disorders.
Transfusion-Associated and Solid Organ Transplant-Associated GVHD
GVHD is rarely associated with the transfusion of cellular blood products containing viable passenger lymphocytes as well as solid organ transplants. In both cases, immunosuppressed individuals are unable to eradicate donor-derived competent T cells. The result is a severe form of GVHD that primarily attacks the bone marrow, leading to aplasia, which is frequently fatal if not identified early. In these cases of transfusion-associated GVHD, prior donor DNA is usually not available, but analysis of the recipient’s blood will reveal foreign DNA [75]. Chimerism studies can show mixed chimerism in these patients, and cases of > 50 % T cell donor chimerism have been described [76]. This topic is described further in Chap. 57.
Methods
Table 56.3
Comparison of methods of chimerism analysis
Method | Sensitivity | Advantages | Disadvantages |
|---|---|---|---|
Karyotyping | 5–10 % | Monitors chromosomal abnormalities as well | Requires dividing cells; low sensitivity; high false-positive rate |
RFLP | 5–10 % | Highly informative | Requires Southern blot analysis; alleles not distinct |
Red blood cell phenotyping | 0.04–3 % | Simple | Not very informative; can be affected by transfusion; not useful for rapidly proliferating diseases |
STR-PCR | 1–5 % | Highly informative; quantitative | Only moderate sensitivity; multiplex amplification may decrease sensitivity |
FISH for X and Y chromosomes | 0.1–0.5 % | Sensitive; feasible to screen large numbers of cells for quantitative result | Only for sex-mismatched transplants |
SNP Analysis | 0.01–1 % | Highly sensitive; quantitative | Less quantitative with higher chimerism; need to evaluate many SNPs to find informative SNP |
Y-chromosome PCR | 0.001–0.1 % | Sensitive; highly informative in sex-mismatched transplants | Only for sex-mismatched transplants with male recipient and female donor |
Cell sorting + STR-PCR | 0.0001–0.1 % | Informative; sensitive; cell lineage information important | Requires large volume of blood; technically demanding |
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) analysis for the X and Y chromosomes (XY-FISH) can be used for chimerism analysis by hybridization of interphase preparations with two differentially labeled probes for sequences on the X and Y chromosomes, respectively [77]. Classic karyotyping methods could be used for detection of X and Y chromosomes but would be labor-intensive, slow, and limited by the number of cells able to be analyzed. For example, to attain a sensitivity of 1 %, more than 300 cells would need to be evaluated. FISH allows the rapid screening of a large number of cells. The method can yield quantitative results by rapidly counting between 500 and 1,000 cells. A sensitivity of approximately 0.1 % or lower can be achieved [59]. Standardization of XY-FISH has made it suitable for clinical use [78]. However, the major limitation of XY-FISH is the applicability only for sex-mismatched transplants.
Tandem Repeats
Tandem repeats are a class of polymorphism consisting of a variable number of tandemly repeated core sequences [79]. Tandem repeats are classified according to the number of nucleotides in each core sequence. Minisatellites have core sequences of approximately 8–80 base pairs (bp) and are also known as VNTR. In contrast, microsatellites, also known as STR loci, have core sequences of up to 7 bp [80]. Allelic variation at each VNTR and STR locus arises from the number of core repeats present. STR alleles have between 3 and 40 tandem core repeats; each STR locus typically has 10–20 different allelic variants. Variant alleles with imperfect repeats also occur [80].
STR loci and some VNTR loci are amenable to PCR amplification using primers that flank the repeating sequence. After amplification, the number of repeats is calculated from the size of the PCR product, or, more simply, the products are compared to an “allelic ladder” constructed to contain the majority of known repeat size alleles in the human population at a particular locus [80]. PCR amplification of tetranucleotide (four bp in each core repeat) and pentanucleotide (five bp in each core repeat) STR loci has become the predominant method for DNA-based human identification [81]. Comprehensive information about STR loci and their application in human identification is available from a Web site sponsored by the National Institute of Standards and Technology (http://www.cstl.nist.gov/div831/strbase , accessed 4/12/2015).
Amplification of STR loci has become the most frequently applied technology for the study of chimerism after HSCT for several reasons [82, 83]. By examining multiple STR loci, which is made easier by multiplex PCR, it is possible to find at least one STR locus for each donor–recipient pair that can be used to distinguish the donor from the recipient; thus, STR-PCR is essentially universally informative, except in identical twins. In addition, STR-PCR can be used to quantitatively discriminate mixtures of DNA based on relative amplification of alleles. The sensitivity of STR-PCR for chimerism analysis can achieve a limit of detection of about 1 % chimerism if performed following recommended guidelines [84]. STR alleles are relatively similar in size due to the small repeat unit; thus, compared with VNTR alleles, which have larger differences, STR-PCR theoretically is better for quantification because preferential amplification of smaller alleles is less likely to occur. The advent of capillary electrophoresis (CE) and automated fluorescent detection has made the analysis of PCR products technically straightforward. In addition, the availability of commercial reagents for STR-PCR simplifies assay development.
Y Chromosome Analysis
Amplification of Y chromosome-specific sequences can be used for assessment of chimerism in sex-mismatched HSCT involving a male patient and a female donor, which represents approximately 25 % of all HSCT cases. Because the targeted sequences are present only on the Y chromosome, the method is very sensitive. Sensitivities as high as 1 male in 100,000 female cells (<0.001 %) have been reported, although the clinical relevance of chimerism detection at such high levels of sensitivity is not clear [45, 85]. Use of quantitative real-time PCR has the advantage of being not only sensitive but also quantitative for very low levels of mixed chimerism, useful for detecting an increasing or decreasing signal over time. Amplification of the amelogenin gene, which is present on both the X and Y chromosomes, but is 6 bp longer on the Y chromosome homolog, is used for sex typing in forensic analysis and also has utility for chimerism analysis [80]. The major disadvantage of all Y chromosome PCR methods is the limited application to only a subset of sex-mismatched transplants.
Single Nucleotide Polymorphism Analysis
Single nucleotide polymorphisms (SNPs) are primarily biallelic variants at a single nucleotide position. SNPs occur on average every 1.3 kilobases (kb) in the human genome. Since SNPs are sequence polymorphisms rather than length polymorphisms, their utility for chimerism analysis is based on the frequency of heterozygosity in the particular population under study. With only three potential genotypes for most SNP loci (AA, AB, BB), the chance of distinguishing two individuals is much lower than for STR loci, which have 10–20 alleles per locus. Consequently multiple SNPs must be analyzed to successfully distinguish all donors and recipients. One investigator statistically calculated the number of SNPs required to identify an informative locus in 96.5 % of cases to be 25 SNPs [86]. In one study using a panel of 51 SNPs, an informative locus was found in 100 % of cases; however, this study used a microarray-based system to enable high-throughput testing [87]. While the number of loci necessary is a disadvantage, an advantage of SNP detection over tandem repeat amplification is that SNPs are less susceptible to preferential amplification of alleles based on length, and stutter peaks (described later) are not a concern.
While there are many methods for analysis of SNPs, real-time PCR technology is commonly used for SNP-based chimerism analysis [82, 88]. Real-time PCR permits sensitive detection of mixed chimerism using SNPs at a level of approximately 0.1 %. Although real-time PCR is quantitative, the coefficient of variation (CV) at chimerism levels above 5 % is as high as 30–50 %, compared with a CV of about 5 % for STR-PCR methods [77]. Thus, while SNP real-time PCR is more sensitive and quantitative, particularly at low levels of mixed chimerism, the variability of the quantitation when the level of recipient is above 5 % does not permit accurate determination of dynamic change in mixed chimerism levels [82]. Real-time PCR SNP analysis of mixed chimerism is a promising method; yet, despite years of evaluation and new technologies for analysis, SNPs have not supplanted STRs for chimerism analysis [82].
Laboratory Testing
PCR amplification of STR loci is currently the most commonly accepted and most widely used method for assessment of mixed chimerism after HSCT [53, 84, 89]. Therefore, the remainder of this chapter details the use and interpretation of PCR testing of STR loci for analysis of mixed chimerism in a clinical laboratory.
STR Amplification Reagents
Chimerism analysis requires the identification of at least one informative STR locus that distinguishes recipient from donor cells prior to transplant, in order to be able to monitor recipient cells, donor cells, or both in the recipient after transplant. A variety of commercial reagents are available for amplification of STR loci. The majority of commercially available STR amplification reagents are designed for forensic analysis in which a maximum amount of information is desired from frequently limited samples. Multiplex PCR, in which multiple loci are amplified simultaneously by including more than one set of PCR primers in the reaction, is ideal for this purpose. The STR locus-specific PCR products from the multiplexed reactions are distinguished by the expected size range of the products for each locus (designed to be nonoverlapping) and by the use of different fluorophores for each locus in a multiplexed set, allowing amplification of loci with overlapping product sizes. High-order multiplex primer sets have been designed that amplify 16 or more STR loci. The two major suppliers of forensic STR primer sets are Promega Corporation (Madison, WI) and Life Technologies (Foster City, CA), each with a variety of multiplex combinations, some designed for detection with specific methods or instruments. At the time of writing of this chapter, enforcement of a patent held by Promega currently prohibits the use of the Life Technologies STR reagents for chimerism analysis in the setting of HSCT and other clinical and research applications. Primer sequences are also available online and in publications for laboratories to develop and validate their own assays; however, the optimization of multiplex amplification is time-consuming and complex.
Selecting a STR Marker Panel
All STR loci are not created equal. Some are more informative than others based on allele frequencies and rates of heterozygosity. The possible constellations of alleles can be grouped into several categories [90, 91]. Some loci have a higher proportion of ideal allele constellations that improve the ability to detect low levels of recipient in a donor background. The Penta E locus was found to have the highest percent of ideal allele constellations in a study that evaluated 27 loci in 203 recipient–donor pairs [90]. Interestingly, in our laboratory’s experience using 15 STR loci for pre-transplant analysis and then selecting a single optimal locus, Penta E was the most commonly selected single locus. In contrast, CSF1PO was the least selected locus and it had the lowest frequency of ideal allele constellations in the study by Thiede et al. [90]. In Europe, the EuroChimerism consortium was established to facilitate harmonization of chimerism diagnostics between European centers because of their assertion that existing forensic commercial primer sets are not ideal for chimerism [92]. This group recommended an STR marker panel specifically for chimerism analysis to maximize informativity, reliability, and sensitivity. Nevertheless, in our experience over 15 years, 12–15 commercially available forensic loci have been used successfully in an active transplant program to perform chimerism analysis. Even though several loci have been informative more often than others, the less informative loci have all been used for some recipient–donor pairs, indicating that they all have potential value; this is especially true in a two-donor transplant case where several loci are usually needed to monitor chimerism.
Detection Method
Amplified STR products can be detected in numerous ways, as described for forensic analysis in Chap. 54. By far the most frequently used method in clinical laboratories for analysis of STR amplification products is fluorescent detection by CE, which has the advantage that a gel does not need to be poured and analysis of samples is rapid [80]. CE can be performed with a single capillary instrument or with a multi-capillary instrument; for example, with 16 or 24 capillaries. The advantage of multiple parallel capillary systems is that analysis can be performed more rapidly than with a single capillary instrument. Capillary arrays with 96 or more capillaries are available; however, their instrument and reagent costs are greater, and thus they are only cost-effective for very high-volume testing. CE instruments that can detect multiple fluorophores permit not only multiplex amplification of loci resulting in PCR products overlapping in size, but also inclusion of a size marker for accurate sizing of the fragments. Of note, not all fluorophores are detected by all CE instruments; therefore it is important to check for compatibility before selecting reagents.
Pre-transplant Testing
The goal of pre-transplant analysis is to identify the informative loci that can be used to monitor chimerism after HSCT. This is accomplished by determining the STR alleles of the recipient and the donor(s) at each STR locus in the marker panel used by the laboratory, and then selecting at least one locus with one or more different donor and/or recipient alleles in a constellation pattern conducive to sensitive and accurate monitoring of post-transplant chimerism.
Samples
Usually peripheral blood samples from the donor and recipient are used to extract DNA for pre-transplant analysis. Analysis of an additional buccal cell sample from the recipient not only serves as a quality control to confirm the identity of the recipient sample in the laboratory, but also as a control for balanced amplification of alleles in a non-hematopoietic lineage. If there is allelic imbalance in the blood sample at a particular locus, comparison with the buccal sample amplification pattern can help determine if the effect is constitutional, stochastic, or due to disease (Fig. 56.2). A locus with significant unbalanced amplification of alleles in either sample should be avoided.
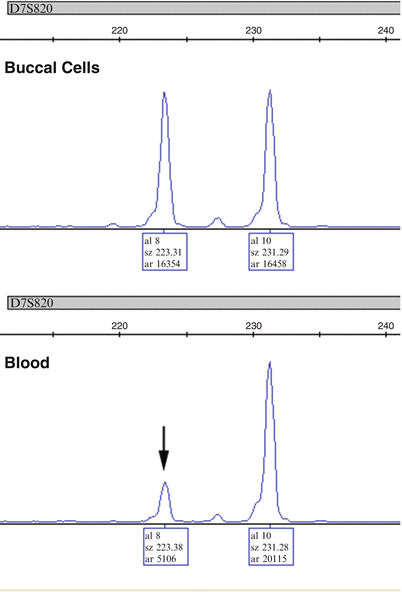

Figure 56.2
Amplification of the D7S820 locus in DNA obtained from buccal cells and blood in a patient with myelodysplastic syndrome. The blood, which contained disease, shows that one allele (8, vertical arrow) has only 25 % of the peak area of the other allele peak (10). The fact that the alleles are not imbalanced in the buccal cells suggests that this is due to a chromosomal abnormality related to the disease and not a constitutional polymorphism affecting amplification of allele 8. Thus, D7S820 should be excluded from calculations of chimerism due to underestimation of the % recipient if the disease relapses
On occasion, a recipient is transplanted prior to the STR evaluation of a pre-transplant sample. In this case, a post-transplant blood sample will not be useful for identification of the recipient’s genotype. Buccal cells are the most commonly used source of recipient DNA if blood is not an option, although any other available historical tissue that is appropriately preserved can be used. While post-transplant buccal samples will be of mostly recipient genetic origin, they will frequently contain a minority of admixed donor DNA, thereby generating a mixed chimeric pattern [93]. Since the recipient’s alleles usually predominate, they can be deduced if the donor alleles are known. A saliva or mouthwash sample is not equivalent to a buccal cell sample since saliva generally contains significantly more leukocytes. In one study, the median amount of donor DNA in the buccal swabs was 21 % whereas it was 74 % in the mouthwash samples [94]. If only donor alleles are detected in a buccal swab DNA sample (and the donor is not an identical twin sibling), complete “engraftment” of buccal epithelial cells should be considered. Several cases of buccal cell “engraftment” after HSCT have been reported [95].
Identification of Informative Loci
To identify ideal informative loci for each recipient–donor pair usually many loci must be examined. This is particularly true when the individuals are related and share many alleles by inheritance. Multiplex amplification increases the speed and decreases the labor associated with the analysis of multiple loci. Of the loci found to be informative, not all will be appropriate for chimerism analysis. This is because the goal is not to differentiate the individuals as in forensic identity testing but to detect small amounts of recipient or donor alleles in a mixed specimen [90].
The main factor limiting the utility of some informative alleles is the presence of a stutter peak which is one repeat smaller (n − 1) and, to a much lesser extent, one repeat larger (n + 1) or two repeats smaller (n − 2) than each major allele peak [96]. When tandem repeats are amplified, stutter peaks are thought to result from slippage of the polymerase, causing mispairing of the template, which is shifted by one repeat unit, resulting in an additional PCR product predominantly one repeat smaller than the expected PCR product. The intensity of the n − 1 stutter peak is usually approximately 2–11 % of the area of the corresponding STR allele [96]. The amount of stutter is influenced by the locus (some loci tend to have more stutter than others), the core repeat unit length (smaller core repeats have more stutter than larger repeats), and the allele size (larger repeat alleles have more stutter than smaller repeat alleles) [80].
For chimerism analysis, optimal informative loci have one or two recipient-specific allele(s) that do not co-localize with a donor allele or stutter peak (Fig. 56.3) [97]. Ideally, the recipient-specific allele should be at least two repeats larger or three repeats smaller than the donor allele(s) to avoid the stutter locations [89]. Having two unique recipient alleles that meet these criteria (recipient is heterozygous) may provide confirmation that a recipient DNA is present; however, at low levels of chimerism a heterozygous allele may be below the limit of detection compared to a homozygous peak. In addition, due to the theoretical possibility for preferential amplification of shorter fragments, the sensitivity for detection of a minor recipient-cell population may be greater if the informative recipient allele(s) have fewer repeats than the donor allele(s) [89]. Because of the many possible combinations of alleles between recipients and donors, each recipient–donor pair presents a unique situation, and each must be evaluated on a case-by-case basis to determine which loci can be used and which should be excluded due to artifacts (like stutter) that will reduce sensitivity and accuracy. These are only guidelines for the selection of informative loci; how useful or accurate a particular locus will be post-transplant ultimately is determined empirically by use in the post-transplant setting. In some instances, an optimal locus cannot be identified. In this case, post-transplant analysis still can be performed using a suboptimal locus with the caveat that the sensitivity of detection will be decreased because of co-localization of the recipient-specific allele with a stutter or other background peak. As described in the analysis section, chimerism calculations can be adjusted with caution to take stutter contributions into account.
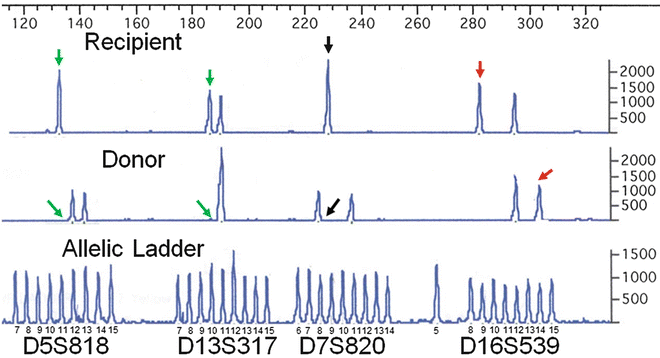

Figure 56.3
Identification of informative loci using PCR amplification of STR loci and CE analysis. The STR alleles at four loci (GammaSTR, Promega Corporation) are compared for a recipient–donor pair to identify the best informative locus. The number of repeats in each allele is determined by comparison to the allelic ladder for which the sizes are indicated. While all four loci are informative for distinguishing the recipient from the donor, the D16S539 locus is best because the recipient-specific allele (red vertical arrow) is separated from the nearest donor allele by at least two repeats. In contrast, for the D5S818 and D13S317 loci, the recipient-specific allele (green vertical arrows) co-localizes with a donor stutter peak, the location of which is indicated by the diagonal green arrows. The D7S820 recipient-specific allele (black vertical arrow) is acceptable, but, if present, an n + 1 stutter (black diagonal arrow) from the smaller size donor allele may interfere with detection. The allele combination at the D16S539 locus also has the advantage of a unique donor allele (red diagonal arrow). PCR polymerase chain reaction, STR short tandem repeat, CE capillary electrophoresis
Another factor to consider when selecting informative loci is whether there are specific alleles for both the recipient and the donor (Fig. 56.3). While this is not essential for myeloablative HSCT, for which the combined donor and recipient allele pattern of included loci must be optimized to enhance detection of a small percentage of recipient DNA in a donor background, it is very important for non-myeloablative HSCT, since any level of mixed chimerism (low donor or low recipient) may be seen. If no such locus can be found, then at least two loci should be selected, one optimized for detection of recipient allele(s) and the other for donor allele(s). As the level of mixed chimerism in the patient changes from low to high donor chimerism or vice versa in case of disease relapse, the loci used may need to be changed to optimize sensitivity (Fig. 56.4). The use of more than one informative locus to generate an average chimerism result will minimize the effect of artifacts and maximize analytic sensitivity. However, even if multiple loci are used the set of loci included in the average should be reevaluated if the percent of recipient DNA is very high.
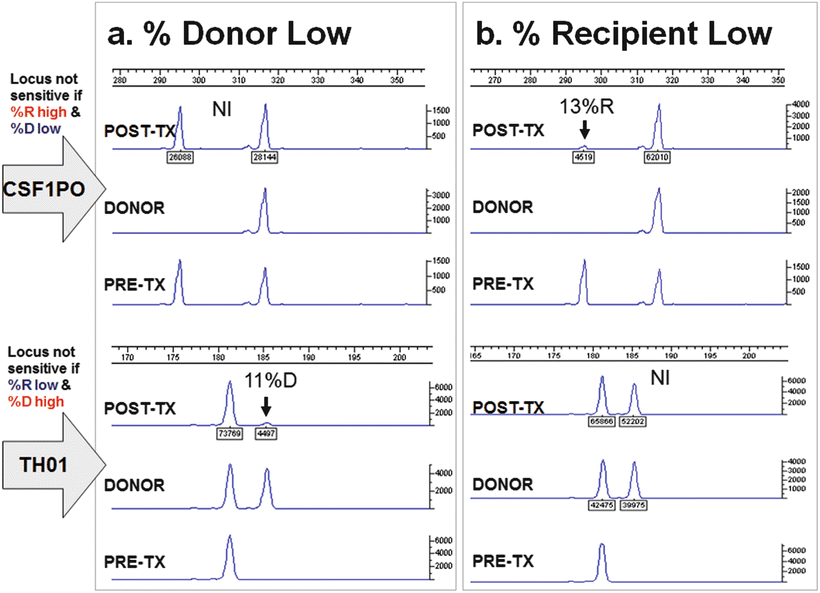

Figure 56.4




If the recipient or donor is homozygous and that allele is shared with the donor or recipient, respectively, this locus will not be equally informative at all levels of chimerism. Electropherograms at two loci (CSF1PO and TH01) are shown for two post-transplant time points (a and b) for a recipient–donor pair. For each time point, the donor and pre-transplant recipient (PRE-TX) alleles are shown as well as a post-transplant (POST-TX) sample showing mixed chimerism. For this recipient–donor pair, when the percentage of donor DNA is low (a), the CSF1PO locus is not informative (NI) for detection of donor because a small change in donor would be smaller than the measurement error of chimerism due to variable amplification of the two alleles. In contrast, the TH01 locus (bottom of panel a), which has a donor-specific allele is informative and sensitive for detection of donor (11 % donor). Similarly, when the percentage of donor DNA is increased (b), the situation is reversed. The CSF1PO locus is informative and sensitive for detection of low amounts of recipient (13 %R). On the other hand, the TH01 locus is not informative (NI) because, as with the CSF1PO locus when the donor was low, the potential variable amplification of the alleles (measurement error) could lead to a false-positive or a false-negative result. When the minor component (recipient or donor) is approximately 25 % or less, loci with this constellation of alleles should be excluded from use. In the ChimerMarker software (SoftGenetics). The use of loci with this constellation of alleles is called “deconvolution;” deconvolution should only be used when absolutely necessary if using this software because it does not distinguish between the two types of deconvolution
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


