Togaviridae
Richard J. Kuhn
The togaviruses are simple enveloped plus-strand RNA viruses that are spherical in appearance and contribute significantly to human disease. Although they were originally classified together with several groups of viruses that are transmitted predominantly by insects, more recent analyses have categorized them into a distinct family with two genera: Alphavirus and the Rubivirus.167,208 The Alphavirus genus is by far the larger of the two, with about 30 recognized members, whereas the Rubivirus genus is composed of a single member, rubella virus. Virus classification into each group is determined by genome organization and nucleotide homologies. The alphaviruses are responsible for a variety of human and animal diseases, involving encephalitis, arthritis, fever, and rash, and are transmitted primarily by arthropod vectors. Viruses such as Chikungunya (CHIKV) and Venezuelan equine encephalitis virus (VEEV) have been responsible for recent human outbreaks and have raised awareness of the significance and potential threat of alphaviruses to human health. Rubella virus is a common childhood illness for which an effective vaccine is available. However, in the absence of immunity, the virus can induce severe congenital defects in the fetuses of infected women.
Sindbis virus (SINV), the type-member of the alphavirus genus, has been studied extensively, in large part due to its facile growth in cell culture and its ability to cause mild or inapparent illness in humans. The virus has an 11.7-kb RNA genome that is capped at its 5′ end and contains a poly A tract at its 3′ terminus.265 Virions have a spherical icosahedral arrangement of proteins that has facilitated their structural analysis. The detailed knowledge of the viral life cycle, which is the focus of this chapter, has been exploited for the development of alphavirus gene expression vectors. Many members of this virus group have been studied for their role in pathogenesis. Rubella virus, as expected from its classification, shares a number of properties with the alphaviruses, yet has several important distinctions that are highlighted throughout the chapter.
Classification of Viruses Within the Togaviridae Family
Viruses transmitted by arthropods have been referred to as arboviruses.208 It was originally noted that many arboviruses had a similar morphologic structure, as observed by electron microscopy, that resembled a Roman cloak (in Latin, toga), hence the name togaviruses. Originally, the family Togaviridae consisted of group A (alphaviruses) and group B (flaviviruses); however, the genera Rubivirus and Pestivirus were later added based on their similar physical properties but despite their lack of arthropod transmission. With the development of sequencing, it became apparent that the original joint classification for these viruses was in error. The togaviruses have nonstructural or replication proteins encoded at the 5′ end of their genome RNA, whereas the 3′ end encodes the proteins that comprise the virus particle, or virion. In the togaviruses, these structural proteins are translated from a subgenomic messenger RNA (mRNA) that derives from, and is co-terminal with, the 3′ end of the genome.219,265
The larger of the two genera within togaviruses, the Alphaviruses, has been classified into seven antigenically related complexes.209 Most phylogenetic analyses support this classification, but several recent additions add complexity to the organization. The alphaviruses have a worldwide geographic distribution, including the continent of Antarctica. The alphaviruses have classically been described as either Old World or New World viruses, depending on their distribution, and it is
likely that several transoceanic exchanges have occurred.209 Most alphaviruses are transmitted by arthropod vectors that probably control their geographic dispersal. However, the recent identification of the salmonid viruses, salmon pancreas disease virus, and sleeping disease virus (infecting rainbow trout), present examples of alphaviruses for which arthropod transmission is unlikely.293,309 These salmonid viruses appear to have diverged from the Old World–New World lineages early in alphavirus evolution, with no present-day close relatives. Another identified alphavirus, southern elephant seal virus, has been isolated from the louse Lepidophthirus macrorhini.151 This isolation demonstrates not only that alphaviruses are transmitted by lice, but that they can also infect marine mammals, and a recent report suggests a marine origin for the alphaviruses.51 Given this wide host range, it seems probable that amphibian- and reptile-specific alphaviruses also await future identification.
likely that several transoceanic exchanges have occurred.209 Most alphaviruses are transmitted by arthropod vectors that probably control their geographic dispersal. However, the recent identification of the salmonid viruses, salmon pancreas disease virus, and sleeping disease virus (infecting rainbow trout), present examples of alphaviruses for which arthropod transmission is unlikely.293,309 These salmonid viruses appear to have diverged from the Old World–New World lineages early in alphavirus evolution, with no present-day close relatives. Another identified alphavirus, southern elephant seal virus, has been isolated from the louse Lepidophthirus macrorhini.151 This isolation demonstrates not only that alphaviruses are transmitted by lice, but that they can also infect marine mammals, and a recent report suggests a marine origin for the alphaviruses.51 Given this wide host range, it seems probable that amphibian- and reptile-specific alphaviruses also await future identification.
Although the alphaviruses and rubella virus have been classified within the same family, the evolutionary relationship between them is obscure.54 They have a similar genome organization, and their virions share physical similarities, yet their replication and assembly strategies are sufficiently diverse to question whether they arose from a direct ancestor.
Virion Structure
Structure of Mature Virion
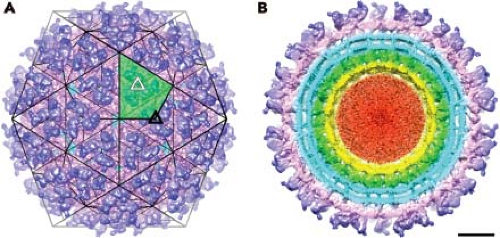
The structure of the alphavirus virion has been extensively studied, and numerous high-resolution structural studies now provide a near-atomic view of the virion. Although a variety of biophysical methods were used to elaborate the alphavirus structure, work that has advanced the field the most has come from cryo-electron microscopy (cryo-EM) and image reconstruction techniques.93,94,287,288 The virion is 70 nm in diameter, with a molecular mass of 52 × 106 D and a density of 1.22 g per cc. It is composed of repeating units of the E1 and E2 transmembrane glycoproteins, the capsid or nucleocapsid protein (C), a host-derived lipid bilayer, and a single molecule of genome RNA. The protein components of the virion are arranged as a T = 4 icosahedral lattice, with 240 copies of each subunit.27,198,201 These subunits interact with one another to form a rigid structure across the membrane in a one-to-one relationship between glycoprotein E2 and the capsid protein. Smaller amounts of another membrane-associated protein, 6K, are also found in the virus particle.64,162 More recently it has been discovered that another small protein, the TransFrame (TF) protein, is found in substoichiometric amounts in the virion.47 The lipid bilayer is derived from the host plasma membrane and is enriched in cholesterol and sphingolipid, molecules that are required for entry and budding.114 Inside the bilayer, the C surrounds the genome RNA and forms an icosahedral shell. Thus, the alphaviruses are composed of multiple organized shells of molecules that effectively protect and deliver the viral RNA to susceptible host cells.
Cryo-EM has been used extensively to study the structure of alphaviruses, including SINV,198 Semliki Forest virus (SFV),283 Ross River virus (RRV),27 VEEV,197,315 Western equine encephalitis virus (WEEV),247 Aura virus,316 CHIKV,298a and Barmah Forest virus (BFV).122 The most recent studies with VEEV, BFV, and SINV are the most advanced, with a resolution between 4.4 and 7.0 Å reported for these viruses.122,315 The surface view of the virion, seen in Figure 22.1 for SINV, reveals a spherical particle with spike protrusions rising 100 Å from the surface. The icosahedral nature of the particle results in an ordered distribution of the petal-like spikes. The asymmetric unit, which is shaded in green in Figure 22.1A, contains four E1–E2 heterodimers. Each spike consists of three heterodimers of E1 and E2 glycoproteins. A total of 80 spikes reside on icosahedral threefold (solid black triangle in Fig. 22.1A) and quasi-threefold axes (open white triangles in Fig. 22.1A). Earlier biochemical studies established the nature of the heterodimer, and this relationship has been confirmed
by cryo-EM reconstructions218,321 and by x-ray crystallography.145,289 Although the protein lattice occupies a substantial surface area, small openings are present in the virion that reveal the underlying lipid bilayer. These openings are most pronounced at the twofold axes, but can also be found at the fivefold axes and around the base of each spike.
by cryo-EM reconstructions218,321 and by x-ray crystallography.145,289 Although the protein lattice occupies a substantial surface area, small openings are present in the virion that reveal the underlying lipid bilayer. These openings are most pronounced at the twofold axes, but can also be found at the fivefold axes and around the base of each spike.
The transmembrane components of the two glycoproteins are clearly seen in the cryo-EM reconstructions (Fig. 22.1B). The shape of density suggests that each transmembrane segment traverses the bilayer as a helix, although the E1 transmembrane domain is better represented by two alpha helices separated by a two amino acid kink.315 For SINV the E1 glycoprotein has five amino acid residues that penetrate across to the inner side of the membrane (cdE1), whereas E2 has 33 amino acids (cdE2) that interact with the nucleocapsid core.14,148,153,271 This interaction is observed in cryo-EM reconstructions and demonstrates that each E2 molecule makes specific contacts with each capsid protein. The nucleocapsid core has a T = 4 arrangement of capsid protein, with the C-terminal protease domain forming pentameric and hexameric projections that appear as capsomeres on the surface of nucleocapsid cores (shown in green in Fig. 22.1B). The genome RNA does not appear to assume regular symmetry within the nucleocapsid core and is not ordered in the reconstructions (red in Fig. 22.1B).
Structure of Immature Virion
The structure of the immature virus containing an uncleaved precursor to E2, called PE2 or p62, has also been solved using cryo-EM.46,200 Mutant versions of both SINV and SFV were used for independent structure determinations resulting in similar structures. The extra density corresponding to the small protein E3 was found predominantly between the petals of the spike resulting in a dual-lobed petal. At a resolution of 25 Å, no apparent differences were found in the skirt or other regions of the spike and suggest that following cleavage of PE2 and release of E3, no significant conformational changes occur in the virus structure. The immature form of the virus that contains PE2 has been proposed to stabilize the fusion protein as it transits the mildly acidic environment of the Golgi.155,157,291
Structural Proteins of the Virion
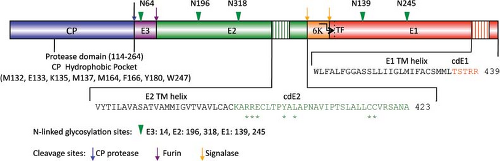
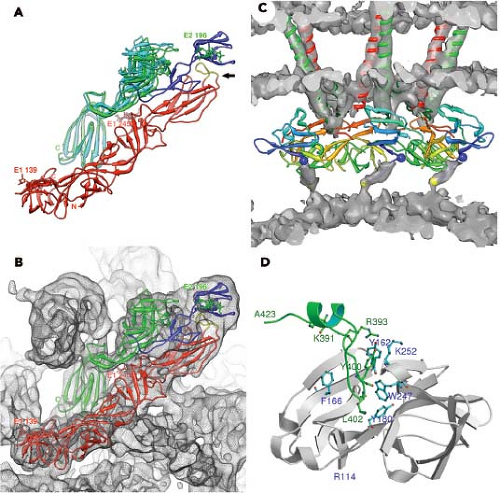
The proteins that comprise the alphavirus virion are synthesized as a polyprotein from a subgenomic RNA and are shown in Figure 22.2. The structures of the three major virion proteins have been determined by x-ray crystallography to high resolution. The capsid protein functions to encapsidate the genome RNA and forms a T = 4 icosahedron prior to release from the cell. The 264 amino acid capsid protein of SINV was crystallized, although the N-terminal 105 residues have been absent in all of the final structures.29 It was suggested that the highly basic N-terminal domain was susceptible to proteases and degraded during crystal growth. A similar observation was made for the SFV capsid protein.28 The C-terminal residues 114 to 264 of the SINV capsid protein form a chymotrypsin-like fold with the ultimate residue tryptophan 264 bound in the active-site protease pocket. The fold contains two Greek key b−barrel domains that bring the catalytic triad of Ser215, His141, and Asp163 into juxtaposition for activity.86 This structure substantiated biochemical and genetic data that proposed that the capsid protein had an autocatalytic serine protease activity.10,85,168 The atomic structure of the capsid protein was used to generate a “pseudoatomic” resolution structure of the alphavirus nucleocapsid core (Figure 22.3) by fitting its coordinates into the cryo-EM density.27,179,228 This was accomplished and it was suggested that the missing residues of the N-terminus would point inward from the core surface and interact with the negatively charged viral RNA. The recent 4.4 Å cryo-EM structure of VEEV provides support for the earlier fitted structures, but the N-terminal basic domain residues (1–110). Remains obscure as ordered density within the nucleocapsid core has not been observed.315
The E1 protein functions as a class II fusion machine to promote the joining of viral and cellular membranes under conditions of low pH. The structure of the E1 molecule from SFV was solved using x-ray crystallography.138 The starting material for crystallization was the SFV spike cleaved with subtilisin, which released the ectodomains of the E1 and E2 glycoproteins.307 Unfortunately, the crystals contained only the E1 polypeptide, and it was assumed that E2 was proteolyzed.
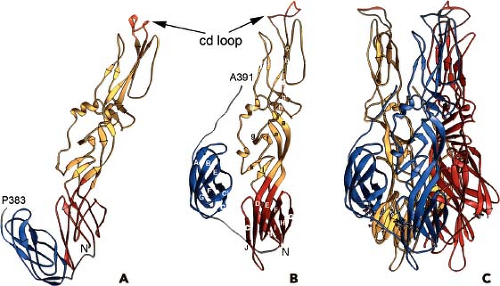
The structure of the E1 ectodomain (residues 1–383) is shown in Figure 22.3 and contains three b−barrel domains. Domain I, the so-called central domain, links domains II and III. The extended domain II contains at its distal end the fusion peptide, a short loop of hydrophobic amino acids that promotes insertion of the protein into the target membrane. Domain III has an immunoglobulin (Ig)-like fold and connects at its C-terminus with the transmembrane domain of the protein.
The structure of the E1 ectodomain (residues 1–383) is shown in Figure 22.3 and contains three b−barrel domains. Domain I, the so-called central domain, links domains II and III. The extended domain II contains at its distal end the fusion peptide, a short loop of hydrophobic amino acids that promotes insertion of the protein into the target membrane. Domain III has an immunoglobulin (Ig)-like fold and connects at its C-terminus with the transmembrane domain of the protein.
The structure of E1 is remarkably similar to that of the flavivirus E protein.217,269 Unlike the alphaviruses, the flavivirus E protein functions both in receptor attachment and in membrane fusion. In the alphaviruses, these two activities are carried out by proteins E2 and E1, respectively. In the flaviviruses, the E protein is oriented roughly parallel to the lipid membrane, and it was anticipated that E1 might assume a similar orientation in the alphavirus virion.124 This was essentially verified once the E1 atomic structure was fitted into the cryo-EM density. This fitting was accomplished for both SFV and SINV, and most recently for CHIKV.138,145,207,289 Furthermore, for SINV, the two sites of E1 glycosylation were mapped onto the cryo-EM structure, and these sites at Asn139 and Asn245 were used as positional markers to fix the position of E1 in the virion.207 E1 lies at an angle of ∼50 degrees relative to the surface of the membrane (Figure 22.3B), and it forms an icosahedral lattice
comprising the region that has been referred to as the skirt.138,207 The crystallographic E1 dimer, which is a back-to-back dimer as opposed to a face-to-face dimer seen in the flaviviruses, is essentially preserved in the arrangement of E1 in the virion. The interface residues that make contact in the crystallographic dimer are presumably those that are important for forming the icosahedral lattice in the virion.
comprising the region that has been referred to as the skirt.138,207 The crystallographic E1 dimer, which is a back-to-back dimer as opposed to a face-to-face dimer seen in the flaviviruses, is essentially preserved in the arrangement of E1 in the virion. The interface residues that make contact in the crystallographic dimer are presumably those that are important for forming the icosahedral lattice in the virion.
Although E1 comprises the skirt region of the alphavirus surface, the majority of the protruding spike is comprised of the E2 molecule. E2 constitutes the petals that make up the spike, and covers the distal end of the E1 molecule that points outward. E2 serves to engage cell surface receptor molecules required for entry of the virion into the cell. The crystal structure of E2 together with both E1 and E3 for CHIKV was solved and reveals a three-immunoglobulin domain protein.289 Domain B is located at the outermost point of the spike and contains the residues that have been implicated in receptor binding and as well as those involved in binding neutralizing antibodies. Domain B exhibits mobility in the context of the spike and was absent from the intact SINV E1–E2 trimer structure done at low pH.145 At the other end of the protein, located closest to the viral membrane, is the C-terminal domain C. The N-terminal domain A (residues 1–132 in SINV) is the central bridge of E2, and connections are made to the B domain by b−ribbon connectors. The b−ribbon connector is flanked by a pair of well-conserved histidine residues that might function as an acid switch to release the connector and B domain under acidic conditions. Although E3 does not directly contact E1, its contact with the b−ribbon connector holds it in place to prevent movement of the B domain while the virus is in the process of egress from the cell in an immature state. The furin cleavage of the p62 to release E3, therefore, releases the clamp holding the B domain and activating the complex for pH-triggered fusion. E1 and E2 have extensive contacts with each other, promoting the spike architecture (Fig. 22.3).
In addition to the three major structural proteins identified by cryo-EM in the virion, two small transmembrane proteins are present in sub-stoichiometric amounts and can be identified using purified virus and mass spectrometry. The existence of the 6K protein in the virion has been well established, but more recently the TF protein (∼8 kD) has also been found within SFV particles.47 Given the low abundance and presumably random distribution of these small transmembrane proteins, they have not been detected using high-resolution cryo-EM, and their membrane architecture has to date prevented x-ray crystal structures. Whether the TF and 6K proteins exist as oligomers is also unknown.
Genome Structure and Organization
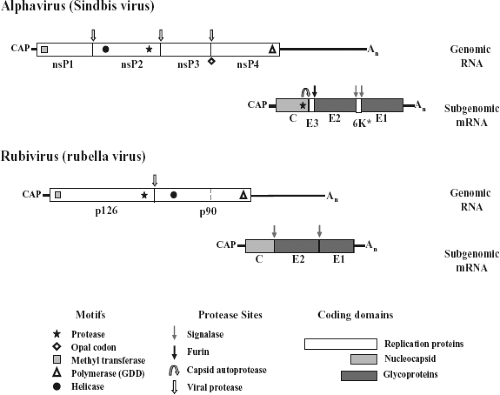
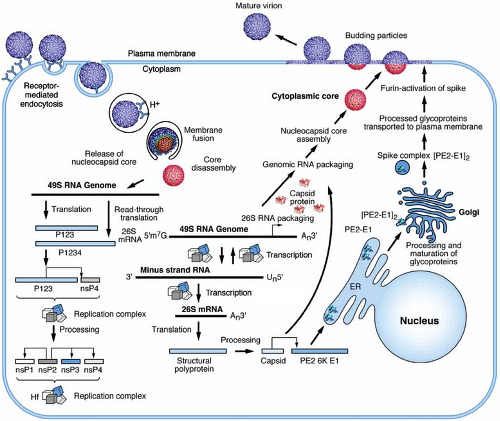
The togavirus genome resides on a positive-strand RNA that contains a 5′ terminal 7-methylguanosine and a 3′ terminus that is polyadenylated. The alphavirus genome, represented in Figure 22.4 by the type virus SINV, is approximately 11.7 kb in length, whereas rubella virus is nearly 2 kb shorter, at 9.8 kb.40,265 The genomes segregate their replication and virion protein coding regions into two segments, with the replication region mapping to the 5′ two-thirds and the structural region mapping to the 3′ one-third. Limited nucleotide homology exists between genomes in the two genera, although there are several sequences in both translated and nontranslated regions that do have homology; however, most evidence suggests that their replication and assembly strategies are quite different.40
The nonstructural or replication proteins are translated from the genome RNA, whereas the structural or virion proteins are translated from a subgenomic mRNA.191,251 In SINV, the 5′ nontranslated region (NTR) is 59 nucleotides, about average for the alphaviruses, whereas the 3′ NTR is also close to the average in length at 322 nucleotides.
The nonstructural or replication proteins are translated from the genome RNA, whereas the structural or virion proteins are translated from a subgenomic mRNA.191,251 In SINV, the 5′ nontranslated region (NTR) is 59 nucleotides, about average for the alphaviruses, whereas the 3′ NTR is also close to the average in length at 322 nucleotides.
Using comparative genome analyses and functional genetic studies of defective interfering particles and viruses, four conserved regions (conserved sequence element [CSE]) of the alphavirus genome were identified as cis-acting elements important for replication.184,185,186 Two conserved regions are found near the 5′ end of the genome, one is found in the junction region between nonstructural and structural genes, and one is found at the 3′ end immediately preceding the poly (A). Three presumably similar functioning CSEs can also be found in the rubella genome.40 In the alphaviruses, each CSE has been shown to interact in a host-dependent manner, suggesting that host factors may play a role in their function.44,97,98,123,187,188 It has been shown that a U-rich region in the 3′ NTR of SINV contains elements responsible for viral RNA stability and that the cellular HuR protein binds this region, thus decreasing the rate of cell-mediated decay of the genome RNA.260 In addition, studies have shown that host proteins bind to the 3′ end of the minus-strand RNA of SINV, and in one case the protein was identified as the mosquito homolog to the La protein.193,194,195
Alphavirus Replication
Mechanism of Attachment and Receptors
Alphaviruses display an extremely broad host range, both in terms of susceptible animal species and in terms of cells in culture. This broad host range has prompted speculation as to the nature of the receptor, with two hypotheses proposed to explain this phenomenon.266 In the first, the virus E2 glycoprotein contains multiple receptor-binding sites so that distinct cellular receptors can bind the viral surface protein. The second hypothesis proposes that the virus uses a ubiquitous receptor that is highly conserved across species, including both mammals and mosquitoes. Data exist to support each model, and it is likely that a combination of the two is the true strategy for alphavirus attachment to the cell.
The variety of molecules that have been implicated in SINV argues for multiple distinct receptor-binding sites on E2.266 The use of the laminin receptor with its high conservation across species suggests that it might serve as a receptor in multiple cell types and multiple host species.295 However, the picture remains obscure for SINV because laminin receptor functions in baby hamster kidney cells but not in chicken embryo fibroblasts, where a 63-kD protein has been implicated.296 In mouse neuroblastoma cells, proteins of 74 and 110 kD have been reported as possible SINV cellular receptors.278 DC-SIGN and L-SIGN, C-type lectins that bind mannose-enriched carbohydrates, have also been implicated as receptors of alphaviruses that have been produced in mosquito cells.118 Very recent studies using a genome-wide RNA interference (RNAi) screen in Drosophila cells identified the natural resistance-associated macrophage protein (NRAMP) as a cellular receptor functioning to permit SINV infection.226 Likewise, NRAMP2, the vertebrate homolog, allowed for SINV, but not RRV, entry into mammalian cells. This new approach and finding raises the possibility that a family of related conserved multipass membrane proteins may serve as receptors for other alphaviruses.
Natural isolates of EEEV utilize cell surface heparan sulfate as an attachment receptor.65 This use of heparan sulfate may direct tropism of EEEV and promote enhanced neurovirulence. In contrast, passage of several other alphaviruses in culture leads to the accumulation of adaptive mutations, some of which introduce basic amino acids in their E2 glycoprotein.18,20,21,119 This increase in positively charged amino acids in E2 leads to high efficiency attachment to cells through heparan sulfate molecules. The importance of this interaction was demonstrated genetically by the generation of a Chinese hamster ovary cell line using retroviral insertional mutagenesis that was deficient in the expression of heparan sulfate and chondroitin sulfate.108 These cells were resistant to SINV infection and defective in binding virus. The substitution of a single residue on the E2 glycoprotein of RRV was sufficient to permit heparan sulfate binding, and this attachment was mapped using cryo-EM to the distal tip of the spike.97,317 The binding of heparan sulfate does not result in conformational changes in the virion, nor does it enhance the fusion process.257,317 Therefore, it likely serves simply as a mechanism to attach the particle to the cell surface with high affinity so an efficient interaction with the entry receptor can occur. In contrast to what is observed in cell culture with SINV, infection of mice results in the development of large-plaque viral mutants with a reduced affinity for heparan sulfate and a greater viremia.21 However, given these diverse observations, the utilization of heparan sulfate and its role in pathogenesis among the different alphaviruses requires further investigation.
Mechanisms of Entry, Membrane Fusion, and Uncoating
Following attachment to cells and engagement with an entry receptor, alphaviruses proceed via the endocytic pathway to gain access to the cell interior (Fig. 22.5).38,98,166 Structural and biochemical experiments demonstrate that binding of RRV to heparan sulfate does not induce conformational changes in the virion.257,317 However, other receptors may induce conformational changes in the particle that might mediate entry such as the reduction of disulfide bonds to disrupt protein–protein interactions.1,11,199,237 Despite these suggestions that disulfide exchange may play a role in alphavirus entry, the use of thiol-blocking reagents failed to show a significant inhibition of infection.77 However, it is clear that the attachment of SINV to cells results in the exposure of new epitopes defined by monoclonal antibodies and suggests that protein rearrangements occur following receptor engagement.48,172 Similar observations have been made using purified virus and treatment with heat, pH, and reducing agents.171 In addition, the recent observations on the role of virion and protein dynamics in the flavivirus life cycle, suggest that other structurally related viruses might employ particle dynamics to sense their environment as they search for an entry receptor.40a In addition to the role of thiol exchange reactions that have been proposed to play a role in alphavirus entry, there has even been a suggestion that cell penetration may occur at the cell surface in the absence of fusion by conformational rearrangements in the envelope glycoproteins.1,199 However, it has been fairly well established that membrane fusion triggered by an exposure to acidic conditions
is the gateway for the release of genome RNA into the cytoplasm (Fig. 22.6).
is the gateway for the release of genome RNA into the cytoplasm (Fig. 22.6).
Receptor-bound viruses undergo endocytosis into coated vesicles using a clathrin-dependent pathway. This pathway was demonstrated by DeTulleo and Kirchhausen38 using dominant-negative mutants of dynamin to block the formation of clathrin-coated pits and prevent entry of SFV and SINV. The vesicles are subsequently acidified, providing the trigger for fusion between viral and cellular membranes. Acid-induced fusion is supported by numerous studies, but most convincingly by the use of lysosomotropic weak bases that raise the pH of endocytic vesicles and prevent entry of alphaviruses.77,99,310 Although it has been argued that viral RNA replication might also be affected by the acidification, pseudotyped viruses containing alphavirus envelope proteins are also inhibited in entry by this treatment.42,100,246
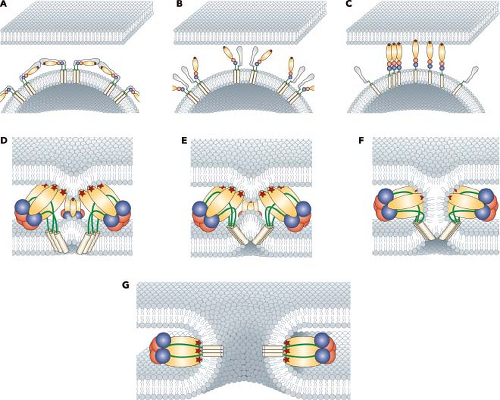
The role of the alphavirus E1 and E2 glycoproteins in the entry process has been firmly established (Figs. 22.6 and 22.7). In the presence of acidic pH, the E1–E2 heterodimer is destabilized and the two proteins dissociate.291,293 The dissociation of the proteins results in the exposure of the fusion peptide that is found on the distal tip of E1.4,74,88,138 The fusion peptide of E1 inserts into the target membrane in a cholesterol-dependent manner followed by the trimerization of E1.3,73,116,189,206,292 A large conformational change in E1 results in domain III and the stem-anchor region of the protein packing against domain II, resulting in the viral and target membranes being brought into close opposition (Fig. 22.6).76 A set of E1 trimers, possibly
resembling what has been seen in liposomes, results in membrane deformation and promotes membrane mixing.74 Finally, a fusion pore will form as the two membranes complete the process, and the nucleocapsid core will be released into the cytoplasm (Fig. 22.7).
resembling what has been seen in liposomes, results in membrane deformation and promotes membrane mixing.74 Finally, a fusion pore will form as the two membranes complete the process, and the nucleocapsid core will be released into the cytoplasm (Fig. 22.7).
Rey et al.138 recognized that the structural features of the alphavirus E1 and flavivirus E proteins were distinct from the structures of other previously identified fusion proteins such as hemagglutinin (HA) from influenza, and proposed that they represented a novel class of fusion machines. They termed these class II membrane fusion proteins and elaborated several distinguishing features that separated the two classes. Class II fusion proteins are composed predominantly of b-strands, contain an internal fusion peptide, and have a companion protein that stabilizes the structure; this companion protein forms an activated metastable structure following proteolytic processing of a precursor protein. In the alphaviruses, PE2 is proteolytically activated by cleavage to generate E3 and E2, with E2 and E1 forming a stable heterodimer.155,156 Mutagenesis experiments have provided insight into the fusion process and the residues that play regulatory and supporting roles in this process. As predicted, a pH-sensitive histidine residue in E1 (SFV H3) appears to regulate the low pH-dependent folding of E1 that is required for fusion.215 Several other well-conserved histidines in E1 were evaluated and found unable to influence the activity. In addition, Kielian and colleagues152 found that a salt bridge is formed within domain II of the E1 trimer core, which appears critical for stabilizing the homotrimer fusion intermediate.
Biochemical experiments with SFV have defined the requirements and steps in the alphavirus fusion process. Kielian et al.116,159,206 demonstrated a strict dependence on cholesterol for fusion. This requirement has been narrowed down to the sterol 3b hydroxyl group in cholesterol.31,176,290,312 A mutant was isolated (named srf-3 for sterol requirement in function) that was cholesterol independent and had an amino acid substitution of a proline to serine at E1 residue 226. This mutation did not affect fusion with normal membranes but significantly enhanced fusion to cholesterol-free membranes.24,280 In mosquito cells, the srf-3 mutant grew better than wild-type virus.5 The importance of this region of E1, which lies in the ij loop, for alphavirus fusion was further demonstrated by mutation of a conserved histidine at position 230 to alanine with the resulting virus particles being noninfectious, but capable of proceeding through E1 homotrimer formation under low pH conditions and suggesting a late step in fusion.23 However, the exact role of cholesterol to promote fusion is not known. The structure of the postfusion form of E1 has been determined using x-ray crystallography.74,75,76 This was accomplished using a soluble form of the E1 ectodomain known as E1* following exposure to low pH in the presence of liposomes, and solubilization with detergents. The structure of this homotrimer reveals the movement of domain III by 37 Å toward the fusion peptide and target membrane (Fig. 22.6)76 and is similar to changes that are induced by class I fusion protein activation.115 It is important to note that the addition of exogenous domain III can inhibit fusion by binding to E1 and preventing fold-back of the endogenous domain III.146 This fold-back process appears to be reversible and is mediated by a series of interactions between the domain I and domain III linker region.320 A complete picture of the virion during the fusion process is not available, despite several attempts using cryo-EM to examine these steps.46,62,84,199
Following the fusion of the viral and cellular membranes, the nucleocapsid core is released into the cytoplasm. The stability of that core is not known, but uncoating has been suggested to require the interaction of the core with ribosomes. This is based on several reports that suggest that ribosomal RNA competes for a site on the capsid protein and that displacement of this site results in disassembly.253,254,305 This site has been identified in SINV capsid protein between residues 94 and 105 and coincides with a predicted site for genome RNA binding.308 It has also been suggested that the core might be primed for uncoating by exposure to low pH, which causes the core to become unstable.303,306 This exposure to low pH could occur in the endosome because both E1 and 6K have been proposed to have ion channel properties.158,169,304 However, studies using pre-formed cores microinjected into naïve cells demonstrated that these cores do uncoat presumably devoid of conditions of low pH.258 Whether these cores interact with ribosomes to promote disassembly and release of genome RNA remains to be shown.
Translation and the Role of Viral-Encoded Replication Proteins
The genome RNA serves as an mRNA for the synthesis of the nonstructural or replication proteins (Fig. 22.4). These are produced by two polyproteins that originate translation at nucleotide 60 in SINV.268 The smaller but more abundant polyprotein P123 terminates translation at an opal codon following 1,897 amino acids. Readthrough of the opal codon occurs with a low frequency (∼10% to 20%) and results in the production of the larger P1234 polyprotein. Not all alphaviruses have a termination codon to control the production of the two polyproteins, but mutagenesis of the opal codon in SINV adversely affects replication.144 Processing of the polyproteins occurs through the action of a virus-encoded protease located within the nonstructural protein 2 (nsP2).39,91,92 The processing of the polyprotein to generate precursor and end-product nsP’s is believed to regulate the synthesis of viral RNAs.35,137,250 Translation of the structural polyprotein proceeds through a subgenomic mRNA that is initiated near the coding region for the C-terminus of P1234. The subgenomic mRNA is 3′ co-terminal with the genome RNA and is produced later in the infection.268
Initial studies of the replication proteins used temperature-sensitive mutants that were conditional lethal for viral
replication. These studies established complementation groups and identified specific functions of the replication proteins.264 Four complementation groups were identified, and these correlated with the four nsP’s. Once sequence information was available, motifs were identified that verified function and permitted phylogenetic relationships between other virus families to be established. This resulted in the suggestion of an alphavirus-like superfamily that contained RNA virus members from several plant families and argued for an evolutionary relationship between them.2,267
replication. These studies established complementation groups and identified specific functions of the replication proteins.264 Four complementation groups were identified, and these correlated with the four nsP’s. Once sequence information was available, motifs were identified that verified function and permitted phylogenetic relationships between other virus families to be established. This resulted in the suggestion of an alphavirus-like superfamily that contained RNA virus members from several plant families and argued for an evolutionary relationship between them.2,267
Table 22.1 Translation Products of Alphaviruses (Sindbis Virus) | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree