2
Structure of Bacterial Cells
CHAPTER CONTENTS
SHAPE & SIZE OF BACTERIA
Bacteria are classified by shape into three basic groups: cocci, bacilli, and spirochetes (Figure 2–1). The cocci are round, the bacilli are rods, and the spirochetes are spiral-shaped. Some bacteria are variable in shape and are said to be pleomorphic (many-shaped). The shape of a bacterium is determined by its rigid cell wall. The microscopic appearance of a bacterium is one of the most important criteria used in its identification.
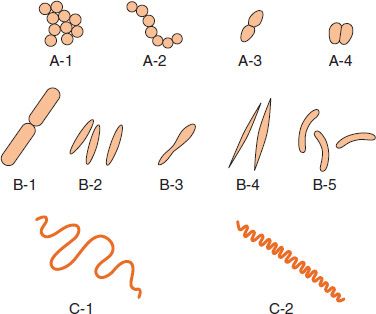
FIGURE 2–1 Bacterial morphology. A: Cocci in clusters (e.g., Staphylococcus; A-1); chains (e.g., Streptococcus; A-2); in pairs with pointed ends (e.g., Streptococcus pneumoniae; A-3); in pairs with kidney bean shape (e.g., Neisseria; A-4). B: Rods (bacilli): with square ends (e.g., Bacillus; B-1); with rounded ends (e.g., Salmonella; B-2); club-shaped (e.g., Corynebacterium; B-3); fusiform (e.g., Fusobacterium; B-4); comma-shaped (e.g., Vibrio; B-5). C: Spirochetes: relaxed coil (e.g., Borrelia; C-1); tightly coiled (e.g., Treponema; C-2). (Modified and reproduced with permission from Joklik WK et al. Zinsser Microbiology. 20th ed. Originally published by Appleton & Lange. Copyright 1992 by McGraw-Hill.)
In addition to their characteristic shapes, the arrangement of bacteria is important. For example, certain cocci occur in pairs (diplococci), some in chains (streptococci), and others in grapelike clusters (staphylococci). These arrangements are determined by the orientation and degree of attachment of the bacteria at the time of cell division. The arrangement of rods and spirochetes is medically less important and is not described in this introductory chapter.
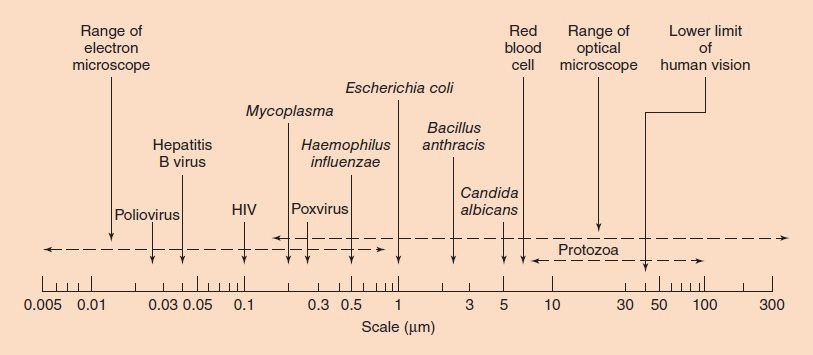
Bacteria range in size from about 0.2 to 5 μm (Figure 2–2). The smallest bacteria (Mycoplasma) are about the same size as the largest viruses (poxviruses) and are the smallest organisms capable of existing outside a host. The longest bacteria rods are the size of some yeasts and human red blood cells (7 μm).
FIGURE 2–2 Sizes of representative bacteria, viruses, yeasts, protozoa, and human red cells. The bacteria range in size from Mycoplasma, the smallest, to Bacillus anthracis, one of the largest. The viruses range from poliovirus, one of the smallest, to poxviruses, the largest. Yeasts, such as Candida albicans, are generally larger than bacteria. Protozoa have many different forms and a broad size range. HIV, human immunodeficiency virus. (Modified and reproduced with permission from Joklik WK et al. Zinsser Microbiology. 20th ed. Originally published by Appleton & Lange. Copyright 1992 McGraw-Hill.)
STRUCTURE OF BACTERIA
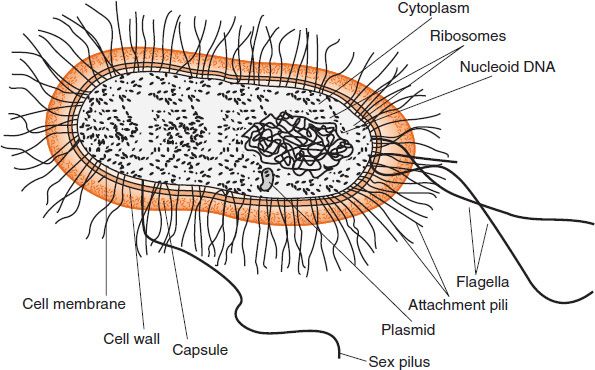
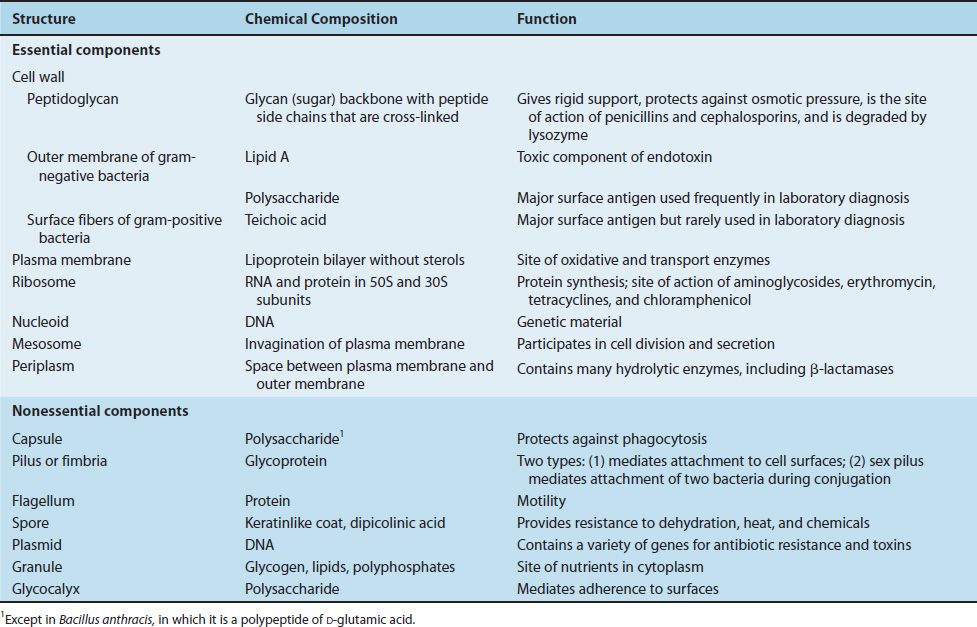
The structure of a typical bacterium is illustrated in Figure 2–3, and the important features of each component are presented in Table 2–1.
FIGURE 2–3 Bacterial structure. (Modified with permission from Ryan K et al. Sherris Medical Microbiology. 4th ed. Copyright 2004 McGraw-Hill.)
TABLE 2–1 Bacterial Structures
Cell Wall
The cell wall is the outermost component common to all bacteria (except Mycoplasma species, which are bounded by a cell membrane, not a cell wall). Some bacteria have surface features external to the cell wall, such as a capsule, flagella, and pili, which are less common components and are discussed next.
The cell wall is located external to the cytoplasmic membrane and is composed of peptidoglycan (see page 6). The peptidoglycan provides structural support and maintains the characteristic shape of the cell.
Cell Walls of Gram-Positive and Gram-Negative Bacteria
The structure, chemical composition, and thickness of the cell wall differ in gram-positive and gram-negative bacteria (Table 2–2, Figure 2–4, and “Gram Stain” box).
TABLE 2–2 Comparison of Cell Walls of Gram-Positive and Gram-Negative Bacteria

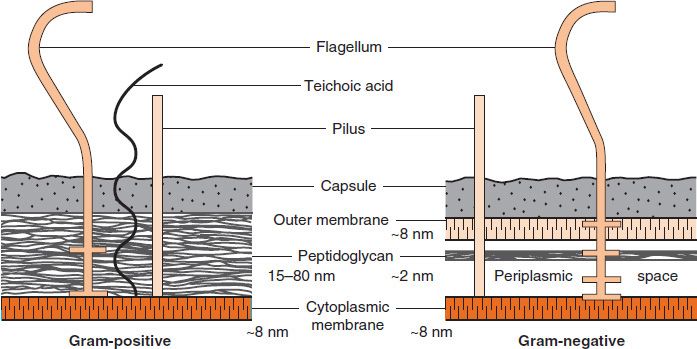
FIGURE 2–4 Cell walls of gram-positive and gram-negative bacteria. Note that the peptidoglycan in gram-positive bacteria is much thicker than in gram-negative bacteria. Note also that only gram-negative bacteria have an outer membrane containing endotoxin (lipopolysaccharide [LPS]) and have a periplasmic space where β-lactamases are found. Several important gram-positive bacteria, such as staphylococci and streptococci, have teichoic acids. (Reproduced with permission from Ingraham JL, Maaløe O, Neidhardt FC. Growth of the Bacterial Cell. Sinauer Associates; 1983.)
(1) The peptidoglycan layer is much thicker in gram-positive than in gram-negative bacteria. Many gram-positive bacteria also have fibers of teichoic acid, which protrude outside the peptidoglycan, whereas gram-negative bacteria do not have teichoic acids.
(2) In contrast, the gram-negative bacteria have a complex outer layer consisting of lipopolysaccharide, lipoprotein, and phospholipid. Lying between the outer-membrane layer and the cytoplasmic membrane in gram-negative bacteria is the periplasmic space, which is the site, in some species, of enzymes called β-lactamases that degrade penicillins and other β-lactam drugs.
The cell wall has several other important properties:
(1) In gram-negative bacteria, it contains endotoxin, a lipopolysaccharide (see pages 8 and 44).
(2) Its polysaccharides and proteins are antigens that are useful in laboratory identification.
(3) Its porin proteins play a role in facilitating the passage of small, hydrophilic molecules into the cell. Porin proteins in the outer membrane of gram-negative bacteria act as a channel to allow the entry of essential substances such as sugars, amino acids, vitamins, and metals as well as many antimicrobial drugs such as penicillins.
Cell Walls of Acid-Fast Bacteria
Mycobacteria (e.g., Mycobacterium tuberculosis) have an unusual cell wall, resulting in their inability to be Gram-stained. These bacteria are said to be acid-fast because they resist decolorization with acid–alcohol after being stained with carbolfuchsin. This property is related to the high concentration of lipids, called mycolic acids, in the cell wall of mycobacteria.
In view of their importance, three components of the cell wall (i.e., peptidoglycan, lipopolysaccharide, and teichoic acid) are discussed in detail here.
Peptidoglycan
Peptidoglycan is a complex, interwoven network that surrounds the entire cell and is composed of a single covalently linked macromolecule. It is found only in bacterial cell walls. It provides rigid support for the cell, is important in maintaining the characteristic shape of the cell, and allows the cell to withstand media of low osmotic pressure, such as water. A representative segment of the peptidoglycan layer is shown in Figure 2–5. The term peptidoglycan is derived from the peptides and the sugars (glycan) that make up the molecule. Synonyms for peptidoglycan are murein and mucopeptide.
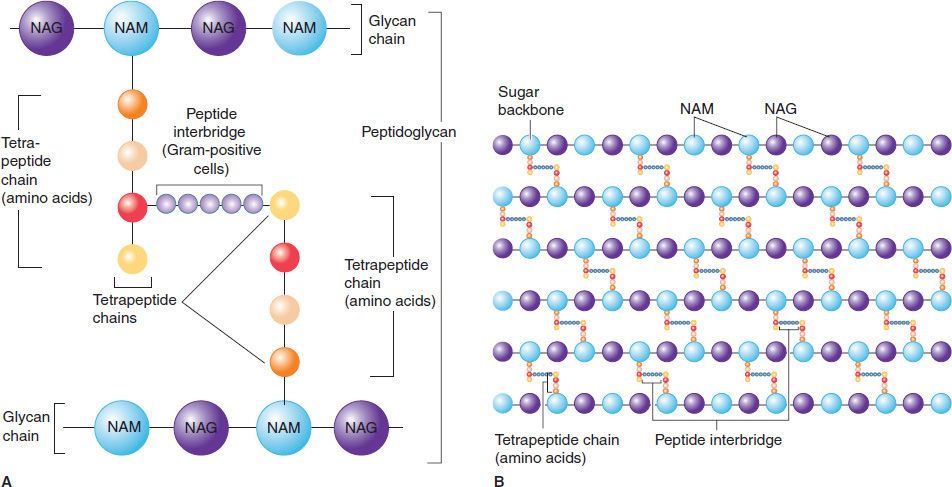
FIGURE 2–5 Peptidoglycan structure. A: Peptidoglycan is composed of a glycan chain (NAM and NAG), a tetrapeptide chain, and a cross-link (peptide interbridge). B: In the cell wall, the peptidoglycan forms a multilayered, three-dimensional structure. NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid. (Modified and reproduced with permission from Nester EW et al. Microbiology: A Human Perspective. 6th ed. Copyright 2009, McGraw-Hill.)
Figure 2–5 illustrates the carbohydrate backbone, which is composed of alternating N-acetylmuramic acid and N-acetylglucosamine molecules. Attached to each of the muramic acid molecules is a tetrapeptide consisting of both D– and L-amino acids, the precise composition of which differs from one bacterium to another. Two of these amino acids are worthy of special mention: diaminopimelic acid, which is unique to bacterial cell walls, and D-alanine, which is involved in the cross-links between the tetrapeptides and in the action of penicillin. Note that this tetrapeptide contains the rare D-isomers of amino acids; most proteins contain the L-isomer. The other important component in this network is the peptide cross-link between the two tetrapeptides. The cross-links vary among species; in Staphylococcus aureus, for example, five glycines link the terminal D-alanine to the penultimate L-lysine.
Because peptidoglycan is present in bacteria but not in human cells, it is a good target for antibacterial drugs. Several of these drugs, such as penicillins, cephalosporins, and vancomycin, inhibit the synthesis of peptidoglycan by inhibiting the transpeptidase that makes the cross-links between the two adjacent tetrapeptides (see Chapter 10).
GRAM STAIN
Lysozyme, an enzyme present in human tears, mucus, and saliva, can cleave the peptidoglycan backbone by breaking its glycosyl bonds, thereby contributing to the natural resistance of the host to microbial infection. Lysozyme-treated bacteria may swell and rupture as a result of the entry of water into the cells, which have a high internal osmotic pressure. However, if the lysozyme-treated cells are in a solution with the same osmotic pressure as that of the bacterial interior, they will survive as spherical forms, called protoplasts, surrounded only by a cytoplasmic membrane.
Lipopolysaccharide
The lipopolysaccharide (LPS) of the outer membrane of the cell wall of gram-negative bacteria is endotoxin. It is responsible for many of the features of disease, such as fever and shock (especially hypotension), caused by these organisms (see page 44). It is called endotoxin because it is an integral part of the cell wall, in contrast to exotoxins, which are actively secreted from the bacteria. The constellation of symptoms caused by the endotoxin of one gram-negative bacteria is similar to another, but the severity of the symptoms can differ greatly. In contrast, the symptoms caused by exotoxins of different bacteria are usually quite different.
The LPS is composed of three distinct units (Figure 2–6):
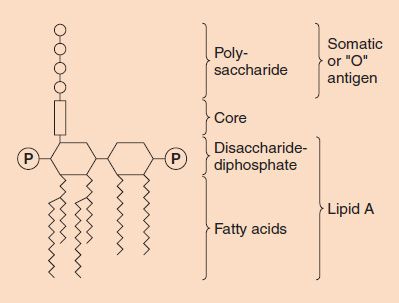
FIGURE 2–6 Endotoxin (lipopolysaccharide [LPS]) structure. The O-antigen polysaccharide is exposed on the exterior of the cell, whereas the lipid A faces the interior. (Modified and reproduced with permission from Brooks GF et al. Medical Microbiology. 19th ed. Originally published by Appleton & Lange. Copyright 1991 McGraw-Hill.)
(1) A phospholipid called lipid A, which is responsible for the toxic effects.
(2) A core polysaccharide of five sugars linked through ketodeoxyoctulonate (KDO) to lipid A.
(3) An outer polysaccharide consisting of up to 25 repeating units of three to five sugars. This outer polymer is the important somatic, or O, antigen of several gram-negative bacteria that is used to identify certain organisms in the clinical laboratory. Some bacteria, notably members of the genus Neisseria, have an outer lipooligosaccharide (LOS) containing very few repeating units of sugars.
Teichoic Acid
Teichoic acids are fibers located in the outer layer of the gram-positive cell wall and extend from it. They are composed of polymers of either glycerol phosphate or ribitol phosphate. Some polymers of glycerol teichoic acid penetrate the peptidoglycan layer and are covalently linked to the lipid in the cytoplasmic membrane, in which case they are called lipoteichoic acid; others anchor to the muramic acid of the peptidoglycan.
The medical importance of teichoic acids lies in their ability to induce septic shock when caused by certain gram-positive bacteria; that is, they activate the same pathways as does endotoxin (LPS) in gram-negative bacteria. Teichoic acids also mediate the attachment of staphylococci to mucosal cells. Gram-negative bacteria do not have teichoic acids.
Cytoplasmic Membrane
Just inside the peptidoglycan layer of the cell wall lies the cytoplasmic membrane, which is composed of a phospholipid bilayer similar in microscopic appearance to that in eukaryotic cells. They are chemically similar, but eukaryotic membranes contain sterols, whereas prokaryotes generally do not. The only prokaryotes that have sterols in their membranes are members of the genus Mycoplasma. The membrane has four important functions: (1) active transport of molecules into the cell, (2) energy generation by oxidative phosphorylation, (3) synthesis of precursors of the cell wall, and (4) secretion of enzymes and toxins.
Cytoplasm
The cytoplasm has two distinct areas when seen in the electron microscope:
(1) An amorphous matrix that contains ribosomes, nutrient granules, metabolites, and plasmids.
(2) An inner, nucleoid region composed of DNA.
Ribosomes
Bacterial ribosomes are the site of protein synthesis as in eukaryotic cells, but they differ from eukaryotic ribosomes in size and chemical composition. Bacterial ribosomes are 70S in size, with 50S and 30S subunits, whereas eukaryotic ribosomes are 80S in size, with 60S and 40S subunits. The differences in both the ribosomal RNAs and proteins constitute the basis of the selective action of several antibiotics that inhibit bacterial, but not human, protein synthesis (see Chapter 10).
Granules
The cytoplasm contains several different types of granules that serve as storage areas for nutrients and stain characteristically with certain dyes. For example, volutin is a reserve of high energy stored in the form of polymerized metaphosphate. It appears as a “metachromatic” granule since it stains red with methylene blue dye instead of blue as one would expect. Metachromatic granules are a characteristic feature of Corynebacterium diphtheriae, the cause of diphtheria.
Nucleoid
The nucleoid is the area of the cytoplasm in which DNA is located. The DNA of prokaryotes is a single, circular molecule that has a molecular weight (MW) of approximately 2 × 109 and contains about 2000 genes. (By contrast, human DNA has approximately 100,000 genes.) Because the nucleoid contains no nuclear membrane, no nucleolus, no mitotic spindle, and no histones, there is little resemblance to the eukaryotic nucleus. One major difference between bacterial DNA and eukaryotic DNA is that bacterial DNA has no introns, whereas eukaryotic DNA does.
Plasmids
Plasmids are extrachromosomal, double-stranded, circular DNA molecules that are capable of replicating independently of the bacterial chromosome. Although plasmids are usually extrachromosomal, they can be integrated into the bacterial chromosome. Plasmids occur in both gram-positive and gram-negative bacteria, and several different types of plasmids can exist in one cell:
(1) Transmissible plasmids can be transferred from cell to cell by conjugation (see Chapter 4 for a discussion of conjugation). They are large (MW 40–100 million), since they contain about a dozen genes responsible for synthesis of the sex pilus and for the enzymes required for transfer. They are usually present in a few (1–3) copies per cell.
(2) Nontransmissible plasmids are small (MW 3–20 million), since they do not contain the transfer genes; they are frequently present in many (10–60) copies per cell.
Plasmids carry the genes for the following functions and structures of medical importance:
(1) Antibiotic resistance, which is mediated by a variety of enzymes.
(2) Resistance to heavy metals, such as mercury, the active component of some antiseptics (e.g., merthiolate and mercurochrome), and silver, which is mediated by a reductase enzyme.
(3) Resistance to ultraviolet light, which is mediated by DNA repair enzymes.
(4) Pili (fimbriae), which mediate the adherence of bacteria to epithelial cells.
(5) Exotoxins, including several enterotoxins.
Other plasmid-encoded products of interest are as follows:
(1) Bacteriocins are toxic proteins produced by certain bacteria that are lethal for other bacteria. Two common mechanisms of action of bacteriocins are (i) degradation of bacterial cell membranes by producing pores in the membrane and (ii) degradation of bacterial DNA by DNAse. Examples of bacteriocins produced by medically important bacteria are colicins made by Escherichia coli and pyocins made by Pseudomonas aeruginosa. Bacteria that produce bacteriocins have a selective advantage in the competition for food sources over those that do not. However, the medical importance of bacteriocins is that they may be useful in treating infections caused by antibiotic-resistant bacteria.
(2) Nitrogen fixation enzymes in Rhizobium in the root nodules of legumes.
(3) Tumors caused by Agrobacterium in plants.
(4) Several antibiotics produced by Streptomyces.
(5) A variety of degradative enzymes that are produced by Pseudomonas and are capable of cleaning up environmental hazards such as oil spills and toxic chemical waste sites.
Transposons
Transposons are pieces of DNA that move readily from one site to another either within or between the DNAs of bacteria, plasmids, and bacteriophages. Because of their unusual ability to move, they are nicknamed “jumping genes.” Some transposons move by replicating their DNA and inserting the new copy into another site (replicative transposition), whereas others are excised from the site without replicating and then inserted into the new site (direct transposition). Transposons can code for drug-resistant enzymes, toxins, or a variety of metabolic enzymes and can either cause mutations in the gene into which they insert or alter the expression of nearby genes.
Transposons typically have four identifiable domains. On each end is a short DNA sequence of inverted repeats, which are involved in the integration of the transposon into the recipient DNA. The second domain is the gene for the transposase, which is the enzyme that mediates the excision and integration processes. The third region is the gene for the repressor that regulates the synthesis of both the transposase and the gene product of the fourth domain, which, in many cases, is an enzyme mediating antibiotic resistance (Figure 2–7).
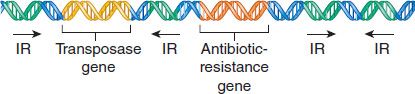
FIGURE 2–7 Transposon genes. This transposon is carrying a drug-resistance gene. IR, inverted repeat. (Modified and reproduced with permission from Willey JM et al. Prescott’s Principles of Microbiology. New York: McGraw-Hill, 2009.)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree