Rotaviruses
Mary K. Estes
Harry B. Greenberg
Introduction and History
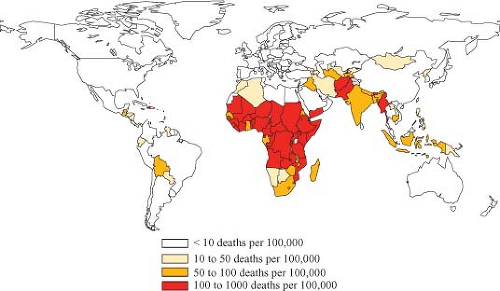
Rotaviruses are the single most important cause of severe diarrheal illness in infants and young children in both developed and developing countries worldwide, accounting for 30% to 50% of these illnesses.45,562,563,564 Regardless of cause, diarrheal diseases (a) are one of the most common illnesses in this age group throughout the world; (b) are one of the six leading causes of 10.6 million deaths that occur annually in children younger than 5 years of age; and (c) account for 18% (i.e., greater than 2 million) of the 10.6 million deaths,89 with the greatest toll being in developing countries.89,788
Viruses were first discovered to be significant causes of diarrheal illness in the 1970s, with Norwalk virus being the first agent discovered in 1972 by Kapikian et al400 from an outbreak of gastroenteritis in a school in Norwalk, Ohio. The Norwalk and related viruses, now known as noroviruses, are members of the genus Norovirus of the family Caliciviridae (see Chapter 20). Human rotaviruses were discovered in 1973, when particles were visualized by Bishop et al59,60 in electron micrographs of thin sections of duodenal mucosa and later virus was identified in feces by electron microscopy (EM).58,246,398,505 Both of these fastidious gastroenteritis viruses were discovered without the benefit of tissue culture technology; their identification relied on direct visualization by EM. The 70-nm particles (Fig. 45.1)388,398 from children’s feces were subsequently designated rotavirus247 and documented to be an important etiologic agent of severe diarrhea of infants and young children during the first 2 years of life56,391 in both developed and developing countries (Fig. 45.2); rotaviruses consistently outranked in importance other known etiologic agents of severe diarrhea.390
Although the human rotaviruses were discovered in 1973, several animal viruses were described during the previous 10 years that were later found to be rotaviruses based on exhibiting characteristic rotavirus morphology and sharing a group antigen with human rotaviruses.247,393 These animal agents included (a) the epizootic diarrhea of infant mice (EDIM) virus seen by Adams and Kraft,4 using thin-section EM in intestinal tissue of mice infected with EDIM virus; (b) a 70-nm simian agent 11 (SA11) that was cultivated in vervet monkey kidney cells from a rectal swab obtained from a healthy vervet monkey470; (c) the O (offal) agent isolated in vervet monkey kidney cell culture from the mixed washings of intestines of cattle and sheep471; and (d) 70-nm virus particles in stools from calves with a diarrheal illness that could be passaged serially in calves and produce disease,498 were also cultivated in primary fetal bovine cell cultures, and were named the Nebraska calf diarrhea virus (NCDV).499 Thus, rotaviruses were excreted in stools of many species and frequently associated with diarrheal disease.
Classification
Rotaviruses are members of the genus Rotavirus within the family Reoviridae, and rotaviruses share common morphologic
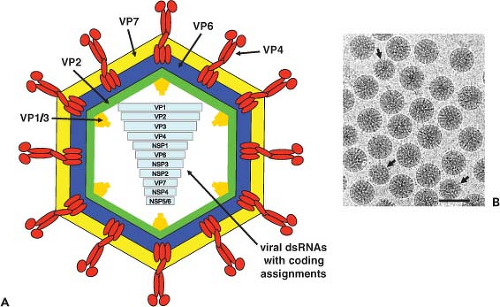
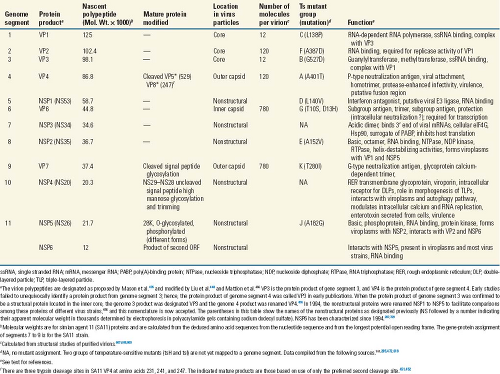
and biochemical properties (Table 45.1). Early studies using negative-stain EM techniques underestimated the particle diameter, and the subsequent cryo-EM studies, in which no stains are used, established the particle diameter to be 100 nm including the spikes. Salient features are that (a) mature virus particles, including spikes, are about 100 nm (1,000 Å) in diameter and possess a triple-layered icosahedral protein capsid composed of an outer layer, an intermediate layer, and an inner core layer; (b) 60 protein spikes extend from the smooth surface of the outer shell; (c) outer capsid integrity requires calcium; (d) particles contain an RNA-dependent RNA polymerase and other enzymes capable of producing capped RNA transcripts; (e) the virus genome contains 11 segments of double-stranded RNA (dsRNA); (f) rotaviruses of the same group (see later) are capable of genetic reassortment; (g) virus replication occurs in the cytoplasm of infected cells; (h) virus cultivation in vitro is facilitated by treatment with proteolytic enzymes enhancing viral infectivity by cleavage of the outer capsid spike polypeptide; and (i) the viruses exhibit a unique morphogenic pathway (transiently enveloped virus particles are formed by budding into the endoplasmic reticulum [ER]) during morphogenesis. Mature particles are nonenveloped,
and virions are liberated from infected cells by cell lysis or by a nonclassic vesicular transport in polarized epithelial cells.
and biochemical properties (Table 45.1). Early studies using negative-stain EM techniques underestimated the particle diameter, and the subsequent cryo-EM studies, in which no stains are used, established the particle diameter to be 100 nm including the spikes. Salient features are that (a) mature virus particles, including spikes, are about 100 nm (1,000 Å) in diameter and possess a triple-layered icosahedral protein capsid composed of an outer layer, an intermediate layer, and an inner core layer; (b) 60 protein spikes extend from the smooth surface of the outer shell; (c) outer capsid integrity requires calcium; (d) particles contain an RNA-dependent RNA polymerase and other enzymes capable of producing capped RNA transcripts; (e) the virus genome contains 11 segments of double-stranded RNA (dsRNA); (f) rotaviruses of the same group (see later) are capable of genetic reassortment; (g) virus replication occurs in the cytoplasm of infected cells; (h) virus cultivation in vitro is facilitated by treatment with proteolytic enzymes enhancing viral infectivity by cleavage of the outer capsid spike polypeptide; and (i) the viruses exhibit a unique morphogenic pathway (transiently enveloped virus particles are formed by budding into the endoplasmic reticulum [ER]) during morphogenesis. Mature particles are nonenveloped,
and virions are liberated from infected cells by cell lysis or by a nonclassic vesicular transport in polarized epithelial cells.
Rotaviruses are classified serologically by a scheme that allows for the presence of multiple groups (serogroups, based on VP6 reactivity) and of multiple serotypes within each group (based on VP4 and VP7 neutralizing epitopes). Rotaviruses are composed of seven distinct groups (A to G, now designated RVA, RVB, RVC, etc.). RVA, RVB, and RVC strains are found in both humans and animals, whereas rotaviruses of groups D, E, F, and G have been found only in animals to date. Viruses within each group are capable of genetic reassortment, but reassortment does not occur among viruses in different groups, and thus RV groups are considered unique species.806 A rotavirus group includes viruses that share cross-reacting antigens detectable by a number of serologic methods, such as immunofluorescence, enzyme-linked immunosorbent assay (ELISA), and immune electron microscopy (IEM). Cross-reactive epitopes on the inner capsid protein (VP6) are those usually detected by diagnostic ELISA, primarily because this protein is highly antigenic and it represents the largest mass of the particle. However, common antigens are found on most (if not all) of the structural proteins and probably on many of the nonstructural proteins as well. This is documented by observing that monospecific antisera and some monoclonal antibodies (mAbs) specific for individual polypeptides cross-react with strains other than those to which they were made.
Table 45.1 General Characteristics of Rotaviruses | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
RVAs cause significant diarrheal disease in infants and in the young of various mammalian and avian species. RVBs have been associated with epidemics of severe diarrhea primarily in adults in Asia.355,655,700 RVCs have been sporadically reported in fecal specimens from children with diarrhea and in several family outbreaks.557 Rapid diagnostic tests (ELISA), mAbs, and polymerase chain reaction (PCR) assays to detect non–group A rotaviruses are available mainly in research laboratories, and these facilitate determining the clinical importance of these viruses.534,807 Very few non–group A rotavirus strains have been successfully propagated in cell culture. The inability to grow most non–group A viruses has hampered obtaining detailed information on these viruses, although gene-coding assignments and sequence data are available. Unless noted otherwise, this chapter focuses on information about the RVA viruses. Reviews on the non–group A rotaviruses and comparisons between the proteins of the group A and non–group A viruses have been published elsewhere.80,374,486,655
Within RVA, viruses are classified into serotypes defined by reactivity in plaque reduction (or fluorescent foci reduction) neutralization assays using hyperimmune serum prepared in antibody-negative animals. With such assays, 27 VP7 (or G [for glycoprotein]) serotypes have been identified (Table 45.2), and strains of animal and human origin may fall within the same G serotype. Neutralization assays can measure reactivity of antibody against the two outer capsid neutralizing antigens (VP7 and VP4, Tables 45.2 and 45.3). In most cases, however, the predominant reactivity measured when using hyperimmune antisera is against the glycoprotein VP7. This may be because VP7 makes up a greater percentage of the virion outer capsid than VP4 does, or alternatively, with hyperimmunization, VP7 selectively induces highly specific antibodies. The protein specificity of neutralizing antibodies after primary and secondary infection is less well defined. Identical classification of the same virus isolates using mAbs to VP7 unequivocally demonstrates that plaque-reduction neutralization assays with hyperimmune serum primarily measure reactivities with VP7.54,155,708
In some cases, a rotavirus strain will not react clearly in reciprocal neutralization assays with hyperimmune antiserum. This is usually because the two viruses being compared possess distinct immunologic forms of VP4 (the second outer capsid protein), which is also a neutralization antigen (Table 45.3). Many mAbs to VP4 possess neutralization activity. Because the genes encoding these two distinct neutralization antigens can segregate (reassort) independently, it is not surprising that some virus isolates possess heterologous neutralization (VP4, VP7) antigens.249 Rotaviruses are classified by a binary system (similar to that used for influenza viruses) in which distinct types of VP4 and VP7 are recognized.296,345,634 A lack of readily available typing serum or mAbs to different VP4 types, however, has hampered classification of VP4 (or P [for protease-sensitive protein]) serotypes. Instead, properties of VP4 have been studied primarily by sequence analysis, and current evidence indicates the existence of at least 35 different genotypes of VP4 (Table 45.3). Genotypes of VP4 and VP7 are determined by sequence analysis, whereas serotypes are determined by reactivity of individual strains or selected reassortants with polyclonal or monoclonal antisera.216,343 For VP7, a correlation between genotype and serotype has been established. Such a correlation is much less clear for VP4, although a variable region on VP8* that spans amino acid (aa) 71 to 204 can define P type–specific epitopes.289 Serotype designation, thus, reflects the expression of neutralization epitopes on both VP4 and VP7. The serology of the epitopes in proteins that interact in the capsid is complicated but is beginning to be understood with the availability of high-resolution structural data on these outer capsid proteins.
In 2008, a comprehensive nucleotide sequence–based, complete genome classification system was developed for
RVAs.483 This system assigns a specific genotype to each of the 11 RV genome segments according to established nucleotide percent cutoff values. The VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 genes of RV strains are described using the abbreviations Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx (x = Arabic numbers starting from 1), respectively (Table 45.4). A Rotavirus Classification Working Group that includes researchers worldwide maintains and evaluates this system. Recent updates are published and new guidelines recommend uniform nomenclature for individual strains to be RV group/species of origin/country of identification/common name/year of identification/G and P type.482 The prototype simian agent 11 is designated RVA/Simian-tc/ZAF/SA11-H96/1958/G3P5B[2] and the full descriptor of genes is indicated by G3-P[2]-I2-R2-C5-M5-A5-N5-T5-E2-H5 using this new system.
RVAs.483 This system assigns a specific genotype to each of the 11 RV genome segments according to established nucleotide percent cutoff values. The VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 genes of RV strains are described using the abbreviations Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx (x = Arabic numbers starting from 1), respectively (Table 45.4). A Rotavirus Classification Working Group that includes researchers worldwide maintains and evaluates this system. Recent updates are published and new guidelines recommend uniform nomenclature for individual strains to be RV group/species of origin/country of identification/common name/year of identification/G and P type.482 The prototype simian agent 11 is designated RVA/Simian-tc/ZAF/SA11-H96/1958/G3P5B[2] and the full descriptor of genes is indicated by G3-P[2]-I2-R2-C5-M5-A5-N5-T5-E2-H5 using this new system.
Virion Structure
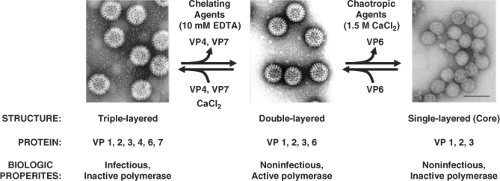
The morphologic appearance of rotavirus particles is distinctive, and three types of particles can be observed by EM (Fig. 45.3). The complete particles resemble a wheel with short spokes and a well-defined, smooth outer rim. The name rotavirus (from the Latin rota, meaning “wheel”) was coined based on this morphology.247 The complete infectious particles (virions) are also called triple-layered particles (TLPs). These particles are ∼100 nm in diameter, which is relatively large for a nonenveloped, icosahedral virus. Double-layered particles (DLPs) lacking the outer shell are described as rough particles because their periphery shows projecting trimeric subunits of the inner capsid. Single-layered particles (SLPs or cores) are seen infrequently; they usually lack genomic RNA and are aggregated.
Table 45.4 Nucleotide Percentage Identity Cut-off Values Defining Genotypes for 11 Rotavirus Gene Segments | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
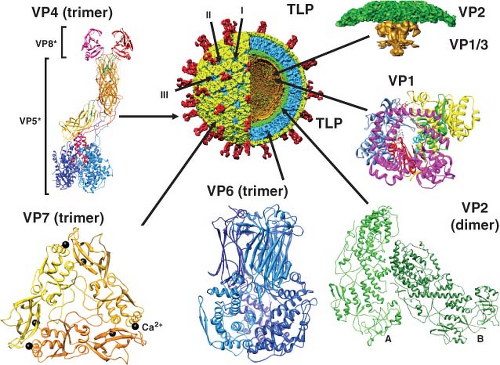
The structures of triple- and double-layered rotavirus particles are solved to near-atomic resolution based on x-ray crystallography and particle reconstructions of cryo-electron microscopy (cryo-EM) images, and these provide a detailed description of particles121,434,435,440,488,605,675,679,803,828 (Fig. 45.4). Particles possess icosahedral symmetry with a T = 13l (levo) icosahedral surface lattice for the two outer layers, while the innermost layer exhibits a unique T = 1 icosahedral organization. Distinguishing features of the TLP structure include 132 large aqueous channels and 60 spikes.
The channels span the two outer layers and link the outer surface with the inner core. Three types of channels are distinguishable based on their position and size. Twelve type I channels are located at the icosahedral fivefold axes, 60 type II channels are at each of the pentavalent positions surrounding the fivefold axes, and a second set of 60 type III channels are at the six-coordinated positions surrounding the icosahedral threefold axes. Type III channels are about 140 Å in depth and about 55 Å wide at the outer surface of the virus. On entering the particle, these channels constrict before widening to their
maximal width, which is close to the surface of the inner shell. Similar features and dimensions are seen in the other two types of channels, except that type I channels have a narrower (∼40 Å) opening at the outer surface of the virus. The type I channels are conduits for the export of messenger RNA (mRNA) that first interacts with the enzyme complexes composed of VP1, the RNA-dependent RNA polymerase, and VP3, the methyltransferase and guanylyltransferase, that are present at the inner surface of the fivefold axes of the SLP432,435,604 (see Fig. 45.4). Atomic structures of the rotavirus polymerase, alone and in complex with RNA, show VP1 is a compact, globular protein of ∼70 Å in diameter that has three domains: an N-terminal domain, a polymerase domain, and a C-terminal domain.460 The polymerase domain exhibits the right-handed architecture (fingers-palm-thumb) typical of polymerases in general as well as canonical motifs (A to F) involved in various aspects of phosphodiester bond formation461,546 (Fig. 45.4). The N-terminal and C-terminal domains envelop the polymerase domain, creating a cage-like enzyme with a hollow catalytic center. Four tunnels lead into the center, serving as conduits for the entry and exit of free nucleotides (nucleoside triphosphates [NTPs]), template RNAs, and RNA products. This solved structure represents a catalytically inactive form of the polymerase captured prior to the initiation of dsRNA synthesis. Models of how this enzyme functions in coordinated genome replication and packaging are discussed later.
maximal width, which is close to the surface of the inner shell. Similar features and dimensions are seen in the other two types of channels, except that type I channels have a narrower (∼40 Å) opening at the outer surface of the virus. The type I channels are conduits for the export of messenger RNA (mRNA) that first interacts with the enzyme complexes composed of VP1, the RNA-dependent RNA polymerase, and VP3, the methyltransferase and guanylyltransferase, that are present at the inner surface of the fivefold axes of the SLP432,435,604 (see Fig. 45.4). Atomic structures of the rotavirus polymerase, alone and in complex with RNA, show VP1 is a compact, globular protein of ∼70 Å in diameter that has three domains: an N-terminal domain, a polymerase domain, and a C-terminal domain.460 The polymerase domain exhibits the right-handed architecture (fingers-palm-thumb) typical of polymerases in general as well as canonical motifs (A to F) involved in various aspects of phosphodiester bond formation461,546 (Fig. 45.4). The N-terminal and C-terminal domains envelop the polymerase domain, creating a cage-like enzyme with a hollow catalytic center. Four tunnels lead into the center, serving as conduits for the entry and exit of free nucleotides (nucleoside triphosphates [NTPs]), template RNAs, and RNA products. This solved structure represents a catalytically inactive form of the polymerase captured prior to the initiation of dsRNA synthesis. Models of how this enzyme functions in coordinated genome replication and packaging are discussed later.
The single-layered particle exhibits a unique T = 1 symmetry and is composed of 120 molecules of VP2 arranged as 60 dimers that surround the genomic dsRNA that is highly ordered434,488,604 (Fig. 45.4). X-ray and cryo-EM structures of DLPs show that VP2 has two structural isoforms that interact extensively. One of the subunits in the asymmetric unit (VP2A) packs around the icosahedral fivefold axis forming a star-shaped complex with a small pore in the middle lined by conserved basic residues. The other subunit (VP2B) fills in space between the VP2A subunits forming a decameric cap structure at the fivefold axis. Twelve of these decameric complexes make up the VP2 layer that is 25 to 30 Å thick. Many segmented dsRNA viruses contain 120 molecules of a core protein (VP2 for rotavirus, VP3 for bluetongue virus, lambda 1 for reoviruses, the single capsid protein for cypovirus) that surrounds an ordered genome.307,337,825 Although the inner shell protein shares similar features among these dsRNA viruses, rotaviruses and orbiviruses are distinguished by housing their enzymatic functions entirely within the inner shell, leading them to be called nonturreted viruses, and nascent mRNA transcripts are released through channels penetrating the two capsid layers at the icosahedral vertices.337,433,434,536 This
capsid architecture contrasts with the structure of the turreted viruses, reoviruses,624,829 aquareovirus,827 and cypovirus,434,536,815 where the polymerase enzyme is housed within the core, but the capping enzymes are incorporated as pentameric turret-like projections that extend through the inner capsid layer at each icosahedral vertex.337
capsid architecture contrasts with the structure of the turreted viruses, reoviruses,624,829 aquareovirus,827 and cypovirus,434,536,815 where the polymerase enzyme is housed within the core, but the capping enzymes are incorporated as pentameric turret-like projections that extend through the inner capsid layer at each icosahedral vertex.337
The VP2 layer is surrounded by 260 trimers of VP6 that form a T = 13 icosahedral lattice (Fig. 45.4). These trimers are located right below the VP7 trimers in the outer layer so that the channels are in register. The double-layered particle is about 705 Å in diameter, and the structure of the VP6 subunit has two domains with an overall structure similar to the VP7 of bluetongue virus307,478 (Fig. 45.4) and to the μ1 protein of orthoreovirus.442 The distal domain of rotavirus VP6 has an eight-stranded jelly-roll β-barrel fold that makes contacts with VP7 and VP4, whereas the proximal domain with a cluster of eight α-helices and a conformationally flexible loop structure in VP6 is involved in establishing optimal contacts with the underlying VP2 subunits.488,828 Interactions of VP6 with the VP7 layer at the top and the VP2 layer at the bottom are important in stabilizing the entire rotavirus capsid and integrating the two essential functions of particles: cell entry and endogenous transcription. Structural integrity of the DLP is an essential requirement for endogenous transcription that takes place within the confines of the DLP with capped transcripts exiting through the aqueous type I channels at the fivefold axes.434
Sixty trimeric spikes extend from the smooth surface of the outer shell (Fig. 45.4). These protein spikes are situated at an edge of the type II channels surrounding the fivefold icosahedral axes. The spikes are composed of the protein VP4, as initially shown by seeing that two Fab subunits of mAb to VP4 bind on the sides near the tips of the spikes.606 Subsequent cryo-EM studies,158,192,587,679 including the most recent study at near-atomic resolution,675 confirmed that VP4 is the spike protein and showed that the spike is multidomained with a unique trimeric organization that projects about 120 Å from the surface of the virus with a total radial length of 200 Å. Spikes with well-defined structural features, two distal globular domains, a central body with an approximate twofold symmetry, and a globular domain called the foot domain are only visible in rotavirus particles grown in the presence of trypsin that enhances virus yield.158,679,802 Proteolysis cleaves VP4 (88K) into VP8* (28 kD, aa 1 to 247) and VP5* (60 kD, aa 248 to 776), and the cleavage products remain noncovalently associated in the virion (Fig. 45.4 and Table 45.5).
Recent crystallographic structures of VP8* and portions of VP5* as well as cryo-EM analyses at about 4.3 Å resolution indicate that the overall conformation of each VP4 subunit is that of a high loop, with the N-terminal helical segment of VP8* anchored against the C-terminal domain of VP5* in the foot domain of the same polypeptide chain.192,194,675 Different conformations including dimers, trimers, and asymmetric domains make up the unique trimeric configuration of the cleaved VP4 spike. Two β-barrel domains of VP5*, at the central body of the spike, adopt a dimeric appearance above the capsid surface, while another VP5* β-barrel of the third VP4 molecule is positioned closer to the capsid surface interacting with the VP7 capsid layer; the globular domains of each of the VP5* polypeptides form a trimeric base anchored inside the type II channels between the VP7 and VP6 capsid layers.440,587,675,802 Two VP8* molecules are present at the top of two upright VP5* molecules and the third VP8* is disordered or may be removed by trypsin cleavage.675 Thus, VP4 subunits undergo extensive rearrangements that resemble conformational transitions of membrane fusion proteins of enveloped viruses during entry into cells.192
The spike is held in place by interactions with both VP6 and VP7. The spike extends inward about 80 Å where it inserts into the lattice of VP6 trimers at the type II channels that surround the icosahedral fivefold axes. This interaction may template trimerization of cytosolic VP4.675 The VP7 shell partly covers the base of the VP4 spike and appears to lock VP4 onto the virion within the type II channel.
The outer 35-Å-thick capsid layer of rotavirus is formed by 260 trimers of the glycoprotein VP7 (37 kD) (Fig. 45.4). VP7 is a calcium-binding protein262,648 and consists of two domains: domain I with a disulphide bridge exhibits a Rossmann fold and domain II with three disulfide bridges exhibits a jelly-roll β-sandwich fold. Two Ca2+ ions are bound at each subunit interface in the trimer.17 Three VP7 subunits interact with each other to form a plate-like trimer that sits on top of the VP6 trimers and the N-terminal arms of three VP7 subunits grip the underlying VP6 trimers and intrude into the VP4 foot cavity. These interactions imply that the VP4 spikes must first be attached to the DLPs prior to the addition of VP7 during virus assembly, and addition of VP7 results in a shift in the underlying VP6 trimers.675 This order of outer capsid assembly is supported by in vitro reconstitution studies where sequential addition of recombinant VP4 followed by VP7 onto DLPs can produce infectious virus.731
It remains to be determined whether the helical heptad repeat or the putative fusion domain in the rotavirus spike protein is important only for virus entry or also during viral morphogenesis in cells, a unique process that involves a budding of particles through the membrane of the ER (see later).
Genome Structure and Organization
The viral genome of 11 segments of dsRNA is contained within the virus core capsid. Deproteinized rotavirus genomic dsRNA is not infectious, reflecting that virus particles contain their own RNA-dependent RNA polymerase to transcribe the individual RNA segments into active mRNA. Hydrodynamic studies of the flexibility or “stiffness” of isolated rotavirus RNA segments in solution indicate that packaging of these RNA segments into the rotavirus capsid requires intimate protein–RNA interactions.387 The proteins directly responsible for segment packaging remain unknown; the structural proteins present in core particles (VP1, VP2, and VP3) are obvious candidates, but nonstructural proteins may play a role (see later). The genome RNA is highly ordered within the particle, with about 25% of the genome making up a dodecahedral structure, and VP2 interacts with the RNA. Several points of contact between the inwardly protruding portion of VP2 as well as VP1 and VP3 interact with the RNA surrounding each fivefold axis430,604 (Fig. 45.4), and VP2 interactions with the VP1 polymerase are required for replicase activity.492,573,824
The nucleotide sequence of all 11 rotavirus RNA segments for many rotavirus strains are known, and this forms the basis for the new classification system discussed earlier.483 The prototype simian SA11 strain was the first genome completely sequenced. The sequences from different rotavirus strains show
general features (Fig. 45.5) of the structure of each genome segment. Each positive-sense RNA segment starts with a 5′-guanidine followed by a set of conserved sequences that are part of the 5′ noncoding sequences. An open reading frame (ORF) coding for the protein product and ending with the stop codon follows, and then another set of noncoding sequences is found containing a subset of conserved terminal 3′ sequences and ending with two 3′ terminal cytidines. Almost all mRNAs end with the consensus sequence 5′-UGUGACC-3′, and these sequences contain important signals for gene expression and genome replication. The last four nucleotides of the mRNAs function as translation enhancers.126 The lengths of the 3′ and 5′ noncoding sequences vary for different genes, but the noncoding sequences of homologous strains are highly conserved. No polyadenylation signal is found at the 3′ end of the genes. All of the sequenced genes possess at least one long ORF after the first initiation codon. This is usually a strong initiation codon based on Kozak’s rules.425 Although some of the genes possess additional in-phase (genes 7, 9, and 10) or out-of-phase (gene 11) ORFs, current evidence indicates that all the genes are monocistronic, except gene 11.487
general features (Fig. 45.5) of the structure of each genome segment. Each positive-sense RNA segment starts with a 5′-guanidine followed by a set of conserved sequences that are part of the 5′ noncoding sequences. An open reading frame (ORF) coding for the protein product and ending with the stop codon follows, and then another set of noncoding sequences is found containing a subset of conserved terminal 3′ sequences and ending with two 3′ terminal cytidines. Almost all mRNAs end with the consensus sequence 5′-UGUGACC-3′, and these sequences contain important signals for gene expression and genome replication. The last four nucleotides of the mRNAs function as translation enhancers.126 The lengths of the 3′ and 5′ noncoding sequences vary for different genes, but the noncoding sequences of homologous strains are highly conserved. No polyadenylation signal is found at the 3′ end of the genes. All of the sequenced genes possess at least one long ORF after the first initiation codon. This is usually a strong initiation codon based on Kozak’s rules.425 Although some of the genes possess additional in-phase (genes 7, 9, and 10) or out-of-phase (gene 11) ORFs, current evidence indicates that all the genes are monocistronic, except gene 11.487
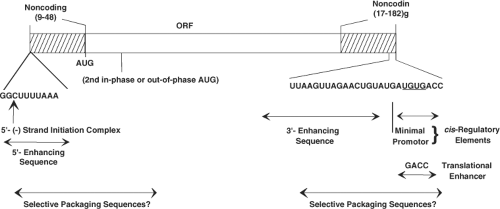 Figure 45.5. Major features of rotavirus gene structure. Schematic shows the overall structure of rotavirus genes derived from published sequences of genes 1 through 11. All 11 rotavirus genes lack a polyadenylation signal, are A + U rich, and contain conserved consensus sequences at their 5′ and 3′ ends. Variations in the conserved ends are also shown. The prototype simian agent 11 (SA11) genome segments range in size from 667 (segment 11) to 3302 (segment 1) with a total of 18,556 base pairs. The bottom arrows show cis-regulatory elements of rotavirus messenger RNA (mRNA) required for replication of transcripts assayed in a cell-free replication system.404,576,730,786 The study of viruses with variations in the sequence at the 3′ termini indicates the minimal promoter is URN0-5CC. The 5′ and 3′ noncoding regions at the termini of the mRNA are predicted to interact and stably base-pair to form a panhandle structure possibly stabilized by the viral polymerase,359,576,730 and interactions between the 3′ terminus with the nonstructural protein NSP3 may promote translation of viral mRNA.748 The penultimate 5′-GACC-3′ is a translation enhancer.126 |
The rotavirus gene sequences are A+U rich (58% to 67%), and this bias against CGN and NCC codons is shared with many eukaryotic and viral genes. The dsRNA segments are base-paired end to end, and the positive-sense strand contains a 5′ cap sequence m7GpppG(m)GC.359,580 Similar features of the RNA termini (capped structures and 5′ and 3′ conserved sequences) are found in the genomes of other segmented viruses (e.g., reovirus, cytoplasmic polyhedrosis virus, orbivirus) in the family Reoviridae and in other virus families with segmented RNA genomes (Orthomyxoviridae, Arenaviridae, and Bunyaviridae). One of the most intriguing aspects of the replication cycle of rotaviruses and all segmented dsRNA viruses relates to mechanism(s) of how these viruses coordinately replicate and package the 11 viral mRNAs. The 11 mRNAs must share common cis-acting signals because they are all replicated by the same polymerase, and the UGUG sequence of the consensus sequence is recognized in a base-specific manner by the polymerase.460,461 In addition, each mRNA must also contain a signal that is unique to it alone, because the 11 mRNAs must be distinguished from one another during packaging. Generally, the conserved terminal sequences in genome segments contain cis-acting signals that are important for transcription, RNA translation, RNA transport, replication, assembly, or encapsidation of the viral genome segments. Some of the cis-acting signals for rotavirus RNA replication and translation have been identified (Fig. 45.5), but assembly or encapsidation signals remain unknown.574 The highly conserved noncoding regions of the RNA may contain the gene-specific packaging signals. Sequence comparisons between RV strains have contributed to identifying conserved sequences and/or secondary structures in the RV genome.439 Conserved secondary structures in the positive-sense RNAs, including long-range interactions at the 5′ and 3′ terminal regions present in all segments, may facilitate RNA circularization, although such structures remain to be detected in cells.439 Computer modeling and RNAse mapping experiments predict that viral (+)RNAs fold into panhandles through 5′ and 3′ base pairing and the 3′ consensus sequence extends from the panhandle as a single-stranded tail.116,439,730 Interactions of this extended RNA with different rotavirus and cellular proteins are thought to regulate whether
the (+)RNA functions in translation, genome replication, or assortment and packaging.491
the (+)RNA functions in translation, genome replication, or assortment and packaging.491
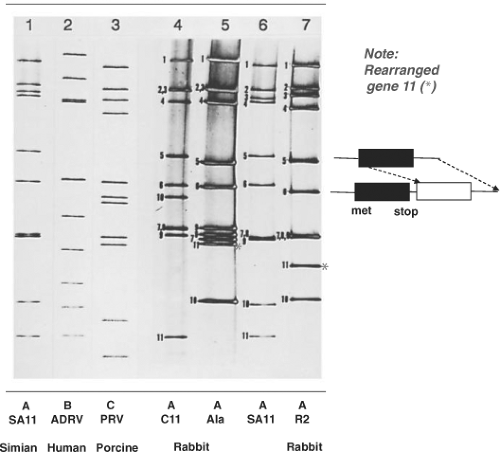 Figure 45.6. Electropherogram of rotavirus RNA segments. The RNA segments were separated by electrophoresis in a 10% polyacrylamide gel and visualized by staining with silver nitrate. The RNA patterns of a group A rotavirus (simian agent 11 [SA11], lane 1), a group B rotavirus (adult diarrhea rotavirus isolate from China, lane 2), and a group C rotavirus (lane 3) are shown. The rearranged RNA patterns of three group A rabbit rotavirus strains (C11, lane 4; Ala, lane 5; and R2, lane 7) and their cognate segments compared with those of SA11 (lane 6) are also shown. The cognate genes were identified by hybridization with complementary DNAs (cDNAs) for each SA11 RNA segment. Red asterisks show rearranged gene 11s in the electropherotypes of Ala and R2 viruses. Schematic on right illustrates features of a rearranged gene 11 that has a duplicated open reading frame but would encode a normal protein because it lacks an initiation site in the duplicated ORF. (From Tanaka et al.706 et al. Molecular characterization of three rabbit rotavirus strains. Arch Virol 1988;98:253–265.) |
Rotaviruses are the only known agents of mammals or birds that contain 11 segments of dsRNA. In most cases, the electrophoretic pattern of the genome of the group A viruses is composed of four high-molecular-weight dsRNA segments (numbered 1 to 4), two middle-sized segments (5 and 6), a distinctive triplet of segments (7 to 9), and two smaller segments (10 and 11). When this basic pattern is not seen, the rotavirus being analyzed may be a group A avian virus, a non–group A virus, a group A virus that contains rearrangements within individual genome segments (Fig. 45.6), or a new unique group A virus. Analysis of genomic electropherotypes is a relatively easy, rapid, and popular technique for virus detection and for molecular epidemiology studies to monitor virus outbreaks and transmission. However, because distinct RNA patterns can arise by different mechanisms (reassortment, mutation, rearrangements) and RNA segments of different sequences may co-migrate, these profiles are not useful as a definitive criterion for classification of a virus strain.109,220 (See also Molecular Epidemiology, later). Nucleic acid hybridization combined with northern blot was initially used to classify viruses based on the relatedness of genome segments225 (e.g., to classify genetically related viruses into different genogroups)531 and to identify the origin of specific RNA segments in virus reassortants. This method characterized viruses involved in cross-species transmission529,530 but is now being replaced by complete genome sequencing, which has clearly identified zoonotic transmission of many rotavirus genes and reassortment.473
In viruses with genome rearrangements, typical RNA segments are missing or are decreased in concentration in an electrophoretic profile and are replaced by additional, more slowly (or rarely more rapidly) migrating bands of dsRNA (Fig. 45.6, lanes 5 and 7). The slowly migrating bands represent concatemeric forms of dsRNA-containing sequences specific for the missing RNA segments, which has been reviewed elsewhere.184 The more rapidly migrating bands appear to represent deletions. Viruses with genome rearrangements of this type have been isolated most frequently during infection from immunodeficient, chronically infected children, asymptomatically infected immunocompetent children, and animals (calves, pigs, or rabbits). Viruses with rearranged genomes have also been obtained in vitro after serial high multiplicity of infection passage of tissue culture–adapted rotaviruses. Virus isolates with rearrangements in segments 5, 6, 8, 10, and 11 have been characterized, with the greatest number having rearrangements detected in segments 5 and 11. Viruses with a rearranged segment 11 may have some selective advantage (better growth in vitro or stability), so they are detected more easily, rather than occurring more frequently.
Viruses that contain rearranged genome segments are generally not defective, and the rearranged segments can reassort and replace normal RNA segments structurally and functionally. These viruses do not have a growth advantage, but they exhibit a selective advantage for being incorporated in viral progeny, indicating a preferential packaging of rearranged segments into progeny.737 Biophysical characterization of such particles has shown that up to 1,800 additional base pairs can be packaged in particles without causing detectable changes in particle diameter or apparent sedimentation values. The density of particles containing rearranged genomes may be increased, however, and the increase in density is directly proportional to
the number of additionally packaged base pairs.493 Thus, rotaviruses have considerable capacity to package additional genomic RNA, although the upper limit is unknown. Whereas a total of 11 RNA segments are invariably packaged, much less constraint appears to exist on the length of individual RNA segments assembled into the maturing virus particle. Because the amount of viral RNA present in virions can vary by a significant amount (∼1,800 nucleotides), a headfull mechanism for RNA packaging, as seen for the dsRNA phage phi6,510 may not operate for rotaviruses.
the number of additionally packaged base pairs.493 Thus, rotaviruses have considerable capacity to package additional genomic RNA, although the upper limit is unknown. Whereas a total of 11 RNA segments are invariably packaged, much less constraint appears to exist on the length of individual RNA segments assembled into the maturing virus particle. Because the amount of viral RNA present in virions can vary by a significant amount (∼1,800 nucleotides), a headfull mechanism for RNA packaging, as seen for the dsRNA phage phi6,510 may not operate for rotaviruses.
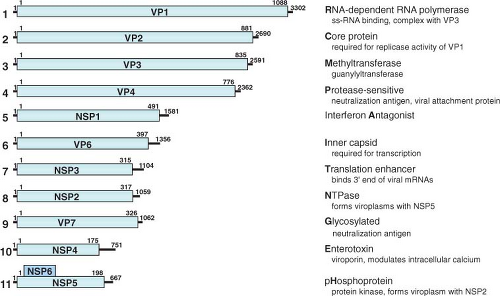 Figure 45.7. Genome structure of rotavirus simian agent 11 (SA11) virus. RNA segments (in nucleotides) shown in the positive sense and their encoded proteins (in amino acids). The lines at the 5′ and 3′ termini represent the noncoding regions. A few key functions of the proteins are listed with the letter bolded that is used in a new classification system based on the entire genome. See the text and Tables 45.4 and 45.5 for information on the classification system and more details on gene-coding assignments and locations of known temperature-sensitive mutations. |
In most cases, the profiles of virus-encoded proteins in cells infected with rotaviruses with rearranged genomes are similar to those seen in cells infected with standard rotavirus strains, indicating that the rearrangement of the sequences in a segment leaves the normal reading frames and their expression unaltered. Sequence analyses of rearranged genome segments confirm this and reveal mechanisms by which the rearrangements arise. In most cases, the rearrangements result from a head-to-tail duplication that occurs immediately downstream from the normal ORF and, hence, the rearranged segment retains the capacity to express its normal protein product. In some cases, gene rearrangements result in truncated proteins. In one case, a gene rearrangement in genome segment 5 introduced a point mutation in the ORF and produced a truncated NSP1 lacking the C-terminal half of the protein.352 This rearranged virus is nondefective in vitro, indicating the C-terminal half of NSP1 is nonessential for rotavirus replication, at least in cell culture.350 Another mutant lacking the cysteine-rich zinc finger motif in the genome segment 5 protein, NSP1, is also viable in cell culture.707 Rearrangements have also been identified that affect the ORF for NSP3, and analysis of these viruses suggests a mechanism for gene rearrangements in which secondary structures facilitate and direct the transfer of the RNA polymerase from the 5′ to the neighboring 3′ end of the template during the replication step.266 Genome rearrangements (concatemerization and deletion) are a third mechanism of evolution (in addition to reassortment and mutation) of rotaviruses. The discovery that rearranged segments are preferentially segregated into progeny virions is helpful to identify sequences critical for replication and RNA packaging.184 While the exact sequences required for packaging remain to be identified, rearranged genomes that are selectively packaged have parts of the 5′ ends duplicated, suggesting packaging signals including coding sequences are located in the 5′ region of the RNAs.737
Coding Assignments
The coding assignments and many properties of the proteins encoded in each of the 11 genome segments are now well established (Fig. 45.7 and Table 45.5), although new protein functions continue to be identified. Initially protein assignments
were determined by in vitro translation using mRNA or denatured dsRNA and by analyses of reassortant viruses. Complete coding assignments were known first for SA11, the type species of the Rotavirus genus. Comparative studies of other rotavirus strains have shown that the absolute order of migration in gels of cognate genes may differ among virus strains. Identification of cognate genes, therefore, is now routinely achieved from sequence information obtained directly from dsRNA or single-stranded RNA (ssRNA) following reverse transcription-polymerase chain reaction (RT-PCR) amplification followed by analysis of sequence homology with accumulating nucleic acid sequence databases.
were determined by in vitro translation using mRNA or denatured dsRNA and by analyses of reassortant viruses. Complete coding assignments were known first for SA11, the type species of the Rotavirus genus. Comparative studies of other rotavirus strains have shown that the absolute order of migration in gels of cognate genes may differ among virus strains. Identification of cognate genes, therefore, is now routinely achieved from sequence information obtained directly from dsRNA or single-stranded RNA (ssRNA) following reverse transcription-polymerase chain reaction (RT-PCR) amplification followed by analysis of sequence homology with accumulating nucleic acid sequence databases.
The rotavirus genome segments code for structural proteins found in virus particles and nonstructural proteins found in infected cells but not present in mature particles (Table 45.5 and Fig. 45.7). Six of the genome segments code for structural proteins found in virus particles. Another six proteins are nonstructural, with a few tissue culture adapted virus strains lacking NSP6. Early studies often presented conflicting conclusions concerning the numbers and locations of the rotavirus proteins. Many of these conflicts were resolved, as reviewed elsewhere,486 when it was recognized that posttranslational modifications (glycosylation, trimming of carbohydrate residues, and proteolytic cleavages) occur after polypeptide synthesis. In addition, strain variations (e.g., the presence of more than one glycosylation site on VP7 in some bovine, equine, porcine, and human rotavirus strains) provide explanations for other differences in polypeptide patterns.
The nomenclature of the viral proteins (as originally proposed for SA11 proteins) designates structural proteins as viral protein (VP) followed by a number, with VP1 being the highest-molecular-weight protein, and proteins generated by cleavage of a larger precursor being indicated by an asterisk (VP4 is cleaved to produce VP5* and VP8*212,221). Initial studies referred to the nonstructural proteins as NS followed by a number indicating the protein’s molecular weight. This nomenclature has been replaced by NSP1 to NSP6 to facilitate comparisons among cognate nonstructural proteins of different molecular weights486 (Table 45.5). In fact, much of the literature before 1988 refers to the genome segment 4 product as “VP3”; before 1994, NS53 and NS35 were the designations used for what is now referred to as NSP1 and NSP2, and so forth (Table 45.5). The new nomenclature is used throughout this chapter.
Stages of Replication
Overview of the Replication Cycle
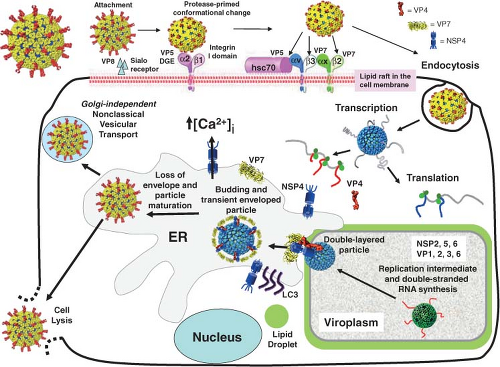
Figure 45.8 shows a schematic of the rotavirus replication cycle. Most details of this cycle have been obtained from studies of rotaviruses infecting monkey kidney cell monolayers or polarized intestinal epithelial cells. Other information has come from assays to probe specific steps in the replication cycle based on the expression and interaction of individual wild-type or mutated proteins and RNA in in vitro systems, and conclusions from these studies are generally confirmed in the context of virus replication systems using confocal microscopy and small interfering RNA (siRNA). One limitation to some current conclusions on protein function is that an efficient reverse genetics system to test results by incorporating any desired mutation into any gene in the rotavirus genome remains to be established. However, some progress has been achieved as discussed under Genetics and Reverse Genetics.
In vivo, the natural cell tropism for rotaviruses is the differentiated enterocyte in the small intestine, suggesting these cells express specific receptor(s) for virus attachment and entry into cells. However, extraintestinal spread of rotavirus also occurs in humans and all animal models studied,27,63,64,65,66,125,159,230,239,519,569,611,701 demonstrating a wider range of host cells than previously thought and possible additional receptors. Rotavirus replication in continuous cell cultures derived from monkey kidneys is fairly rapid, with maximal yields of virus being found after 10 to 12 hours at 37°C or 18 hours at 33°C when cells are infected at high multiplicities of infection (10 to 20 plaque-forming units [pfu]/cell).137,489,615 Rotavirus replication in differentiated human intestinal cell lines (Caco-2 cells) grown on permeable filter membranes is slower, with maximal yields of virus detected on the apical surface of cells 20 to 24 hours after infection.131,132,381 Significantly, EM studies of virus replication in polarized intestinal cells indicate that the replication process in these cells has some distinct differences from the virus replication cycle in nonpolarized cell cultures. These emerging differences are described further later.
The general features of rotavirus replication (based on studies in cultures of monkey kidney cells) are as follows:
Cultivation of most virus strains requires the addition of exogenous proteases to the culture medium. This ensures activation of viral infectivity by cleaving the outer capsid protein VP4.
Replication is totally cytoplasmic.
Cells do not contain enzymes to replicate dsRNA; hence, the virus must supply the necessary enzymes.
Transcripts function both to produce proteins and as a template for production of negative-strand RNA. Once the complementary negative strand is synthesized, it remains associated with the positive strand.
The dsRNA segments are formed within nascent subviral particles, and free dsRNA or free negative-stranded ssRNA is generally not found in infected cells.
RNA replication occurs within cytoplasmic viroplasms.
Subviral particles form in association with viroplasms, and these particles mature by budding through the membrane of the ER. In this process, particles acquire their outer capsid proteins.
Levels of intracellular calcium are important for controlling virus assembly and particle integrity.
Cell lysis releases particles from infected cells grown on monolayers.
In polarized intestinal cells, virus entry occurs almost exclusively through the apical membrane, although some strains enter through both the apical and basolateral membranes (see later). In addition, virus replication in polarized enterocytes alters differentiated enterocyte cell function by perturbing cellular protein trafficking, the cytoskeleton, and tight junctions, and by triggering epithelial cell signaling pathways that can activate innate responses and secretion of various chemokines or cytokines86,380,550 (see later). Finally, virus is released apically from polarized enterocytes by a novel, Golgi-independent, vesicular transport that does not result in extensive cytopathic effects or cell lysis.381
Attachment
The molecular details of rotavirus adsorption, entry, and uncoating are complex and remain incompletely understood, but progress is being made because of new molecular and structural information on the outer capsid proteins and an understanding of differences in virus strains. Rotavirus attachment and entry, highlighted here primarily based on extensive structural and molecular studies using the rhesus rotavirus strain RRV, is a multistep process that involves sialic acid–containing receptors in the initial cell attachment step and coordinated interactions with multiple receptors during the postattachment steps31,194,450,650 (Fig. 45.8). As expected from their locations in the virus structure, VP4 and VP7 are implicated in initial interactions with host cells. A broad range of cells bind rotaviruses and are infected with different efficiencies,133,159 suggesting that the initial binding (attachment) is a generally promiscuous interaction with a common receptor and postattachment co-receptors are critical for virus entry into the cell. Virus attachment is by VP4157,644 or its cleavage product VP8* on TLPs. Binding to cells does not require cleaved VP4138,259 or glycosylated VP7,589 but efficient cell entry requires proteolytic cleavage of VP4. Subsequent cell entry is mediated by VP5*.821
RRV and some other rotavirus strains hemagglutinate red blood cells, and neuraminidase treatment of red blood cells reduces virus binding, indicating a role of sialic acid (SA) in virus attachment.42,693 Neuraminidase (NA) treatment of cultured
cells reduces the infectivity of hemagglutinin (HA)-positive virus strains, SA-containing compounds such as fetuin and mucin inhibit virus binding to cells,406,812 and such strains are named NA sensitive.360 The most easily cultivatable animal rotaviruses bind SA and can infect both surfaces of polarized cells, but they preferentially infect polarized cells apically because of the presence of terminal SA.131,622 Many animal rotaviruses and most strains isolated from humans do not hemagglutinate RBCs, bind to and infect NA-treated cells, and are now called NA-resistant viruses132; these were previously called SA-independent viruses but were renamed after some NA-resistant strains were shown to bind to NA-insensitive internal SA moieties on glycolipids or to modified SA moieties in oligosaccharide structures, such as those present in the GM1 ganglioside, that are resistant to NA treatment.179,321 NA-resistant viruses preferentially infect polarized intestinal cells through the basolateral surface.132,622 The consequences of efficient infection through the basolateral membrane are not fully understood, but further studies on rotavirus entry into cells likely will reveal other virus–host interactions that are relevant for pathogenesis, especially given the extraintestinal spread of virus.
cells reduces the infectivity of hemagglutinin (HA)-positive virus strains, SA-containing compounds such as fetuin and mucin inhibit virus binding to cells,406,812 and such strains are named NA sensitive.360 The most easily cultivatable animal rotaviruses bind SA and can infect both surfaces of polarized cells, but they preferentially infect polarized cells apically because of the presence of terminal SA.131,622 Many animal rotaviruses and most strains isolated from humans do not hemagglutinate RBCs, bind to and infect NA-treated cells, and are now called NA-resistant viruses132; these were previously called SA-independent viruses but were renamed after some NA-resistant strains were shown to bind to NA-insensitive internal SA moieties on glycolipids or to modified SA moieties in oligosaccharide structures, such as those present in the GM1 ganglioside, that are resistant to NA treatment.179,321 NA-resistant viruses preferentially infect polarized intestinal cells through the basolateral surface.132,622 The consequences of efficient infection through the basolateral membrane are not fully understood, but further studies on rotavirus entry into cells likely will reveal other virus–host interactions that are relevant for pathogenesis, especially given the extraintestinal spread of virus.
VP4 is the HA based on studies of (a) reassortants showing that HA activity segregates with the rotavirus gene 4, (b) mutants that lose their HA activity are selected with VP4 monoclonal antibodies, and (c) recombinant VP4 produced in insect cells agglutinates red blood cells. X-ray crystallographic structures of RRV VP8* complexed with SA show the VP8* domain exhibits a β-sandwich fold of the galactins, a family of proteins whose natural ligands are carbohydrates, despite a lack of sequence similarity193 (Figs. 45.4 and 45.9). The SA moiety binds within a shallow groove on the VP8* surface using residues that are conserved in SA-binding strains. The VP8* structure represents one of the first observed cases of a rotavirus protein taking on a fold seen among host proteins and, based on this structural result, it is proposed that VP4 arose from the insertion of a host carbohydrate-binding domain into a viral membrane interaction protein.193 The structures of the VP8* core from two NA-resistant human rotaviruses (DS-1 and Wa strains) show conservation of the same galactin-like fold but exhibit other structural differences including a widening of the cleft.62,514 It has been suggested from NMR, infectivity assays, and modeling that such a wider cleft accommodates cellular glycans with an internal sialic acid moiety.330
Recent data indicate that at least one NA-resistant rotavirus has a narrower VP8* glycan-binding cleft that cannot bind to SA, but instead binds to a nonsialylated histo-blood group antigen.348 This binding may be associated with the ability of this virus to cause zoonotic infections by switching receptors; these results indicate that our understanding of rotavirus–glycan interactions remains incomplete and glycan binding may differ significantly among human rotaviruses.
The identification of cellular co-receptor(s) for rotavirus is an active area of research. Early studies and conflicting results are explained by the use of different receptors by different viruses on different cell types.31,194,451,650 Several integrins, including α2β1, αvβ3, αxβ2, and α4β1, are implicated as possible receptors for rotavirus cell entry, with evidence for α2β1 being the strongest.153,297,323,820,822 Antibodies to these integrins or synthetic peptides that represent integrin-ligand motifs that are present in VP5* or VP7 block virus infectivity, and some integrins influence cell binding in an additive manner, suggesting they may play a role in different stages of the cell entry process.319 Neutralizing antibodies to VP5* and VP7, but not to VP8* and VP6, inhibit virus binding to integrins.245 Involvement of integrins at a postattachment step is consistent with observations that integrin expression increases infectivity of rotaviruses in poorly permissive cells.133,335 Currently, rotaviruses are characterized by their integrin usage in addition to their NA sensitivity.297,323,360 Integrin usage has been characterized in nonpolarized monkey kidney cells, endothelial and polarized kidney cells, and intestinal epithelial cells. Integrin and postbinding receptor usage can differ depending on the cell type(s) analyzed, and the interactions between NA-sensitive and NA-resistant rotaviruses with intestinal or extraintestinal cells are distinct and may affect the pathogenesis and outcome of infections. Integrins have a polarized distribution and are located at the basolateral plasma membrane, so this may be why NA-resistant rotaviruses preferentially infect through the basolateral surface.131,622 Interesting studies suggested that the addition of VP8* peptides to polarized cells can trigger the movement of basolateral proteins to the apical surface, resulting in another possible mechanism for how rotaviruses might interact with integrins.537 A DGE sequence in VP5* binds to the α-subunit I domain on activated α2β1, and rotavirus binding is eliminated by mutations in activation-responsive helices of this integrin.244 The residues that bind to rotavirus overlap with those used by type I collagen but are distinct from those that bind echovirus 1. However, rotavirus is distinguished from collagen by its specific α2β1 binding site requirements, and rotavirus does not activate α2β1 or induce p38 signaling as occurs with collagen-integrin binding.244 Despite demonstrated binding of VP5* to integrins, the lack of signaling after this binding leaves open the question of how virus actually penetrates the membrane. VP7 is proposed to also interact with αXβ2 and αvβ3 though GPR and CNP motifs, respectively,297,822 but the proposed integrin-binding motif (GPR, residues 253 to 255 on VP7) is questionable because it is located on the inward-facing surface of the trimer and would not be available to interact with integrins until after uncoating.17,297 However, the CPN residues are potentially available for integrin binding. Evaluation of the role of integrin α2 and β3 in rotavirus cell entry using RNA silencing in permissive cells also concluded that these integrins may not play a major role in the rotavirus cell entry process.362 Other receptors remain to be identified as a porcine rotavirus CRW-8 does not use human or monkey α2β1 as a cellular receptor.298 Thus, the crucial molecules involved in cell entry may still remain to be identified.
Heat shock cognate protein 70 (hsc70) is another putative co-receptor proposed to bind virus and mediate virus entry into cells based on experiments in which rotavirus entry is blocked by a VP5* synthetic peptide and by antibodies against human recombinant hsc70.318,451,584,819 The role of hsc70 is proposed based on observing enhanced virus entry into cells and subsequent enhanced virus infectivity after heating some cells at 45°C.454 Binding of purified hsc70 to two domains in VP5* as well as to a domain in VP6 are also detected in cell-free assays and may modify the conformation of virus particles to help the virus enter cells.313,584 To date, however, binding affinities to the proposed post-SA binding receptor molecules have not been determined, and no structures of rotavirus–protein co-receptor complexes are solved. All the implicated RV receptors on cells (GM1 ganglioside, integrin subunits, and hsc70)
are associated with detergent-resistant lipid microdomains, and infectious rotavirus associates with these domains. Thus, lipid rafts are thought to provide a platform to facilitate efficient interaction of rotavirus receptors with virus particles.361
are associated with detergent-resistant lipid microdomains, and infectious rotavirus associates with these domains. Thus, lipid rafts are thought to provide a platform to facilitate efficient interaction of rotavirus receptors with virus particles.361
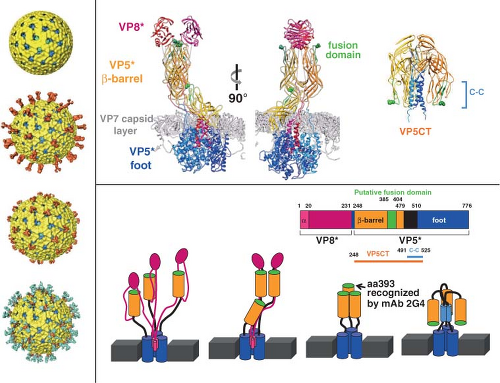 Figure 45.9. Conformational transitions of rotavirus particles and the spike protein VP4. Left-side panel: Images of cryo-electron microscopy (cryo-EM) reconstructions of rotavirus particles grown in the absence of trypsin (top), grown in the presence of trypsin (second from the top), and treated with high pH (third from the top), and high-pH-treated virus with a monoclonal antibody (2G4) bound to VP5* (bottom). Trypsin treatment causes a conformational change in the spikes and primes the cleavage products for further conformational changes. pH-treated particles provided initial evidence that the spike is a trimer and this structure may represent an intermediate that binds to co-receptors. Escape mutants indicate that mAb 2G4 binds to amino acid 393. Top right panel: High-resolution structures illustrating different domains and conformations of VP4. Domains in VP4 are color coded and show VP8* (magenta) and subdomains within VP5* including the β-barrel (orange), the fusion domain (green), and the foot domain (blue). The VP7 capsid layer is illustrated in gray. The mAb 2G4 binding site is indicated by green balls within the fusion domain. The left two figures show two orientations of an atomic structure of a trypsinized spike with dimeric and trimeric and asymmetric domains. Two of the subunits of VP5* associate to form a dimer-looking central body and the third VP5* subunit traverses the VP7 layer, forming a stalk between the body and the foot domains. Rotation of the body clearly illustrates its dimeric structure above the VP7 layer. The right figure shows a structure of the three color-coded domains of a cleaved form of VP5* (VP5*CT, solved from a cleaved, recombinant VP5*). These subunits associate into a coiled-coil (C-C) stabilized trimer that is shaped like an umbrella; this fold-back is thought to be the stable postmembrane penetration conformation of VP5*. Bottom panel: Top row: Schematic of the linear structure of VP4 showing color-coded domains. Bottom row: A color-coded linear diagram and cartoon representation of the proposed conformations of VP4. The colors depicted in the linear diagram correspond to the VP4 domains in the cartoon representation; VP8* (the α-helix [α] located within the foot domain [pink] and the rest of the VP8* domain [magenta]) and VP5* (the β-barrel domain [orange] containing the putative membrane fusion domain [green] and the stretch of amino acids [black] that link the β-barrel to the foot domain [blue]). The VP5*CT and coil-coil domain indicated in the panel above are indicated below the linear diagram. The numbers indicate amino acids. The cartoon represents the disordered VP4 spike on particles grown in the absence of trypsin; the ordered VP4 spike on virus grown in the presence of trypsin; the membrane-penetrating V5* (VP8* may be retained on the particle and interact with cellular receptors but is not shown in the cartoon); and the postmembrane penetration, fold-back conformation of VP5*.412,675,732,791 |
Penetration and Uncoating
After initial binding to cells, rotavirus cell entry is a coordinated, multistep process involving a series of conformational changes in the capsid proteins (Figs. 45.8 and 45.9) and involves endocytic pathways. Because the initial VP8* glycan binding has a relatively low affinity, the virus entry event is probably responsible, in part, for cell type and host specificity.193,360,501 Crystallographic structures of VP8*–SA complexes indicate minimal conformational changes in VP8* upon SA binding,514 but whether this is true in the context of the entire rotavirus structure remains to be determined. Instead, postattachment conformational changes involve VP5* and VP7.
Efficient entry of rotavirus into cells requires conformational rearrangements of the spike protein that facilitate membrane penetration. Trypsinized particles enter cells more rapidly than those not trypsinized.385,405 The body of the spike (VP5*) has lipophilic activity thought to be important for entry into cells,84,180,196,644,647,820,821 and cell permeabilization properties in the C-terminal VP5* are thought to depend on the exposure of three hydrophobic loops in the VP5* apex normally located below the VP8* lobes.412 Before trypsin cleavage, VP4 is flexible and spikes are not visible by cryo-EM, although they are present on particles158 (Fig. 45.9). Trypsinization of VP4 stabilizes the spike structure, inducing an initial disorder-to-order or flexible form–to–rigid spike transition that results in a unique, elongated, asymmetrical shape and primes VP4 for additional conformational changes158 (Fig. 45.9). Exposure of the specific cleavage site in the asymmetric spike structure and masking of nonspecific trypsin sites in VP4 by VP7 regulates the cleavage of VP4.675 The trypsin-primed VP4 intermediate is held within the particle and ready to undergo further molecular rearrangements during virus entry into cells that includes promoting membrane permeability associated with virus entry.675 The 3.2-Å crystal structure of the main part of the VP5* stalk generated in vitro reveals a folded-back rearrangement that translocates three clustered hydrophobic loops from one end of the molecule to the other to form a coiled-coil stabilized trimer that is shaped like an umbrella; this fold-back form is thought to be the stable “postpenetration” conformation of VP5*192 (Fig. 45.9). An entry-associated event, proposed to be dissociation of VP7 by exposure to low calcium levels in the endosome, triggers the transition of the trypsin-primed VP5* spike to the fold-back trimeric umbrella conformation that interacts with the uncoating membrane and refolds to destabilize the membrane.192,514,805 The VP5* fold-back depends on both membrane interactions and virus uncoating, suggesting that this rearrangement is one of the final steps of RV entry.732,805
Studies with pH treatment of a NA-sensitive virus provide support for this proposal.192,514,586,587,805 At elevated pH, the spike undergoes a dramatic irreversible conformational change and becomes stunted with a pronounced trilobed appearance, although the amount of VP4 on particles remains unchanged (Fig. 45.9). Three Fab fragments of the VP5*-specific mAb, 2G4, can then bind to these altered spikes, indicating that VP4 has undergone a dimer-to-trimer transition (Fig. 45.9). Particles with altered spikes no longer hemagglutinate red blood cells or infect mammalian cells. They retain the ability to bind to mammalian cells, but in an NA-resistant manner, different from untreated particles that bind cells in an NA-sensitive manner. High-pH treatment may trigger a conformational change that mimics the transition in VP4 that occurs with the post-SA attachment step. These particles resemble a mutant virus of an NA-sensitive rhesus rotavirus that exhibits NA-resistant cell binding, in contrast to its parental strain, and attaches to cells by interacting with the integrin α2β1 through a DGE motif in VP5*.820 Of interest, the conformational rearrangements of VP5* translocate this DGE motif to the external surface of the trypsin-primed structural forms of VP5*, making it accessible to bind to an integrin.805 Recent analysis of integrin binding of virus strains with distinct VP5* sequences has identified sequence variation in VP5* amino acids that parallel rotavirus strain–specific differences in the effects of virus binding to the α2 I domain. These results indicate VP5* amino acids 335 to 380 that are surface exposed and near the DGE sequence may also influence rotavirus recognition of α2β1 in addition to the DGE site.244
Internalization does not take place at 0°C to 4°C, indicating that this step requires active cellular processes.406,590 All virus is internalized by 60 to 90 minutes after binding.406 The mechanism of internalization (penetration) into cells remains unclear. Both morphologic and biochemical approaches have been used to investigate the mode of entry of rotaviruses into cells, and both receptor-mediated endocytosis and direct membrane penetration have been suggested as mechanisms of rotavirus entry into cells; trypsin-treated and non–trypsin-treated virus may enter cells by different mechanisms as reviewed previously.215 Recent studies indicate that different rotavirus strains enter cells through different endocytic pathways.323
Other viruses that initiate infection by mechanisms involving receptor-mediated endocytosis often depend on the acidification of endosomes for partial uncoating or entry into the cell. The importance of acidification of endosomes for the initiation of infection of rotaviruses has been studied by several groups.260,385,405,462,791 In all cases, lysosomotropic agents (ammonium chloride, chloroquine, methylamine, and amantadine) do not affect rotavirus entry. Energy inhibitors (sodium azide and dinitrophenol) have a minimal effect on rotavirus infection, and this has been taken to suggest that rotaviruses do not use endocytosis to enter cells. Other endocytosis inhibitors, such as dansylcadaverine and cytochalasin D, and in some, but not all, cases the vacuolar proton–adenosine triphosphatase (ATPase) inhibitor bafilomycin A1, also do not block rotavirus entry. These results indicate that neither endocytosis nor an intraendosomal acidic pH or a proton gradient is required for rotavirus entry into cells.
While the passage of rotaviruses from endocytic vesicles to the cytoplasm does not occur by a pH-dependent fusion mechanism, other data indicate that rotaviruses are still taken up by endocytosis. Direct demonstration of virus fusion with membranes or hemolysis is lacking. Protease cleavage of VP4 is important for rapid entry into cells, and particles containing cleaved VP4 possess lipophilic activity and can affect release of fluorescent dyes from liposomes and isolated membrane vesicles. Rotavirus entry into cells can also be monitored by co-entry of toxins, such as α-sarcin, into cells162,446 and by a cell-to-cell fusion from without assay.207,227 Most observations are consistent with the hypothesis that virus enters cells by
endocytoses after direct interactions with a series of receptors on the plasma membrane.535,647
endocytoses after direct interactions with a series of receptors on the plasma membrane.535,647
The outer capsid proteins of rotavirus that are solubilized from virus particles are able to permeabilize cellular membranes,646 and it has been proposed that the outer capsid proteins are solubilized within an endocytic vesicle because of low Ca2+ concentrations. The decrease in calcium concentrations within the endosomal vesicle might trigger conformational changes in the capsid, capsid solubilization, and vesicle lysis.646 In this Ca2+-dependent endocytosis model, acidification of the endosome would not be needed for the infectious process. Use of the calcium ionophore A23187 to increase the intracellular Ca2+ concentration during the early stages of replication can block uncoating.462 These results support the hypothesis that low Ca2+ concentrations in the intracellular microenvironment may be responsible for uncoating. This idea was originally proposed because it was known that removal of the outer capsid of particles and activation of the endogenous polymerase could be accomplished by calcium chelation.143,341
It is also possible that more than one mechanism, including endocytosis and direct entry, is operative for rotaviruses, as has been proposed for polioviruses and reoviruses.68,189 Further studies are needed to determine whether the common endocytosis-mediated entry pathway exists for all rotaviruses and in all cell types. Studies with drugs and dominant-negative mutants suggest that virus enters cells through a non–clathrin-, non–caveolin-dependent mechanism that depends on the presence of cholesterol on the cell membrane and on a functional dynamin.659 Trypsin also has been detected associated with the rotavirus outer capsid and is activated by solubilization of the outer capsid proteins.43 This activated trypsin is proposed to cleave VP7 and VP4 into fragments capable of disrupting membranes, and this may allow DLPs to gain access to the cytoplasm to begin actively transcribing viral mRNA to complete the next step in the viral life cycle.
The entry of RRV, which is NA sensitive and binds to α2β1 and of αvβ3 integrins and hsp70, is the most extensively studied by biochemical, structural, and molecular methods. RRV entry into polarized cells is through an endocytic pathway but reportedly does not require cholesterol or a functional dynamin.791 Use of imaging and unique mAbs that detect regional or conformation-dependent epitopes on VP8* and VP5* or soluble, unassembled protein to follow the proteins on incoming RRV particles at very early time points prior to the onset of viral replication indicates that internalization and decapsidation occur directly after cell membrane penetration as assessed by disappearance of trimeric VP7.791 In addition, virus entry into cells involves endocytosis, calcium-dependent uncoating, and several VP4 conformational changes; VP8* staining is lost at the time of cell penetration and is not found in the cytoplasm, while VP5* is detected in the cytoplasm within 1 hour of infection. The fold-back conformation of VP5* is only detectable at the entry step.791 VP5* and VP7 co-localize with early endosome markers Rab4 and 5, indicating RRV uses an endocytic route limited to the early endosomes to enter cells. Bafilomycin A1 and concanamycin A, two pharmacologic inhibitors of the vacuolar-type H+-ATPase, reduced cytoplasmic staining of VP6 indicative of blocking entry and reduced the appearance of the folded-back VP5*, suggesting that the appearance of this epitope is specific to entry. Elevating endosomal Ca2+ concentration also blocked entry. This study supports a model of RRV entry in which, after membrane binding and internalization, the low Ca2+ concentration in the endosome triggers VP7 decapsidation and the appearance of the VP5* fold-back, ultimately leading to the release of DLPs into the cell cytoplasm. The results also suggest that the primary effect of BafA1 on RRV infection is mediated indirectly through changes in the endosomal Ca2+ gradient.791 Unexpectedly, these studies did not find a role for dynamin or cholesterol in RRV entry, which had been implicated in RRV entry into nonpolarized cells through a non–clathrin-, non–caveolin-mediated endocytosis pathway that depends on a functional dynamin and on the presence of cholesterol on the cell surface.323,659 Entry of other RV strains with different NA sensitivity and integrin dependence into MA104 cells is reported to be dependent on hsc70, dynamin, and cholesterol, but these distinct strains enter cells through clathrin-mediated endocytosis pathways.323 The reasons for these differences are not known but might result from the different virus strains or the heterogeneity of raft-type membrane microdomains on different cell types in different differentiation states.177
RNA Synthesis
Overview
Incoming RV particles containing the dsRNA genome segments must synthesize mRNAs that direct the synthesis of viral proteins and also serve as templates for the synthesis of the dsRNA genome that becomes encapsidated into newly made particles. The virion polymerase performs these functions as a transcriptase and as a replicase at different times during the replication cycle. Synthesis of viral transcripts is mediated by the endogenous viral RNA-dependent RNA polymerase complex (PC), consisting of VP1 and VP3, which is latent in the virion, where it appears as a flower-shaped feature in the icosahedrally averaged cryo-EM reconstruction of the virion attached to the inner surface of the VP2 layer at all the fivefold axes (Fig. 45.4). The PC contains the enzymatic activities needed for synthesis of capped messenger RNA, including transcriptase, nucleotide phosphohydrolase, guanylyltransferase, and methylases. Each genome segment is transcribed simultaneously and repeatedly by a specific polymerase complex within the confines of the capsid architecture, and the resulting transcripts exit through the type I channel system at the axis adjacent to its site of synthesis. This mechanism of transcription offers an explanation of why no dsRNA virus contains more than 12 genome segments.
The PC must be activated for transcription to occur. Transcription begins following removal of the VP7 outer layer and can also be studied in vitro by treatment of TLPs with a chelating agent or by heat shock treatment that removes the outer capsid proteins.143,694 Transcribing particles will continuously synthesize milligram quantities of mRNAs in vitro as long as fresh precursors and an energy-generating system are provided. Rotavirus transcription requires a hydrolyzable form of adenosine triphosphate (ATP). Studies with analogs that inhibit transcription suggest that ATP is required in reactions other than polymerization694 and may be used for initiation or elongation of RNA molecules.
Ultimately, transcription must be inhibited to allow RNA replication to proceed and virus assembly to be completed. While not completely understood, transcription can be inhibited by several mechanisms. Cryo-EM studies of DLPs complexed with
some monoclonal antibodies to VP6, or the addition of VP7 onto DLPs, indicate a conformational change at the interface of the VP2–VP6 layers or in the VP6 trimers can inhibit sustained elongation and translocation of transcripts.235,435,723 It is also possible that binding of VP6 to NSP4, which serves as an intracellular receptor for particle assembly (see later), is the key interaction that inhibits transcription. This hypothesis is consistent with the observation that knockdown of NSP4 by siRNA increases viral mRNA synthesis.684 NSP4 can form concentration-dependent pentamers, and such structures in the ER may interact with VP6 molecules by a fivefold axis on the surface of the DLPs.106
some monoclonal antibodies to VP6, or the addition of VP7 onto DLPs, indicate a conformational change at the interface of the VP2–VP6 layers or in the VP6 trimers can inhibit sustained elongation and translocation of transcripts.235,435,723 It is also possible that binding of VP6 to NSP4, which serves as an intracellular receptor for particle assembly (see later), is the key interaction that inhibits transcription. This hypothesis is consistent with the observation that knockdown of NSP4 by siRNA increases viral mRNA synthesis.684 NSP4 can form concentration-dependent pentamers, and such structures in the ER may interact with VP6 molecules by a fivefold axis on the surface of the DLPs.106
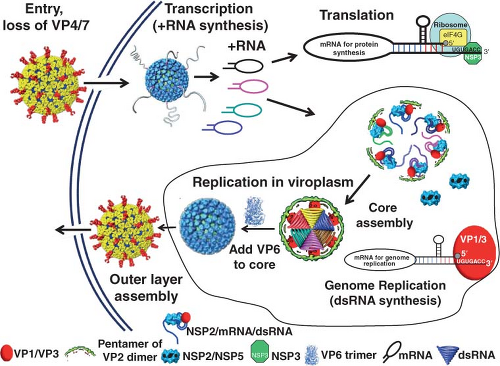 Figure 45.10. Virion disassembly and functions of RNAs for transcription, translation, genome replication, and packaging. With virus entry into cells, the double-layered particles (DLPs) lose the outer layer proteins and the DLPs become transcriptionally active and release (+)RNAs that are thought to form panhandle structures that are translated into viral proteins and replicated into genomic double-stranded RNA (dsRNA) in viroplasms. A pentamer of VP2 dimers and VP1/VP3 form the initial assembly unit. NSP2 is likely involved in feeding (+)RNA into the enzyme complex for (−)RNA synthesis and formation of the duplex RNA.320 Twelve such units assemble together to encapsidate the genome segments to form the virion core, which subsequently is coated with VP6 to make new DLPs. (Modified from Hu L, Crawford SE, Estes MK, et al. 2010. Rotavirus structure and functional implications. In Hein NT, Baldacci G, Haenni AL, Benedetti EL, eds. Viruses Responsible for Emerging Diseases in South-East Asia, Universite Paris Diderot-Paris 7, 2011.) |
The structure of the polymerase must provide mechanisms for transcription and subsequent RNA replication where the minus-strand RNA as well as duplex formation is achieved to produce new infectious virions. Transcription to produce (+)mRNAs requires several functions, including unwinding the genomic dsRNA, entry of the (−)RNA templates for synthesis of nascent (+)RNA, exit of the (−)RNA for subsequent annealing and further rounds of transcription, and exit of the nascent (+)RNA for capping and extrusion from the DLP. In the replication mode, the polymerase must provide a mechanism for entry of (+)RNA template and exit of the duplex. There also should be easy access for the NTPs into the catalytic center of VP1 for (−)RNA synthesis.
The Viral RNA Polymerase, (+) Strand, and (−) Strand RNA Synthesis
Insight into how the rotavirus polymerase mediates both transcription and replication is now available based on the structure of VP1.460 VP1 has a compact cage-like structure with three domains similar to that of the reovirus polymerase.709 VP1 contains a putative cap-binding site to anchor the capped 5′ end of the (+)RNA, and the structure has four distinct tunnels that lead to the central catalytic core of VP1 (Fig. 45.10). Having a
fourth tunnel, a distinction from other known RNA-dependent RNA polymerases, is needed for the exit site of the (+)RNA transcripts. Based on information from reovirus polymerase-RNA elongation complexes,709 the four distinct tunnels in VP1 are implicated in (a) the entry of the templates ([+]RNA or [−]RNA), (b) the entry of NTPs, (c) the exit of dsRNA/(−)RNA, and (d) the exit of the (+)RNA. Having two distinct product exit tunnels ensures that (+)RNAs are effectively shuttled out of the core, while nascent dsRNA gene segments are directed toward the particle interior. In addition, the (−)RNA exits proximal to the template entry tunnel, which should facilitate its reuse in subsequent rounds of (+)RNA synthesis. Finally, the exiting of the (−)RNA template and (+)RNA product of transcription through separate tunnels suggests that the capping enzyme and polymerase are capable of internally separating dsRNA duplexes.
fourth tunnel, a distinction from other known RNA-dependent RNA polymerases, is needed for the exit site of the (+)RNA transcripts. Based on information from reovirus polymerase-RNA elongation complexes,709 the four distinct tunnels in VP1 are implicated in (a) the entry of the templates ([+]RNA or [−]RNA), (b) the entry of NTPs, (c) the exit of dsRNA/(−)RNA, and (d) the exit of the (+)RNA. Having two distinct product exit tunnels ensures that (+)RNAs are effectively shuttled out of the core, while nascent dsRNA gene segments are directed toward the particle interior. In addition, the (−)RNA exits proximal to the template entry tunnel, which should facilitate its reuse in subsequent rounds of (+)RNA synthesis. Finally, the exiting of the (−)RNA template and (+)RNA product of transcription through separate tunnels suggests that the capping enzyme and polymerase are capable of internally separating dsRNA duplexes.
Crystallographic structures of VP1 in complex with the consensus sequences (CSs) in the 3′ end of the minus strand (template for [+]RNA synthesis) and in the 3′ end of the (+)RNA (template for [−]RNA synthesis) are also available.460 These structures indicate that for transcription, sequence-specific recognition of the 3′ CS of the minus strand for positive-strand synthesis is not critical as it takes place within the confines of the capsid.460 In contrast, sequence-specific interactions are made only with the 3′ CS of the plus stand that is anchored to the template entry site in VP1. This sequence-specific interaction involving the UGUG motif in the 3′ CS (+)RNA confirms biochemical studies that showed this motif is the polymerase recognition signal and is required for high-affinity interactions with VP1 and subsequent replicase activity.112,115,116 Of interest, the VP1 3′ CS (+)RNA complex structure is in an auto-inhibited state with a single nucleotide at the 3′ end overshooting the initiation register. For VP1 to initiate (−)RNA synthesis, the overshot 3′ end of the template must be realigned, a priming loop must be repositioned to allow binding of the priming nucleotide, and a plug formed from the C-terminal domain of the polymerase that is in the dsRNA exit tunnel must be dislodged. These conformational changes in VP1 require VP2, GTP, the template, and Mg2+. Interactions with VP2 are known based on biochemical studies showing that interaction of RNA with VP2 is necessary for the initiation of rotavirus genome replication,576,824 but the details of how VP2 interacts with VP1 and how this interaction activates VP1 to initiate genome replication remain to be understood. They may require interactions with other viral proteins within assembling replication intermediates. Genome replication, RNA assortment and encapsidation, and particle assembly are highly coordinated events.
Kinetics and Cellular Sites of Transcription and Replication
Transcription in cells occurs following the release of DLPs from the endosome. Consistent with this idea, cells are susceptible to infection by liposome-mediated transfection of DLPs, indicating that simple delivery of these particles into the cell cytoplasm permits transcription to proceed.41 Transcription is asymmetric, and all transcripts are full-length positive strands made off the dsRNA negative strand.490 Primary transcription must occur before RNA replication. The synthesis of negative-strand RNA occurs in perinuclear nonmembranous, electron-dense cytoplasmic inclusions known as viroplasms (see later), concurrently with the packaging of positive-strand RNA into core replication intermediate (RI) particles.26,683
The kinetics of synthesis of positive- and negative-stranded RNAs has been studied in rotavirus-infected cells,697 in a cell-free system using extracts from infected cells,570 in an electrophoretic system that allows separation of the positive and negative strands of rotavirus RNAs in acid urea agarose gels,571,575 and by quantitative RT-PCR.26 Positive- and negative-stranded RNAs are initially detected during the first 4 hours after infection.26,697 A small linear increment of plus- and minus-strand RNA synthesis is detected followed by a logarithmic increase at later times of infection. This quantitation indicates that the entering DLPs produce a small amount of mRNA, which is then translated and replicated, producing new DLPs. When new DLPs are assembled, these particles then transcribe their genomes, initiating a secondary wave of transcription that significantly increases the amount of viral mRNA and dsRNA. Newly assembled DLPs are required for this second wave of transcription as knock-down of any of the viral proteins that constitute the DLP (VP1, VP2, VP3, and VP6) ablates the logarithmic increase in RNA synthesis.26 The amount of mRNA accumulated at late time points postinfection is significantly greater (at least six times more) than the dsRNA accumulated during the same period of time. The assembly of infectious virus particles parallels the replication of the viral genome.
Viroplasms, the sites of incorporation of (+)RNA into replication intermediates and virus assembly, first appear 2 to 3 hours after infection. The number of viroplasms initially increases and then decreases with time after infection, whereas the area of each viroplasm increases, suggesting fusions of viroplasms.204,226 Viroplasms contain viral proteins (VP1, VP2, VP3, VP6, NSP2, NSP5, and in some strains NSP6).287,591,592,628 NSP2 and NSP5 are major components of viroplasms, and expression of these two proteins alone is sufficient to induce the formation of empty viroplasm-like structures. Viroplasm nucleation may be due to NSP2–tubulin interactions as NSP2 sequesters free tubulin molecules and induces microtubule depolymerization in RV-infected cells.474 Microtubules also may play a role in the growth and fusion of viroplasms later in infection based on inhibition of these processes by nocodazole.95 Viroplasms associate with lipids and proteins (perilipin, adipocyte differentiation-related protein [ADRP]) characteristic of cytoplasmic lipid droplets (LDs), and blocking or interfering with LD formation reduces the number of functional viroplasms and production of infectious virus.122 The proteosome is also essential for early assembly of viroplasms.147,456 Inhibition of proteosome activity following virus entry and uncoating reduces accumulation of virus proteins, viroplasm formation, and RNA replication. The requirements of LDs and proteosomes for viroplasm formation represent examples of a virus hijacking cellular pathways for its own replication, and further information of how RV proteins interact or regulate these pathways should help understand the early stages of viroplasm formation and particle assembly.
The key role of NSP2 and NSP5 in viroplasm functions of genome replication and packaging has been demonstrated by studies of temperature-sensitive mutants or knocking down the expression of NSP2 or NSP5 by RNA interference or antibody treatment, which results in inhibition of viroplasm formation, genome replication, virion assembly, and a general decrease of viral protein synthesis.96,455,619,684,714,744
NSP2 is an essential multifunctional protein with sequence-independent ssRNA binding as well as enzymatic activities including nucleoside triphosphatase (NTPase), nucleoside diphosphate (NDP) kinase,428 RNA triphosphatase (RTPase),745 and nucleic acid helix destabilizing activities.710,711 NSP2 is the most abundant protein of viroplasms,592 is essential for viroplasm formation,619 and exists as an octamer. The monomeric subunit has two distinct domains separated by a deep, catalytic cleft. The association of monomers results in a doughnut-shaped octamer with a 35-Å central hole along the fourfold axes and grooves that run diagonally across the twofold axes and are lined by basic residues370 (Fig. 45.11). The C-terminal domain of NSP2 resembles the cellular histidine triad (HIT) family of proteins that hydrolyze nucleotides. The NTP-binding residues are located within the cleft between the two domains.100 Mutation of the catalytic residue (H225A) abrogates hydrolysis of the γ-phosphate from the 5′ end of RNA and dsRNA synthesis.712,746 Although the NTPase activity is localized in the monomeric subunit, the ability to bind RNA and other proteins requires the formation of the octamer.746 Cryo-EM structures of NSP2 octamer complexes show that both ssRNA and NSP5 (the other key component required for viroplasm formation) share the same binding site, the grooves in the NSP2 octamer, and NSP5 competes with ssRNA binding.376 Tubulin also binds to these charged grooves.475 In virus-infected cells, NSP2 is associated with polymerase complexes, including VP1 and VP2 and partially replicated viral RNA.18 Taken together, these results indicate that NSP2 is critical for RNA replication and suggest that competitive binding of different ligands to the groove may regulate NSP2 function during genome replication and virus assembly (Fig. 45.11).
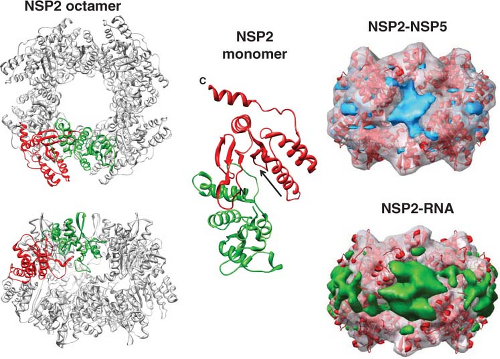 Figure 45.11. Structures of NSP2 and interactions with RNA and NSP5. Left: Octameric structure of NSP2. The N- and C-terminal domains of two subunits are colored green and red, respectively, and the rest of the subunits in the functional octamer are shown in gray. The donut-shaped octamer is viewed along the fourfold axis (top left) and along one of the two twofold axes perpendicular to the groove lined by positively charged residues (bottom left).370 Middle: NSP2 monomer showing the two subunits and catalytic cleft indicated by arrow. Right: Cryo-electron microscopy (cryo-EM) reconstruction of NSP2 binding to NSP5 (top with NSP5 in blue) and NSP2 binding to RNA (bottom with RNA in green) viewed perpendicular to the groove. These two ligands both bind to the grooves of the NSP2 octamer. (From Jiang et al.376 Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J Virol 2006;80:10829–10835.) |
NSP5 is a dimeric phosphoprotein rich in Ser and Thr residues that undergoes O-linked glycosylation,6,601,785 as well as phosphorylation that occurs when NSP5 is co-expressed with VP2.146 NSP5 reportedly exists in several oligomeric forms, and biophysical and structural analyses suggest that the
biologically relevant form in cells required for viroplasm formation is a decamer.475 The self-association of NSP5 may be regulated by NSP6, which is encoded by an alternative reading frame of gene segment 11 in most virus strains and is also associated with viroplasms.729 The precise roles of the modifications in NSP5 function remain unknown. However, the formation of viroplasm-like structures is also calcium regulated, and a pseudo-EF-hand motif in NSP5 (DxDxD) located upstream of a C-terminal helical domain possibly triggers calcium-dependent viroplasm formation.672 Although NSP5 binds to the grooves in NSP2, steric hindrance is proposed to prevent NSP2 from binding to all NSP5 promoters, and therefore, some NSP2 is thought to remain free to interact with other binding partners such as viral RNAs and the viral polymerase to perform functions other than viroplasm organization.475
biologically relevant form in cells required for viroplasm formation is a decamer.475 The self-association of NSP5 may be regulated by NSP6, which is encoded by an alternative reading frame of gene segment 11 in most virus strains and is also associated with viroplasms.729 The precise roles of the modifications in NSP5 function remain unknown. However, the formation of viroplasm-like structures is also calcium regulated, and a pseudo-EF-hand motif in NSP5 (DxDxD) located upstream of a C-terminal helical domain possibly triggers calcium-dependent viroplasm formation.672 Although NSP5 binds to the grooves in NSP2, steric hindrance is proposed to prevent NSP2 from binding to all NSP5 promoters, and therefore, some NSP2 is thought to remain free to interact with other binding partners such as viral RNAs and the viral polymerase to perform functions other than viroplasm organization.475
Viroplasm maturation and function also require NSP4, the only nonstructural protein that does not bind RNA. Based on siRNA experiments that knocked down NSP4 expression, NSP4 plays a role in the intracellular accumulation and the cellular distribution of several viral proteins, influencing the development of viroplasms, linking genome packaging with particle assembly, and acting as a modulator of viral transcription.453,684 A vesicular form of NSP4 is modulated by the levels of intracellular calcium, and that NSP4 also forms caps on viroplasms and co-localizes with the autophagy protein LC3.44 Precisely how NSP4 regulates RNA synthesis remains to be determined. Formation of the punctate vesicular structures requires the elevation of cytoplasmic calcium (Ca2+ cyto), which is mediated by NSP4 functioning as a viroporin in the ER membrane.356 NSP4 regulation of Ca2+ cyto may serve as a viroplasm assembly trigger that directs NSP5 to form viroplasms during viral infection, and later NSP4 binding to VP6 functions to inhibit polymerase activity, leading to a switch to complete genome replication and encapsidation.
Genomic RNA Replication and Encapsidation (Packaging)
After its synthesis, dsRNA remains associated with subviral particles, suggesting that free dsRNA is not found in cells. Because of its inherent stiffness,387 dsRNA is not packaged. Instead, (+)RNAs are assorted, replicated, and packaged within complexes that remain poorly understood, but subviral particles (complexes separable by sedimentation through sucrose gradients and by equilibrium centrifugation in CsCl gradients) in which dsRNA synthesis occurs have been characterized both in infected cells and in a cell-free system. Based on structural and biochemical studies, a model of rotavirus replication includes genome encapsidation and DLP assembly occurring concurrently with the formation of a replication intermediate composed of a pentamer of VP2 interacting with VP1 and VP3.488,585,586 In this model, 12 units each composed of pentamers of VP2 dimers, a VP1/VP3 complex, and a dsRNA segment associate to form the VP2 capsid layer, which provides a platform for the subsequent addition of VP6 trimers resulting in the formation of the DLP. Protein components in these units perhaps represent the replication complex in which the (+)RNA, brought in with the aid of NSP2/NSP5, is fed into the enzyme complex for (−)RNA synthesis and the formation of the duplex RNA, which is spooled around the enzyme complex. VP2 may interact with NSP549 and NSP5 with the NSP2 octamer. This multimeric complex would then provide a platform or scaffold for the replication complex.369 The NSP2 octamer binds (+)RNA and mediates unwinding of the RNA secondary structure via its nucleic acid helix destabilizing activity. The NSP2 octamer also binds the viral polymerase VP1 and may feed unwound RNA to the polymerase.403 The octamer is also an NTPase, but how this activity is used in rotavirus genome replication remains unclear. Structural studies to investigate how NSP2 interacts with nucleotides and hydrolyzes NTP indicate that the NTPase activity of NSP2 is associated with a phosphoryl-transfer function similar to that seen for cellular nucleoside diphosphate kinases. This kinase-like activity of NSP2 may have a role in the homeostasis of nucleotide pools within the viroplasm during genome replication.428 Overall, VP1 replicates the RNA, and the (−)RNA exits VP1 and interacts with an octamer of NSP2 so that NSP2, functioning as an RTPase, cleaves the γ-phosphate from the 5′(−)RNA.745 This accounts for the absence of the γ-phosphate at the 5′ end of the double-stranded genomic RNA.359,490 However, how NSP2 recognizes the 5′ CS of the (−)RNA of the dsRNA products exiting from VP1 is not understood, and this knowledge may be important to understand how NSP2 may play a role in facilitating genome packaging.
The selective packaging mechanism that leads to the presence of equimolar genome segments within rotaviruses, or any of the other members of the family Reoviridae, remains a challenging puzzle. Several models have been proposed.574,586 A currently favored model proposed earlier is based on structural data indicating that the core represents a collection of functionally separate pentameric units, with each unit containing its own RNA-dependent RNA polymerase activity and capping enzyme complex and being responsible for transcription of one of the genome segments.433 In this model, encapsidation would be concurrent with capsid assembly, and each VP1–VP3 enzymatic complex would associate with a specific mRNA and attract the VP2 core protein and assemble into pentamers (Fig. 45.12). RNA–RNA interactions between the mRNA of the distinct pentameric units would then drive the assembly of the icosahedral core from the pentameric units. Structural changes in the core lattice protein VP2, as a consequence of pentamer–pentamer binding, may activate the RNA-dependent RNA polymerase and stimulate negative-stranded RNA synthesis to form the genome. Another model, based on the ability of the rotavirus capsid proteins to self-assemble into empty VLPs157,431 and on data on the dsRNA bacteriophage phi6,510 suggests that empty cores are first made and that mRNAs would be replicated and subsequently inserted into these cores. Future work will determine which of these models, if any, may be correct.
Virion Maturation
A distinctive feature of rotavirus morphogenesis is that subviral particles, which assemble in the cytoplasmic viroplasms, bud through the membrane of the ER, and maturing particles are transiently enveloped (Fig. 45.8). This is one of the most interesting aspects of rotavirus replication, differing from members of other genera in the family Reoviridae and from any other virus. The envelope acquired in this process is lost as particles move toward the interior of the ER, and the envelope is replaced by a thin layer of protein that ultimately constitutes the outer capsid of mature virions. Rotavirus particle transport, maturation, and assembly remain an interesting model to understand the transport of protein complexes across the ER membrane as well as envelope particle formation.
Morphologic and biochemical data are consistent with rapidly assembling DLPs serving as an intermediate stage in the formation of triple-layered virions. The sites and precise details of RNA replication are beginning to be understood, and the viroplasms are the sites of synthesis of the double-layered particles that contain RNA (see earlier). This conclusion is based on the localization of several of the viral proteins (VP2, NSP2, NSP5, NSP6) to viroplasms, and of VP4 and VP6 to the space between the periphery of the viroplasm and the outside of the ER,287,591 and on the observation that particles emerging from these viroplasms bud directly into the ER that contains the glycoproteins VP7 and NSP4.
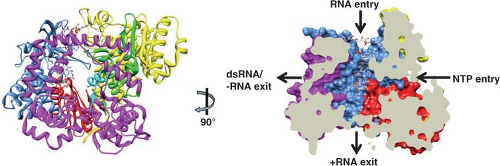 Figure 45.12. Structural features of VP1 polymerase. On the left, the structure of VP1 in complex with RNA oligonucleotide is shown.460 Ribbon diagram of VP1/RNA complex (PDB ID: 2R7R) is shown on the left. The N-terminal domain is in yellow, the C-terminal domain is in magenta, and the C-terminal plug is in cyan. The subdomains of the polymerase domain are in light blue (fingers), red (palm), and green (thumb). On the right is a cutaway of the surface rendering of the complex showing the four tunnels extending into the central cavity, the catalytic center. |
A fraction of VP4 can be detected in a filamentous array287 and at the plasma membrane associated with microtubules.178,538 Although the function of VP4 in lipid rafts at the plasma membrane in viral morphogenesis remains unclear, it has been proposed that VP4 is added to particles as an extra-ER event.178 Particles lacking VP4 can be formed in cells treated with siRNA to gene 4, suggesting that VP4 is not essential for assembly or for release of DLPs from the ER.176 Infectious particles are found associated with lipid rafts at the cell surface.163,165,361,664 Silencing VP4 and NSP4 (but not VP7) reduces rotavirus–raft interaction associations apparently by reducing targeting of VP4 to the rafts; these results support the idea that the primary association of VP4 with rafts occurs during the initial stages of particle assembly in the ER.163 Taken together, two pools of VP4 appear to exist, with one associating with particles in the ER and one being found independently at the plasma membrane.
In the ER of SA11-infected cells, two pools of VP7 exist that can be distinguished using two classes of antibodies.384 One pool, found only on intact particles, is detected only by a neutralizing mAb. The second pool of VP7 is unassembled, remains associated with the ER membrane, and is detected by a polyclonal antibody made to denatured VP7.384 Distinction of these two forms of VP7 permitted a kinetic study of the assembly of VP7 and of other structural proteins into particles. The incorporation of the inner capsid proteins into double-layered particles was found to occur rapidly, whereas VP4 and VP7 appear in mature TLPs with a lag time of 10 to 15 minutes. Kinetic analyses of the processing of the oligosaccharides on the two pools of VP7 show that the virus-associated VP7 oligosaccharides have a 15-minute lag compared with that of the membrane-associated form, suggesting that the latter is the precursor to virion VP7. This lag appears to represent the time required for virus budding and outer capsid assembly.384
NSP4 plays a key role in the assembly of TLPs. NSP4 is the only nonstructural protein that does not bind to RNA. NSP4 has been studied extensively because it plays a role in viral morphogenesis and functions as an enterotoxin (see later). NSP4 has multiple domains and an increasing number of functions.32,215 NSP4 is a 20K primary translation product; it is co-translationally glycosylated to become a 29K species, and oligosaccharide processing yields the mature 28K protein that is a transmembrane protein of the ER.209,383 The 175-aa polypeptide backbone of NSP4 consists of an uncleaved signal sequence, three hydrophobic domains with two N-linked high mannose glycosylation sites being in the first hydrophobic domain, a predicted amphipathic α-helix (AAH) that overlaps a folded coiled-coil region, the H2 transmembrane domain that traverses the ER bilayer, and the C-terminus, which is hydrophilic and forms an extended cytoplasmic domain.71,108,210,383 The carbohydrate moieties remain sensitive to endoglycosidase H digestion, and processing of the Man9GlcNAc carbohydrate added to NSP4 stops at Man8GlcNAc with the mannose-9 species predominating,71,383 indicating that no further trimming occurs in the Golgi apparatus.
The C-terminal cytoplasmic domain (aa 161 to 175) of NSP4 functions in viral morphogenesis by acting as an intracellular receptor on the ER membrane.24,718,719 NSP4 binds newly made DLPs and mediates the budding of these particles into the ER lumen (see later). A receptor role for NSP4 is supported by the observation that DLPs bind to ER membranes containing only NSP4.23,24,502 The AAH region, distinct from the receptor domain, is predicted to adopt an α-helical coiled-coil structure
and is thought to mediate oligomerization of the virus-binding domains into a homotetramer.720 A crystal structure of the oligomerization domain of NSP4, which spans aa residues 95 to 137 (NSP4 95 to 137), self-associates into a homotetrameric coiled-coil, with the hydrophobic core interrupted by three polar layers and two of the four Glu120 residues coordinating a divalent cation72 (Fig. 45.11). Recently, crystallization of a similar domain revealed a pentamer that lacks a cation-binding site.106 Sequence analyses have identified 14 types (14E types) of NSP4 in RVA strains (Table 45.4). The highest sequence diversity of NSP4 is located in the cytoplasmic domain. It is unclear whether this sequence diversity is important and driven by interactions with specific residues on VP6 or divergent regions on VP4, or both. However, specific combinations of types of VP6 and NSP4 have been found in natural reassortants, suggesting these interactions are biologically important.365 There also may be host-specific interactions important for function, but these remain to be fully characterized.
and is thought to mediate oligomerization of the virus-binding domains into a homotetramer.720 A crystal structure of the oligomerization domain of NSP4, which spans aa residues 95 to 137 (NSP4 95 to 137), self-associates into a homotetrameric coiled-coil, with the hydrophobic core interrupted by three polar layers and two of the four Glu120 residues coordinating a divalent cation72 (Fig. 45.11). Recently, crystallization of a similar domain revealed a pentamer that lacks a cation-binding site.106 Sequence analyses have identified 14 types (14E types) of NSP4 in RVA strains (Table 45.4). The highest sequence diversity of NSP4 is located in the cytoplasmic domain. It is unclear whether this sequence diversity is important and driven by interactions with specific residues on VP6 or divergent regions on VP4, or both. However, specific combinations of types of VP6 and NSP4 have been found in natural reassortants, suggesting these interactions are biologically important.365 There also may be host-specific interactions important for function, but these remain to be fully characterized.
Glycosylation of NSP4 is not required for its binding activity to DLP or for oligomerization, but it is required for interaction with calnexin.24,511,719 NSP4 also has a binding site for VP5* within VP424,357 and may play a role in removing the transient envelope.725 Heterooligomers of NSP4, VP4, and VP7 have been detected in enveloped particles,465,602 and calcium has been shown to be important for oligomerization of these proteins in the ER603 as well as for proper folding of VP7 epitopes and outer capsid assembly.17,190,677 The precise mechanisms of how (a) the envelope on particles is removed, (b) the hetero-oligomeric complexes function in particle budding through the ER, and (c) the outer capsid is assembled onto the newly made DLPs remain poorly understood. However, siRNA experiments indicate that VP4 and VP7 are assembled onto the particle in the ER and VP7 is involved in removal of the transient envelope.163,453
Rotavirus maturation is a calcium-dependent process, and virus yields are decreased when virus is produced in cells maintained in calcium-depleted medium.678 Viruses produced in the absence of calcium are almost exclusively DLPs, and budding of virus particles into the ER is not observed.677 The crystal structure of VP7 provides new understanding of the requirement of calcium in forming TLPs. VP7 forms calcium-dependent trimers (Fig. 45.4); two calcium ions coordinate between each VP7 monomer and stabilize the trimeric interactions. Electron cryomicroscopy and single-particle reconstruction of VP7-recoated DLPs show that the N-termini (residues 51 to 70) of VP7 interact with VP6 and the VP7 trimers clamp onto the VP6 trimer.121 Interestingly, unglycosylated VP7 made in the presence of tunicamycin is relatively stable in a calcium-free environment. The budding process can occur in the absence of calcium, but VP7 is retained within the ER.8 VP7 does not fold properly unless it is expressed with other rotavirus proteins, and calcium must be present in cells for correct epitope formation.190,191 Outer capsid assembly also requires proper formation of disulfide bonds on VP7.703 Earlier studies showed treatment of cells with various agents (tunicamycin, dithiothreitol, or calcium-blocking drugs, such as thapsigargin) results in a build-up of enveloped particles in the ER.341,503,589 These agents may disrupt the proper folding of VP7 that is required for removing the envelope.
NSP4 is a novel calcium agonist; it mobilizes intracellular calcium when expressed intracellularly by a phospholipase C (PLC)-independent mechanism or by a PLC-dependent mechanism when it is added to cells from the outside.518 Mobilization of intracellular calcium by a PLC-independent mechanism occurs through NSP4 functioning as a viroporin356 that is important for viral replication and assembly. Binding of VP6 on DLPs to NSP4 that occurs when viroplasms are capped by NSP4 is thought to trigger the budding process. Recent reports indicate that siRNA silencing of NSP4 expression in rotavirus-infected cells affects the distribution of other viral proteins, mRNA synthesis, and the formation of viroplasms where viral RNA replicates, suggesting global regulatory functions of NSP4 in rotavirus replication453,684 with molecular mechanisms that remain to be understood.
At least three pools of intracellular NSP4 exist in rotavirus-infected cells dependent on the level of NSP4 protein expression.44 The first pool is represented by NSP4 localized in the ER membrane and is present throughout the course of infection. This pool serves as a receptor for the budding of immature viral particles into the ER, as described earlier, at the peak of viral infection, when all viral proteins are abundant (after 6 hours after infection). A second minor pool of NSP4 molecules enters the ER-Golgi intermediate compartment (ERGIC) and can be recycled back to the ER or may be a part of the nonclassical secretion pathway for delivery and cleavage of an NSP4 peptide into the medium of infected cells at early time points after infection826 when the levels of viral proteins are relatively low. The third pool of NSP4 molecules, distributed in cytoplasmic vesicular structures associated with the autophagosomal marker LC3 and viroplasms, is regulated by calcium levels and appears at 6 hours after infection, when there is an increase of intracellular calcium levels because of increased expression of viral proteins.504 NSP4 also interacts with calveolin-1, and this may be involved in the secretion of NSP4.566,826 Inhibition of NSP4 expression interferes with the formation of large viroplasms and affects viral protein expression455 and secondary viral mRNA synthesis in rotavirus-infected cells.684 The NSP4- and autophagic marker LC3-positive vesicles may serve as a lipid membrane scaffold for the formation of large viroplasms by recruiting early viroplasms or viroplasm-like structures formed by NSP2 and NSP5.226 These NSP4-positive membranes may also function to regulate packaging of the rotavirus genome and transcription through NSP4 association with VP6 on DLPs. The calcium-dependent compartmentalization of NSP4 into an autophagosomal pathway raises questions regarding the involvement of autophagosomal membranes in rotavirus replication and release of infectious virus from cells.
Understanding viral morphogenesis has been facilitated by the expression of the rotavirus structural proteins individually or in combinations in insect cells using recombinant baculoviruses.157,431,623,823 This approach first showed that the single-layered VP2 particle shell self-assembles when VP2 is expressed alone, and that all of the other capsid proteins can self-assemble into virus-like particles when co-expressed in the proper combinations. Virus-like particles composed of VP2, VP1/2, VP1/2/3, VP2/3, VP2/6, VP2/6/7, VP2/4/6/7, and VP1/2/3/6 can be made.157,823,824 The outer and inner capsid proteins of different virus strains can also reassociate and are able to be transcapsidated onto other virus strains117,118 and infectious virus is produced.731 These results demonstrate that the structural proteins contain the intrinsic information required to form particles and that co-expression of mutant
proteins is a feasible approach to analyze the domains responsible for the structural interactions between the proteins composing the virus particles. These particles have been useful to (a) analyze the role of cleavage sites in the spike protein in infectivity,272 (b) investigate the role of individual structural proteins in inducing protective immunity,130,156,371,543,544 (c) probe the inner structure of particles by analyzing difference maps of particles with distinct protein compositions or by x-ray analysis,211,604,675 and (d) analyze RNA transcription and replication110,114,434,435 and RNA packaging and assembly. Future studies should address questions of which mechanisms control packaging of the viral genome and virus assembly.
proteins is a feasible approach to analyze the domains responsible for the structural interactions between the proteins composing the virus particles. These particles have been useful to (a) analyze the role of cleavage sites in the spike protein in infectivity,272 (b) investigate the role of individual structural proteins in inducing protective immunity,130,156,371,543,544 (c) probe the inner structure of particles by analyzing difference maps of particles with distinct protein compositions or by x-ray analysis,211,604,675 and (d) analyze RNA transcription and replication110,114,434,435 and RNA packaging and assembly. Future studies should address questions of which mechanisms control packaging of the viral genome and virus assembly.
Virus Release
Electron microscopy studies have shown that the infectious cycle ends when progeny virus is released by host cell lysis in nonpolarized cells.12,111,497 Extensive cytolysis late during infection and drastic alterations in the permeability of the plasma membrane of infected cells result in the release of cellular and viral proteins.527 Despite cell lysis, most DLPs and many TLPs remain associated with the cellular debris, suggesting that these particles interact with structures within cells. Interactions with cell membranes and the cell cytoskeleton have been suggested,527 and virus purification procedures generally use Freon extraction to release particles from cellular debris. Whether the cytoskeleton provides a means of transport of viral proteins and particles to discrete sites in the cell for assembly or acts as a stabilizing element at the assembly site and in the newly budded virions or whether particles are simply trapped by the cytoskeleton remains to be determined. VP4 interacts with actin and lipid rafts and can remodel microfilaments, and this has been suggested as a mechanism by which the brush border membrane of polarized epithelial cells is destabilized to facilitate rotavirus exit from cells.264
Rotavirus Effects on the Host Cell
Inhibition of Translation of Cellular mRNAs by NSP3
Viral mRNAs are capped but not polyadenylated, and viral proteins are translated by the cellular translation machinery. Most of the rotavirus structural proteins and the nonstructural proteins are synthesized on free ribosomes, although nascent proteins on free ribosomes have not been analyzed. Instead, this conclusion has been drawn based on the absence of signal sequences that would indicate targeting to the ER and lack of protection from digestion in in vitro protease protection studies.210,383 The viral glycoproteins VP7 and NSP4 are synthesized on ribosomes associated with the membrane of the ER and are co-translationally inserted into the ER membrane as a result of signal sequences at their N-termini. VP7 has a signal sequence that is co-translationally cleaved, whereas the signal sequence on NSP4 is not cleaved.
Translation of the viral mRNAs that are capped but not polyadenylated is facilitated by the action of the nonstructural protein NSP3, which is one of the five nonstructural proteins (NSP1, 2, 3, 5, and 6) that bind nucleic acid.671,713 NSP3 function parallels that of the cellular poly(A)-binding protein (PABP). The N-terminus of NSP3 interacts with the 3′ consensus sequence (UGACC) of viral mRNAs and the C-terminus of NSP3 interacts with eIF4G as does PAPB, but with higher affinity.311,596,597,600,748 These events lead to NSP3 evicting PABP from eIF4G, to the enhancement of translation of rotavirus mRNAs, and to the concomitant impairment of translation of cellular mRNAs.558,748 PABP evicted from eIF4G accumulates in the nucleus of rotavirus-infected cells, and this relocalization of PABP from the cytoplasm to the nucleus occurs relatively early in the infection cycle (∼3 hpi) and requires a limited amount of NSP3.328,516 The complete depletion of PABP from the cell cytoplasm can reinforce the shutoff of translation of cellular polyadenylated mRNAs and may also enhance translation of viral mRNAs by making available other cellular factors involved in translation termination, RNA stability, or subcellular localization of host mRNAs that would normally be bound to cytoplasmic PABP.328
In vivo, NSP3 stimulates the translation of mRNAs in synergy with the cap structure, possibly enabling circularization of viral mRNAs and its delivery to ribosomes for viral protein synthesis126,748 (Fig. 45.12). Atomic structures of both domains of NSP3 complexed with bound ligands indicate that both domains have novel folds and NSP3 functions as a dimer (Fig. 45.13).182,311 Whereas the RNA-binding domain forms a rod-shaped symmetric dimer, the N-terminal domain tightly binds to the consensus 3′ end of the mRNAs inside a tunnel formed at the dimeric interface. NSP3 and eIF4G also interact with a novel cellular protein, named RoXaN, which has a role in translation regulation that remains to be completely understood.765 However, the nuclear localization of PABP seen in rotavirus-infected cells is dependent on the capacity of NSP3 to interact with eIF4G and also requires the interaction of NSP3 with a specific region in RoXaN. This domain functions as a nuclear export signal and RoXaN tethers PABP with RNA. Thus, RoXaN is a cellular partner of NSP3 involved in the nucleocytoplasmic localization of PABP.328 The cellular chaperone heat shock protein 90 also binds to NSP3, prevents proteosomal degradation of NSP3, and increases dimerization of NSP3 required for its functioning to bind eIF4G and effect translocation of PABP to the nucleus.199 This chaperone is induced by virus infection though activation of phosphoinositide 3-kinase (PI3K)/Akt and nuclear factor-κB (NF-κB) signaling that positively regulate rotavirus infection.198,199
The binding of NSP3 to viral mRNAs has also been proposed as a possible mechanism to transport newly made mRNAs to viroplasms for subsequent replication. The cytoskeleton-binding function of NSP3288,487 might be involved in this process directly or NSP3 may interact with other host proteins that link the translational machinery and the cytoskeleton.
While NSP3 indeed inhibits translation of cellular mRNAs, a second mechanism of rotavirus-induced inhibition of cellular mRNA translation involves eIF2α that is phosphorylated early after infection and is maintained throughout the virus replication cycle.515,516 This phosphorylation depends on the synthesis of VP2, NSP2, and NSP5, and the continuous phosphorylated status of eIF2α is beneficial for the virus because viral mRNAs are preferentially translated efficiently while translation of most cellular proteins is stopped. Protein kinase R (PKR) is apparently responsible for this phosphorylation event that is triggered by viral double-stranded RNA detected in the cell cytoplasm outside viroplasms, which is a paradigm-shifting result as traditionally it has been assumed that rotaviral dsRNA is hidden from the interferon system by ensuring that genome replication takes place within replicative intermediate particles, such that single-stranded RNA is replicated as it enters these
particles.572 While further work is needed to characterize the nature of the viral dsRNA present in the cytoplasm of infected cells, the multiple mechanisms used by rotaviruses to remodel the host translation machinery in novel ways to ensure efficient translation of viral proteins will likely continue to reveal unique regulatory systems.636
particles.572 While further work is needed to characterize the nature of the viral dsRNA present in the cytoplasm of infected cells, the multiple mechanisms used by rotaviruses to remodel the host translation machinery in novel ways to ensure efficient translation of viral proteins will likely continue to reveal unique regulatory systems.636
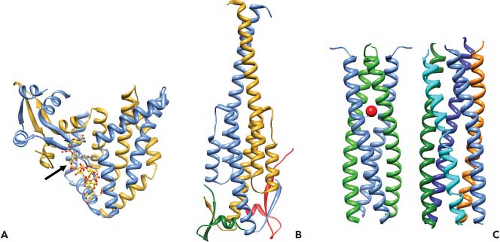 Figure 45.13. X-ray structures of NSP3 and NSP4. A: Structure of the NSP3 N-terminal domain homodimer in complex with the 3′ consensus sequence of rotavirus messenger RNA (mRNA).182 The dimeric subunits are shown in blue and yellow. A single 5′ RNA segment (shown in ball and stick representation and indicated by an arrow) is buried within a basic deep tunnel formed in the asymmetric homodimer (shown in blue and golden yellow). Each subunit participates in different interactions with the mRNA segments. B: Structure of NSP3 C-terminal domain homodimer (shown in blue and golden yellow) in complex with the dimer of the short peptide of eIF4G (shown in green and red).311 C: Ribbon representation of the homotetramer (left) with each dimer shown in blue and green and the homopentamer (right) of NSP4 95–137. The red circle indicates the location of a divalent metal binding site, possibly binding calcium in the tetramer.72,106 |
Rotavirus Effects on Cellular Signaling Pathways and Apoptosis
The effects of rotavirus replication on cultured cells are cell-type specific. In nonpolarized tissue-cultured cells, rotaviruses are normally cytocidal viruses that rapidly kill the permissive cells they infect. Adaptation of rotaviruses to culture can be difficult and may require initial passage of virus in primary monkey kidney cells before adaptation to growth in continuous monkey kidney cell lines. Fully permissive cells generally exhibit cytopathic effect, and cell death is preceded by the shut-off of host RNA, DNA, and protein synthesis.99,209,489 Cell death seems to result from the function of a viral gene on a specific target rather than from cumulative effects on host metabolism, because certain ts mutants (ts groups F [VP2], G [VP6], H, and I [unassigned NSP3 and NSP4] [Table 45.4]) do not efficiently shut off host cell protein synthesis.216,619 NSP4 is a viroporin and a novel calcium agonist that mediates cell death by causing intracellular calcium levels to increase as well as by affecting the plasma membrane permeability and tight junctions of cells.504,540,704,726 Rotavirus induces the unfolded protein response of the cell but also controls it through NSP3 that reduces host protein synthesis.597,738 Rotavirus has been reported to induce apoptosis in polarized epithelial cell lines as well as in mice infected with a murine strain of virus.69,107,702 Replication-competent virus is required to induce apoptosis, indicating that viral gene expression is a prerequisite and, when examined, intracellular calcium levels play a role, suggesting an effect of NSP4.107
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree