Retroviridae
Stephen P. Goff
The retrovirus family, the Retroviridae, are a large and diverse group of viruses found in all vertebrates. These viruses replicate through an extraordinary and unique life cycle, differentiating them sharply from other viruses. The virion particles generally contain a genomic RNA, but upon entry into the host cell, this RNA is reverse transcribed into a DNA form of the genome that is integrated into the host chromosomal DNA. The integrated form of the viral DNA, the provirus, then serves as the template for the formation of viral RNAs and proteins that assemble progeny virions. These features of life cycle—especially the reverse flow of genetic information from RNA to DNA, and the establishment of the DNA in an integrated form in the host genome—are the defining hallmarks of the retroviruses. This life cycle also accounts for many of their diverse biological activities. The creation of the proviral DNA confers on the viruses a powerful ability to maintain a persistent infection in the face of a host immune response and to enter the germ line, permitting the vertical transmission of virus.
The retroviruses have played a unique role in the history of molecular biology. They have attracted attention on several grounds.
Biochemistry: The viral replication enzymes, including the reverse transcriptase (RT) and integrase (IN), are extraordinarily useful tools in manipulating nucleic acids in vitro and in vivo. Through the preparation of complementary DNAs (cDNAs), RT has been crucial for studies of messenger RNA (mRNA) synthesis and gene regulation.
Pathogenicity: Retroviruses are known as major pathogens affecting nearly all vertebrates. HIV-1, the agent of the AIDS pandemic, will probably cause more human death and suffering than all but a handful of pathogens in recorded history.
Markers of evolutionary history: The insertion of a provirus into the germ line provides a Mendelian tag that marks an event at a particular time in evolution. The inheritance of that tag can then be used to follow speciation, population migrations, and evolution of species.
Insertional activation of oncogenes: The integration of retroviral DNA is inherently mutagenic; retrovirus replication thus causes gross alterations of host genes and patterns of gene expression. When insertions lead to tumor formation, the locations serve to identify new oncogenes.
Transduction: Retroviruses can acquire host sequences in the formation of acutely transforming genomes. The identity, structure, and expression of these genes has provided much of our current knowledge of the routes by which normal growth control can be subverted by genetic alterations.
Gene delivery vectors: The structure of transforming viruses provided a model for the use of retroviruses to deliver therapeutic genes efficiently and cleanly into cells. Retroviruses now serve as major tools in the medical black bag of gene therapists.
This chapter will describe the replication and molecular biology of the retroviruses, concentrating on the most broadly conserved aspects of the life cycle. Because of the magnitude of the retroviral literature, citations here cannot be comprehensive, and referencing has been selective and concentrated on more recent publications. The distinctive features of the human retroviruses, especially the lentiviruses and spumaviruses, will be addressed in much more detail in other chapters. A comprehensive review of retroviral biology (called the Retroviruses; [108]) is still current, and should be consulted for additional details of almost all aspects of their replication.
Taxonomic Classification
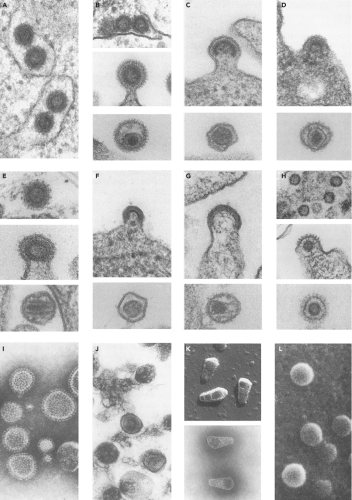
The retroviruses were originally classified by the morphology of the virion core as visualized in the electron microscope. Examples of the appearance of the virions in these micrographs are presented in Figure 47.1. The virion particles are spherical, and are surrounded by an envelope consisting of a lipid membrane bilayer. The surface is studded by projections of an envelope glycoprotein. There is a spherical layer of protein under the membrane, and an internal nucleocapsid (or nucleoid) whose shape varies characteristically from virus to virus. The shape and position of the nucleocapsid core was historically used as the major classifying feature of the retroviral genera. A-type viruses were defined as those forming intracellular structures with a characteristic morphology, a thick shell with a hollow, electron-lucent center. These particles are now appreciated as representing an immature capsid on route toward the formation of other structures. This term is thus no longer in use to denote a virus classification, though it is used to describe the structures formed by some virus-related intracellular retrotransposons (the intracisternal A-type particles, or IAPs).307,349 B-type viruses show a round but eccentrically positioned inner core. C-type viruses assemble at the plasma membrane, and contain a central, symmetrically placed, spherical inner core. The D-type viruses assemble in the cytoplasm, via an A-type intermediate, and upon budding exhibit a distinctive cylindrical core.
These older classifications have been useful in partially defining the various genera of the family Retroviridae, but the number of genera have now been expanded on the basis of new criteria. The genera have recently been formalized and given new names by the International Committee on Taxonomy of Viruses. The alpharetroviruses, betaretroviruses, and gammaretroviruses are considered “simple” retroviruses, while the deltaretroviruses, epsilonretroviruses, lentiviruses, and spumaviruses are
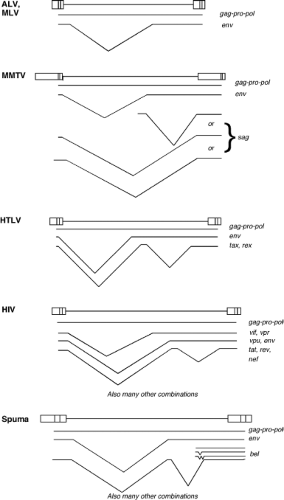
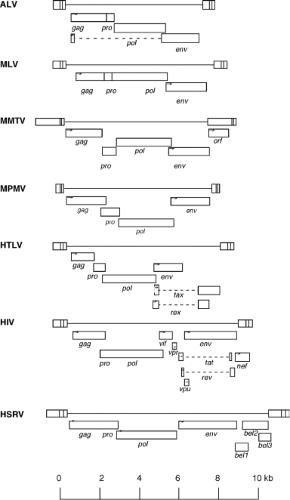
considered “complex.” The simple viruses encode only the Gag, Pro, Pol, and Env gene products; the complex viruses encode these same gene products but also an array of small regulatory proteins with a range of functions. The properties of the viruses belonging to each of these genera are summarized briefly in the following section. Representative members of each genus are listed in Table 47.1.
considered “complex.” The simple viruses encode only the Gag, Pro, Pol, and Env gene products; the complex viruses encode these same gene products but also an array of small regulatory proteins with a range of functions. The properties of the viruses belonging to each of these genera are summarized briefly in the following section. Representative members of each genus are listed in Table 47.1.
Table 47.1 Retrovirus Genera | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Alpharetroviruses
The alpharetroviruses are simple retroviruses characterized by a C-type morphology, and are typified by the avian sarcoma and leukosis viruses (ALSV). The genome contains gag, pro, pol, and env genes, with no additional known genes; pro is at the 3′ end of gag and in the same reading frame. The transfer RNA (tRNA) primer is tRNAtrp. The viruses are widespread in many avian host species. The ALSV members are classified into 10 subgroups (termed A–J) by their distinct receptor utilization. The first four subgroups represent exogenous viruses of chickens; the subgroup E includes a family of endogenous chicken viruses; and subgroups F and G include endogenous viruses of pheasants.
Betaretroviruses
The betaretroviruses are simple retroviruses characterized by either a “B-type” morphology, with a round eccentric core, or “D-type” morphology, with a cylindrical core. The best-known examples are the mouse mammary tumor virus (MMTV) and the Mason-Pfizer monkey virus (MPMV). Assembly occurs in the cytoplasm via an “A-type” intermediate, and the completed immature particle is then transported to the plasma membrane
and budded. The genomes contain gag, pro, pol, and env genes, and the gag, pro, and pol genes are all in different reading frames. The genome of MMTV contains an additional gene termed the sag gene for superantigen. The viruses also contain a dUTPase region as part of the pro open reading frame (ORF).156 The tRNA primer is tRNALys-3 or tRNALys-1,2. There are both exogenous and endogenous viruses in this genus. Examples are found in mice, primates, and sheep.
and budded. The genomes contain gag, pro, pol, and env genes, and the gag, pro, and pol genes are all in different reading frames. The genome of MMTV contains an additional gene termed the sag gene for superantigen. The viruses also contain a dUTPase region as part of the pro open reading frame (ORF).156 The tRNA primer is tRNALys-3 or tRNALys-1,2. There are both exogenous and endogenous viruses in this genus. Examples are found in mice, primates, and sheep.
Gammaretroviruses
The gammaretroviruses are simple viruses characterized by a C-type morphology. This genus has the largest number of members known, including the murine leukemia viruses (MuLVs), the feline leukemia viruses (FeLVs), and the gibbon ape leukemia virus (GALV). The genome contains only gag, pro, pol, and env genes; the gag, pro, and pol sequences are in the same reading frame, and the Gag-Pro-Pol protein is expressed by translational readthrough of a stop codon at the end of gag. The genome primer is most often tRNApro or tRNAglu. The murine viruses are divided into subgroups by their distinct receptor utilization. Many exogenous and endogenous viruses are found in diverse mammals; examples have been isolated from reptiles and birds. A novel gammaretrovirus termed XMRV (for xenotropic murine leukemia virus-like virus) was identified in human prostate cancer tumors,604 but recent work strongly suggests that the virus was a recombinant derived during tumor passage in nude mice.452
Deltaretroviruses
The deltaretroviruses are complex viruses characterized by a C-type morphology. The most famous examples are the human T-lymphotropic viruses (HTLVs) and the bovine leukemia virus (BLV). The genome contains gag, pro, pol, and env genes; the gag, pro, and pol genes are present in three different reading frames, and expression of the Gag-Pro-Pol protein requires two successive frameshifts. In addition, the genomes contain regulatory genes termed rex and tax that are expressed from an alternatively spliced mRNA. These gene products control the synthesis and processing of the viral RNAs. The tRNA primer is tRNApro. No closely related endogenous viruses are known, and the exogenous viruses are only rarely found in a few mammals.
Epsilonretroviruses
The epsilonretroviruses are complex viruses characterized by a C-type morphology. The prototype is the walleye dermal sarcoma virus (WDSV). The genomes contain gag, pro, pol, and env genes; the gag, pro, and pol genes are in the same reading frame. They also contain one to three additional genes termed ORFs A, B, and C. The ORFa gene is a viral homolog of the host cyclin D gene, and so may regulate the cell cycle. The viruses use tRNAHis or Arg as primers. The only known examples are exogenous viruses in fish and reptiles.
Lentiviruses
The lentiviruses are complex viruses characterized by a unique virion morphology, with cylindrical or conical cores. The most important example is the human immunodeficiency virus type 1 (HIV-1), but nonprimate viruses in the genus include the caprine arthritis encephalitis virus (CAEV) and visna virus. The genomes express gag, pro, pol, and env genes; gag is in one reading frame, and pro–pol in another. A single frameshift is used to express Gag-Pro-Pol. The Pol region of the nonprimate lentiviruses includes a domain for dUTPase. A number of accessory genes are also expressed. In HIV-1, these genes are vif, vpr, vpu, tat, rev, and nef; these genes control transcription, RNA processing, virion assembly, and host gene expression, and inactivate host restriction systems. The tRNA primer is tRNALys1,2. A large number of exogenous viruses in this genus have been found in diverse mammals, but the only endogenous sequences are relatively distant from these viruses.
Spumaviruses
The spumaviruses are complex viruses with a unique virion morphology, containing prominent spikes on the surface and a central but uncondensed core. The prototype example is the human foamy virus. The virion is assembled in the cytoplasm and budded into the ER and plasma membrane. There is probably only a single cleavage of the Gag protein near the C-terminus, and no major change in morphology during maturation. The genomes express gag, pro, pol, and env genes, and also at least two accessory genes known as tas/bel-1, and bet.177,380 The tas gene encodes a transcriptional transactivator. Unique features are the separate expression of the Pol protein from a spliced mRNA and the presence of large amounts of reverse transcribed DNA in the virion.390 The genome contains a second transcriptional start site near the 3′ end of the env gene. The tRNA primer is tRNALys1,2. A number of exogenous viruses have been found in diverse mammals, and distantly related sequences are present as endogenous elements in the human genome.
Evolutionary Relationships
The sequences of the various retroviral genomes have been compared and used to determine the relatedness of any pair.375 A number of phylogenetic trees can be constructed using gag, pro, pol, or env genes, and in most aspects these trees are similar. A tree based on comparisons of the pol gene (Fig. 47.2) shows the clustering of viruses within each of the main genera. However, it is important to realize that a phylogenetic tree is not necessarily identical to an evolutionary history, and that the history that led to the formation of the known genera is not necessarily simple. It is noteworthy that there is no obvious clustering of all the simple viruses into a group apart from all the complex viruses. Thus, complex viruses probably arose from the simple ones more than once, with many evolving through the independent acquisition of separate genes.
The retroviruses are related to viruses of other families. The retroviral RTs show close sequence similarity to the polymerases of the hepadnaviruses and the caulimoviruses, which also replicate by reverse transcription. The retroviruses also show extensive similarity in both gag and pol gene sequences to the retrotransposons, endogenous mobile elements with long terminal repeats (LTRs), and to retroposons, elements without LTRs. Retroviral RTs show more distant similarity to proteins encoded by the group II mitochondrial introns and by the retrons, elements in myxobacteria and rare isolates of E. coli; to telomerase, an RT responsible for maintenance of the chromosomal termini in eukaryotes; and slight similarity to the DNA polymerases of viruses and hosts.374
Transforming Viruses
During the replication of any retrovirus, replication-defective variants can arise through deletion or recombination events.
Such mutants or variants can be propagated as a mixed virus culture along with the wild-type parent. In these mixtures of two genomes, the replication-competent parent acts as a helper virus to provide the missing replication functions in trans for the replication-defective virus. If a newly acquired gene product is mitogenic or antiapoptotic for the host cell, or in more subtle ways alters the growth of the cell, the recombinant may become a potent oncogenic virus. A large number of such transducing viruses have been isolated and characterized as derivatives of one or another of the replication-competent parent viruses. A partial listing of the most intensely studied of these viruses is presented in Table 47.2.
Such mutants or variants can be propagated as a mixed virus culture along with the wild-type parent. In these mixtures of two genomes, the replication-competent parent acts as a helper virus to provide the missing replication functions in trans for the replication-defective virus. If a newly acquired gene product is mitogenic or antiapoptotic for the host cell, or in more subtle ways alters the growth of the cell, the recombinant may become a potent oncogenic virus. A large number of such transducing viruses have been isolated and characterized as derivatives of one or another of the replication-competent parent viruses. A partial listing of the most intensely studied of these viruses is presented in Table 47.2.
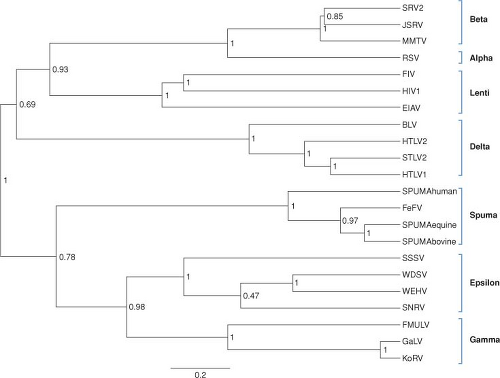 Figure 47.2. Phylogenetic reconstruction of representative exogenous retroviruses using reverse transcriptase sequences. The BEASTv1.6.1 tree144 was created using two independent Bayesian MCMC chains (length of 1 million, 20% burn) run under relaxed clock (uncorrelated exp; 143) and rate heterogeneity among sites (gamma distribution with 8 categories). Monophyletic taxon sets consisting of alpha, beta, delta, epsilon, gamma, lenti, and spuma were also used in the model. The posterior probabilities label each node and branch lengths are scaled to expected substitutions per site. (Prepared by Marcella McClure, Montana State University, Bozeman, MT.) |
Virion Structure
Retrovirus virions are initially assembled and released from infected cells as immature particles containing unprocessed Gag and Gag-Pol precursors of the proteins that eventually make up the mature virus. The immature virion morphology is spherical, with a characteristic electron-lucent center. The virions have been described as a “protein vesicle,” to suggest some fluidity in the interactions between the individual Gag proteins that make up the particle. Upon maturation, the precursor proteins are cleaved, and the structure and morphology of the virion change drastically. The mature retrovirus particle is a spherical structure, roughly 100 nm in diameter. The size of the virions in a given preparation is not highly homogeneous but rather varies over a fairly wide range, suggesting that a discrete, highly ordered structure may not exist. After processing of the Gag precursor during virion maturation, the CA protein collapses to form a more ordered paracrystalline core, but even then the overall diameter of the virion is heterogeneous and suggestive of considerable disorder. The virions exhibit a buoyant density in sucrose in the range of 1.16 to 1.18 g/ml. The sedimentation rate of the particles is typically about 600 S. The virions are sensitive to heat, detergent, and formaldehyde.
Virion Proteins
The stoichiometry of the various viral gene products in the virion is not very firmly established, but estimates suggest that about 1500 Gag precursors are present per particle. After processing, all cleavage products are thought to be retained, suggesting equimolar presence of these proteins in the mature virions. The levels of the Pol proteins are typically about one-tenth to one-twentieth those of the Gag proteins, corresponding to about 100 to 200 molecules per virion. The levels of the Env proteins are highly variable among the viruses. For the gammaretroviruses, the levels of Env are close to that of Gag; perhaps 1200 monomers, or 400 trimers, are present per virion. For the lentiviruses, the levels of Env per virion are much lower, possibly as low as 10 trimers per virion.671
Table 47.2 Examples of Acute Transforming Retroviruses | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||
Nomenclature
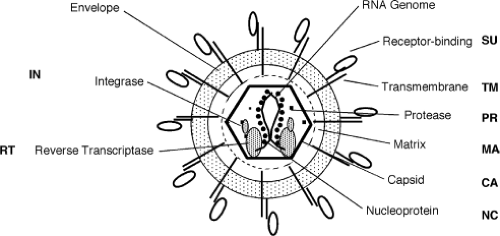
The cleavage of Gag, Pol and Env precursors forms the products in the mature infectious virions. These proteins are named by convention by a two-letter code: MA for matrix or membrane-associated protein; CA for capsid; NC for nucleocapsid; PR for protease; DU for dUTPase; RT for reverse transcriptase; IN for integrase; SU for surface protein; and TM for transmembrane protein.323 The localization of these proteins in the mature virion is not known with great precision, but a highly schematic version of the generic retrovirion can be drawn (Fig. 47.3).
Arrangement of Virion Components
The genomic RNA is highly condensed in the virion by its association with the nucleocapsid protein, NC. The complex is contained within a protein core largely composed of the capsid protein CA, another Gag gene product. The shape of the core is different among the various retroviral genera, and is a distinguishing feature of the genera. In most of the viruses the core is roughly spherical, but in some cases can be either conical or cylindrical. In all the viruses the core is surrounded by a roughly spherical shell consisting of MA, which in turn is surrounded by the lipid bilayer of the virion envelope. The virion membrane contains the envelope glycoprotein, with the TM subunit present as a single-pass transmembrane protein anchor, and the SU subunit as an entirely extravirion protein bound to TM. The envelope proteins for those viruses examined closely have been found to reside in the membrane as trimers.
Organization of the RNA Genome
The viral genome is a dimer of linear, positive-sense, single-stranded RNA (ssRNA), with each monomer 7 to 13 kb in size. The viral genomic RNA is present as a homodimer of two identical sequences, and thus the virions are functionally diploid. The dimer is maintained by interactions between the two 5′ ends of the RNAs in a self-complementary region termed the dimer linkage structure (DLS). The RNA genome is generated by normal host transcriptional machinery, and thus exhibits many of the features of a normal mRNA. The RNA is capped at the 5′ end, using the common m7G5′ppp5′Gmp
structure; and contains a string of poly(A) sequence, about 200 long, at the 3′ end.
structure; and contains a string of poly(A) sequence, about 200 long, at the 3′ end.
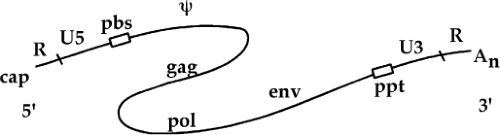
A number of sequence blocks are so important that they have been named to facilitate descriptions of their functions in the life cycle (Fig. 47.4). These key sequences are clustered at the termini of the RNA. A short sequence, the R (for repeated) region, is so called because it is present twice in the RNA: once immediately after the cap at the 5′ end and again at the 3′ end, just before the poly(A) tail. Downstream of the 5′ R lies another sequence, termed U5 for unique 5′ sequence, which includes one of the att sites required for proviral integration. The U5 region is followed by the primer binding site, an 18-nt sequence at which a host tRNA is hybridized to the genome and the site of initiation of minus-strand DNA (msDNA) synthesis.
The region downstream from the primer binding site (pbs) often contains the major signals for the encapsidation of viral RNA into the virion particle, in sequences called the Psi element. The region also often contains a major splice donor site for the formation of subgenomic mRNAs. The bulk of the RNA sequences that follow are coding regions for the viral proteins. The genomes of all the replication-competent retroviruses contain at a minimum three large genes, or open reading frames: from 5′ to 3′ along the genome, the genes are termed gag, for group-specific antigen; pol, for polymerase; and env, for envelope. The three genes in the simple retroviruses occupy nearly all the available space in the center of the genome.
Downstream of the genes lies a short polypurine tract (ppt), a run of at least nine A and G residues. The ppt is the site of initiation of plus strand DNA (psDNA) synthesis. The ppt is followed by a sequence block termed U3 for unique 3′ sequence; this region contains a number of key cis-acting elements for viral gene expression, and one of the att sites required for DNA integration. The U3 abuts the 3′ copy of the R region, which is followed by the poly(A) tail. As will be demonstrated, the R, U5, U3, pbs, and ppt sequences all play important roles in reverse transcription.
Overview of the Life Cycle
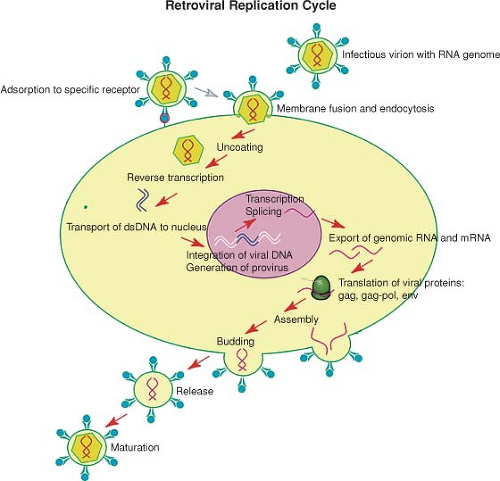
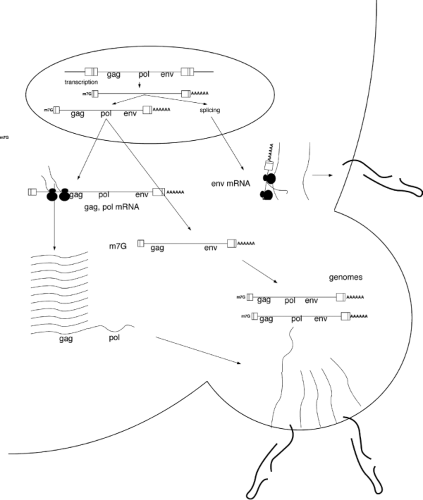
The retroviruses replicate through a complex life cycle. A short summary of the steps of the cycle is as follows (a schematic view is shown in Fig. 47.5):
Receptor binding and membrane fusion
Internalization and uncoating
Reverse transcription of the RNA genome to form double-stranded linear DNA
Nuclear entry of the DNA
Integration of the linear DNA to form the provirus
Transcription of the provirus to form viral RNAs
Splicing and nuclear export of the RNAs
Translation of the RNAs to form precursor proteins
Assembly of the virion and packaging of the viral RNA genome
Budding and release of the virions
Proteolytic processing of the precursors and maturation of the virions
Changes in the Viral Genome
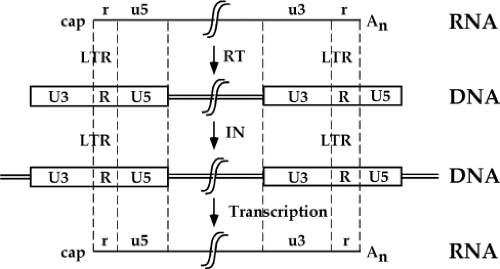
A quick perusal of this list reveals that the life cycle begins with an RNA genome, passes through an intracellular DNA intermediate, and is completed with a return to an RNA form in the progeny virus particle. An overview of the structures of the genome at various times in this cycle is presented in Figure 47.6. The RNA genome of the virion contains short terminal repeats (the R region) at its termini. During reverse transcription, to be seen below, sequence blocks termed U5 and U3 are duplicated, so that the resulting dsDNA is longer at both ends than the RNA template. This DNA thus contains long terminal repeats (the LTRs, consisting of sequence blocks U3, R, and U5) at both ends. The next step is the integration of the DNA to form the provirus; the integrated provirus is collinear with the preintegrative DNA, and retains the LTRs (except for one or two base pairs lost at the termini during the course of integration). Finally, the DNA is forward transcribed by the RNA polymerase II system to produce the progeny RNA genome. Transcription is initiated at the U3-R boundary of the 5′ LTR, and the transcripts are processed and polyadenylated at the R-U5 boundary of the 3′ LTR, recreating the exact structure of the input RNA, complete with its short terminal repeats. This RNA is packaged and exported in virion particles. Each step is described in more detail in the next section.
The Virus Receptors
To enter a cell and initiate infection, all retroviruses require an interaction between a cell surface molecule—a receptor—and
the envelope protein on the virion surface. The interactions are complex, involving an initial binding, drastic conformational changes in the envelope protein, an induced fusion of the viral and cellular membranes, and the internalization of the virion core into the cytoplasm. The SU subunit of Env is thought to make the major initial contacts with receptor, and the TM subunit is thought to be most important for membrane fusion. The reorganization of the two lipid bilayers—one on the virion and one on the cell—to join them and evert the core into the cell is a remarkable process. The details of these complex processes are not understood for any retrovirus, and the whole Env protein is likely to be involved in efficient entry. However, there is a great deal of information about the identity and structures of the receptors used by various retroviruses. It is apparent that these viruses utilize an extraordinarily diverse set of cell surface molecules as receptors (Table 47.3; see 41,581,624 for reviews).
the envelope protein on the virion surface. The interactions are complex, involving an initial binding, drastic conformational changes in the envelope protein, an induced fusion of the viral and cellular membranes, and the internalization of the virion core into the cytoplasm. The SU subunit of Env is thought to make the major initial contacts with receptor, and the TM subunit is thought to be most important for membrane fusion. The reorganization of the two lipid bilayers—one on the virion and one on the cell—to join them and evert the core into the cell is a remarkable process. The details of these complex processes are not understood for any retrovirus, and the whole Env protein is likely to be involved in efficient entry. However, there is a great deal of information about the identity and structures of the receptors used by various retroviruses. It is apparent that these viruses utilize an extraordinarily diverse set of cell surface molecules as receptors (Table 47.3; see 41,581,624 for reviews).
An important tool in the analysis of receptor utilization is the phenomenon of virus interference, or superinfection resistance. Cells chronically infected by a particular virus cannot be infected by any virus that must enter by the same receptor as used by the first virus though they are readily infected by viruses that utilize a distinct receptor. The reason is that the expression of Env protein by the first virus binds to the receptor intracellularly, preventing its export to the cell surface or its function as a receptor for newly applied virus. The phenomenon allows for the rapid classification of those viruses that use a common receptor.
Table 47.3 Retrovirus Receptors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The properties of the receptors of the major retroviral genera are summarized in the following section.
Alpharetrovirus Receptors
The receptor for the A subgroup of avian viruses was identified as encoding a membrane-anchored glycoprotein with sequence similarity to the ligand-binding repeat of the low-density lipoprotein receptor (LDLR).33,653 Its identity as the true receptor has been confirmed by correlating its genetic map position with the tv-a locus.32 The tv-b locus, encoding the receptor for both the B and D subgroups of the ASLV, encodes a protein termed CAR1, unrelated to tv-a but with sequence similarity to the receptors for tumor necrosis factor (TNF) and the Fas death receptors.73 The intracellular portion of the molecule contains the sequence of a “death domain,” present on other cytotoxic receptors, and can trigger the apoptotic death of the cell upon ligand binding. The tv-c locus is closely linked to tv-a but encodes an unrelated surface protein, one with strong sequence similarity to mammalian butyrophilins, members of the immunoglobulin family.157 The tv-e locus is present in turkey but not chicken, and allows for infection by the subgroup E viruses. The gene was cloned by its sequence similarity to the chicken tv-b locus.4
Betaretrovirus Receptors
The receptor for MMTV was cloned by co-segregation of DNA markers with virus susceptibility in mouse/hamster radiation chimeric cell lines, and so identified as the transferrin receptor tfr1 on mouse chromosome 16.520 A second receptor for the betaretroviruses was also identified. The type D simian viruses, including MPMV and SRV-1, −2, −4, and −5, show cross-interference with three type-C viruses: feline endogenous virus (RD114), baboon endogenous virus (BaEV), and avian reticuloendotheliosis virus (REV), suggesting that they all utilize a common cell-surface receptor. Gene transfer of a human cDNA library into nonpermissive mouse cells was used to identify a gene that conferred susceptibility to infection by RD114.583 The cDNA encoded a protein nearly identical to the previously cloned human Na+-dependent neutral-amino-acid transporter named B°.288,498 Consistent with this similarity, expression of
the RD114 receptor in NIH 3T3 cells resulted in enhanced cellular uptake of L-{3H}alanine and L-{3H}glutamine.
the RD114 receptor in NIH 3T3 cells resulted in enhanced cellular uptake of L-{3H}alanine and L-{3H}glutamine.
Gammaretrovirus Receptors
Several receptors for various gammaretroviruses are known.581 The first example, the mouse receptor used by the ecotropic MuLVs, was identified by gene transfer to nonpermissive human cells, selecting for susceptibility to MuLV infection.8 The gene encodes a membrane glycoprotein of 67 kDa containing a total of 14 membrane spanning domains. The normal function of the protein has been identified as a transporter or permease for cationic, basic amino acids.292 The receptor, termed mCAT-1, was shown to be identical to y+, the previously characterized transporter in mammalian cells. The gene for mCAT-1 is now known as Atrc1.
The amphotropic receptor is utilized by a group of MuLVs derived from wild mice able to infect a wide range of mammalian species, including humans. The receptor was cloned by selection for susceptibility to virus infection after transfection of cDNA libraries into nonpermissive CHO cells,154,386 and by its homology to the gene for the previously identified GALV receptor.607 The gene, known variously as Ram1 or GLVR2 or rPiT-2, encodes a 652–amino acid protein that functions as a sodium-dependent phosphate symporter.282 The synthesis and stability of the receptor is regulated by phosphate levels, and its downregulation by virus infection results in substantial reduction in phosphate uptake by cells.
The receptor utilized in common by GALV, simian sarcoma–associated helper virus (SSAV), and FeLV-B is widely expressed in many mammals, including primates, cat, dog, mink, rabbit, and rat (but not mouse), as well as in some avian species. The human receptor is termed GLVR1 or hPiT-1.265,426 The sequence of the gene predicts the existence of 10 membrane-spanning segments, and a large third intracellular loop. The protein is a sodium-dependent phosphate symporter.282,428 Specific amino acid changes introduced into the fourth extracellular loop can block FeLV-B and SSAV infection without affecting GALV, suggesting that these various viruses interact in slightly different ways with the receptor. A remarkable feature of infection by FeLV-B via feline PiT-1 is a requirement for the co-expression of an endogenous Env-like protein dubbed FeLIX.13
The xenotropic MuLVs are viruses present as proviruses in the mouse germ line but unable to infect inbred mouse cells. The polytropic MuLVs are also endogenous viruses with a wide host range that includes many mammalian species. Xenotropic and polytropic MuLVs cross-interfere to various extents in nonmouse species and in wild Asian mice, suggesting that they might use a common receptor for infection. The mouse receptor for the polytropic viruses was cloned by gene transfer, and was identified with the Rmc1 gene.650 The human xenotropic receptor mediates infection by both the xenotropic and polytropic viruses, as well by the XMRV isolate.647 The gene encodes a membrane protein related to the yeast Syg1p protein (suppressor of yeast G alpha deletion). Its function is unknown, but its multiple membrane-spanning segments and its sequence suggests that it may act as a G-coupled receptor.
The receptor utilized by the subgroup C feline leukemia viruses (FeLV-C) encodes a protein with 12 membrane-spanning domains with significant sequence similarity to the D-glucarate transporters of bacteria and nematodes.494 The binding of virus to this receptor may be responsible for its pathogenesis, a block in erythroid differentiation.
Additional receptors for other gammaretroviruses are known to exist. Three newly characterized porcine endogenous retroviruses (PERV-A, -B, and C) have been tested in interference assays with each other and with murine viruses using the known receptors; all three apparently utilize distinct and novel receptors.585 The PERV-A receptor has been identified and is likely a G protein-coupled receptor.160
Deltaretrovirus Receptors
The receptor for the bovine leukemia virus(BLV) is highly similar to the delta subunit of the AP-3 complex.26,576 AP-3 is involved in intracellular trafficking of clathrin-coated vesicles and is not thought to be present on the cell surface. The properties of the receptor are not yet well established.
Lentivirus Receptors
The first receptor identified for any retrovirus was the CD4 molecule, established as essential for infection by HIV-1.122,294,357 CD4 is an important surface protein on T cells, and with few exceptions serves to define the helper subset of T cells. CD4 is also expressed at significant levels on dendritic cells, macrophages, and on certain cells in the brain, likely astrocytes rather than cells of neural origin. The limited distribution of expression of CD4 accounts well for the tropism of HIV-1, largely restricted to helper T cells and macrophages. There may be other routes of entry utilized at lower efficiency: antibody to virus, for example, can promote virus entry into cells by the Fc receptor. Receptor-negative dendritic cells can take up virions via binding to the DC-SIGN molecule and deliver them efficiently to T cells to promote their infection, but even here infection of the recipient cells requires their expression of the CD4 receptor.199,309
Early work established that although CD4 was sufficient to mediate virus binding to a cell surface, it was not sufficient to mediate virus infection and entry. For example, rodent cells and other cells of nonprimate origin could not be successfully infected by HIV-1 even if they were engineered to express human CD4. Searches for genes that would render such cells sensitive to virus infection ultimately led to the identification of various members of the chemokine receptor family, notably CCR5 and CXCR4, as coreceptors needed to mediate the postbinding steps of membrane fusion and virus entry.152,171,552 Antibodies to the coreceptor as well as the natural ligand for these molecules, the chemokines themselves, can block virus entry. Variants of SIV and HIV-1 have been identified that are CD4-independent, needing only a chemokine receptor for infection; the existence of these viruses suggests that the chemokine receptors might have been the primary receptor for a primordial virus. Further proof of the importance of the chemokine receptor is the existence of a mutant allele of the gene encoding CCR5 in the human population, a 32-bp deletion, that confers dramatic virus resistance to homozygous individuals. More discussion of the roles of CD4 and the co-receptors in virus entry will be presented in Chapter 49 on HIV-1.
Penetration and Uncoating
Once virus particles have bound to the receptor, the virion and host membranes fuse together, and the virion core is delivered
into the cytoplasm of the infected cell. Entry may require, or be promoted by, membrane regions of special lipid composition termed lipid “rafts”.334,364,484 Virus particles may “surf” or slide across the outside of the cell to preferred locations where fusion or entry inside the cell can occur.322 For most retroviruses, the processes of fusion and entry are thought to be pH independent: that is, they are not dependent on an endosomal acidification step to induce a pH-dependent change in the conformation of the envelope. Thus, for these viruses fusion can occur at the cell surface. However, the ecotropic and amphotropic MuLVs and the subgroup A avian viruses are inhibited by drugs that block acidification; these viruses thus likely enter by passage through endosomes.
into the cytoplasm of the infected cell. Entry may require, or be promoted by, membrane regions of special lipid composition termed lipid “rafts”.334,364,484 Virus particles may “surf” or slide across the outside of the cell to preferred locations where fusion or entry inside the cell can occur.322 For most retroviruses, the processes of fusion and entry are thought to be pH independent: that is, they are not dependent on an endosomal acidification step to induce a pH-dependent change in the conformation of the envelope. Thus, for these viruses fusion can occur at the cell surface. However, the ecotropic and amphotropic MuLVs and the subgroup A avian viruses are inhibited by drugs that block acidification; these viruses thus likely enter by passage through endosomes.
The process of fusion involves major rearrangements of the Env proteins, and especially includes the exchange of disulfide bonds that exist within or between the TM and SU subunits of Env. The process for the MuLVs seems to be controlled by Ca+2 levels, and involves TM–SU intersubunit disulphide-bond isomerization and SU dissociation.617 Entry by HIV-1 probably also involves the removal or shedding of SU.
The processes of uncoating or opening of the core to permit reverse transcription to begin are poorly understood. It is clear that the previous processing of the Gag precursor to the mature Gag proteins is required; immature virions are uninfectious and cannot initiate reverse transcription, and mutants that prevent particular cleavages of the Gag protein are similarly blocked. A large number of mutant viruses with other alterations in the gag gene have been shown to be defective in early steps of infection, before reverse transcription, but the functions of Gag proteins at this stage remain uncertain. Mutant virions that are fragile and uncoat prematurely or, conversely, are resistant to disassembly, are poorly infectious, suggesting that the timing of uncoating may be critical.178 There are indications that host factors are important in these early stages. In the case of HIV-1, the host protein cyclophilin A, which interacts with CA, is required for the efficient initiation of reverse transcription.559 A plausible role for this protein is to facilitate virion disassembly.229 The TRIM proteins restrict virus infection at this time (see Early block to infection by Trim5a section).
Small molecule inhibitors have been used to demonstrate a role of the cytoskeleton in virus entry, and furthermore to suggest that viruses may utilize different entry pathways in different cell lines.293 Biochemical analyses of these early events are made difficult by the presence of large numbers of defective particles that are probably not on the infectious pathway and that tend to obscure the properties of the rare particles that are on this pathway. Nevertheless, examination by fluorescence microscopy of GFP-tagged virion particles during infection has indicated that intracellular movement likely occurs along cytoskeletal fibers.377
Reverse Transcription
The reverse transcription of the viral RNA genome into a dsDNA form is the defining hallmark of the retroviruses, and the step from which these viruses derive their name. The course of reverse transcription is complex and highly ordered, involving the initiation of DNA synthesis at precise positions and translocations of DNA intermediates that result in duplication of sequence blocks in the final product (for reviews see 201,590). The major steps in the reaction are relatively well established, largely through the analysis of reactions carried out in vitro in purified virion particles (the so-called “endogenous reaction”).
Reverse transcription normally begins soon after entry of the virion core into the cytoplasm of the infected cell. The reaction takes place in a large complex, roughly resembling the virion core, and containing Gag proteins including NC, RT, IN, and the viral RNA.66 The signal that triggers the onset of DNA synthesis is not known, though it may be as simple as the exposure of the viral core to the relatively high levels of deoxyribonucleotides present in the cytoplasm. This notion is consistent with the observation that simply stripping or permeabilizing the virion membrane with detergents in the presence of deoxyribonucleotides is sufficient to induce DNA synthesis. This may also be at least part of the explanation for the difficulty HIV has in completing reverse transcription and infection in quiescent cells. In some cells, notably cells arrested by starvation, triphosphate levels may be low and limiting for RT, so that addition of exogenous nucleosides can stimulate viral DNA synthesis. But the signal may be more complicated. Conformational changes in the RNA genome at the tRNA primer site may trigger DNA synthesis.37
DNA synthesis can be initiated prematurely during virion assembly and release, such that virion preparations can be shown to contain small amounts of the early DNA intermediates, such as minus-strand strong-stop DNA. In most cases the levels of these DNAs are very low, indicating that only a very small minority of the virion particles have carried out any significant synthesis. However, some circumstances affecting the rate of production and release of virions may enhance this synthesis. In addition, in some particular retroviruses, notably the spumaviruses, substantial DNA synthesis occurs during assembly such that the major form of the genome found in mature virions is a partially or even completely reverse transcribed DNA molecule.390,656 These viruses thus resemble the hepadnaviruses more closely than the conventional retroviruses in the relative timing of assembly and reverse transcription.
Steps in Reverse Transcription of the Retroviral Genome
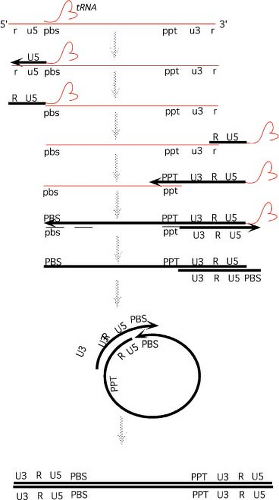
The course of reverse transcription is complex. The reaction can be broken down into a series of discrete steps,201 as presented in Figure 47.7.
Formation of Minus-Strand Strong-Stop DNA
The process of reverse transcription is initiated from the paired 3′ OH of a primer tRNA annealed to the viral RNA genome at a complementary sequence termed the primer binding site (pbs). DNA is first synthesized from this primer, using the plus strand RNA genome as template, to form minus strand DNA sequences. Synthesis occurs toward the 5′ end of the RNA to generate U5 and R sequences. The intermediate formed in this step is termed minus-strand strong-stop DNA. The primer tRNA remains attached to its 5′ end.
First Translocation
The next step involves the translocation, or “jump,” of the strong-stop DNA from the 5′ to the 3′ end of the genome. This translocation requires the degradation of those 5′ RNA sequences that were placed in RNA:DNA hybrid form by the formation of strong-stop DNA. The degradation is mediated
by the RNase H activity of RT; mutants with altered RNase H activity do not mediate the translocation. This step exposes the ssDNA and facilitates its annealing to the r sequences at the 3′ end of the genome.95 Normally a full-length strong-stop DNA, synthesized by copying to the 5′ cap of the RNA, performs the translocation, though incomplete molecules can jump at low efficiency. The NC protein may facilitate the transfer step. Although there have been reports that jumping is always in trans, from one RNA template to the other RNA in the virion, the best evidence is that minus-strand strong-stop jumping goes randomly to either RNA.
by the RNase H activity of RT; mutants with altered RNase H activity do not mediate the translocation. This step exposes the ssDNA and facilitates its annealing to the r sequences at the 3′ end of the genome.95 Normally a full-length strong-stop DNA, synthesized by copying to the 5′ cap of the RNA, performs the translocation, though incomplete molecules can jump at low efficiency. The NC protein may facilitate the transfer step. Although there have been reports that jumping is always in trans, from one RNA template to the other RNA in the virion, the best evidence is that minus-strand strong-stop jumping goes randomly to either RNA.
Long Minus-Strand DNA Synthesis
The annealing of minus-strand strong-stop DNA recreates a suitable primer-template structure for DNA synthesis, and RT can now continue to elongate the minus-strand strong-stop DNA to form long minus-strand products. Synthesis ends in the vicinity of the pbs. As the genome enters RNA:DNA hybrid form, the RNA becomes susceptible to RNase H action and is degraded.
Initiation of Plus Strand DNA Synthesis
The primer for plus-strand synthesis is created by the digestion of the genomic RNA by RNase H. A particular short purine-rich sequence near the 3′ end of the genome, the polypurine tract or ppt, is relatively resistant to the activity of RNase H. The oligonucleotide remains hybridized to minus strand DNA and serves as the primer for synthesis of plus strand DNA, using minus strand DNA as template. The sequence of the PPT, an unusual structure of the nucleic acid at the PPT, and residues of the RNase H domain of RT have all been implicated in defining the cleavages that form the primer. Sequences upstream of the polypurine tract, an AT-rich region called the T-box, are also important for proper priming. The primer, once it has served to initiate DNA synthesis, is removed from the DNA. Synthesis proceeds toward the 5′ end of the minus strand, first copying the U3, R, and U5 sequences, then extending further to copy a portion of the primer tRNA still present at its 5′ end. Elongation stops at a modified base normally found at position 19 of the tRNA. The resulting intermediate is termed plus-strand strong-stop DNA.
In some viruses, secondary plus-strand initiation sites are used. There may be multiple RNA primers generated from the RNA genome by the nuclease action of RNase H that can initiate DNA synthesis at dispersed heterogeneous sites. In the case of the lentiviruses and spumaviruses, a second copy of the ppt sequence near the center of the genome is used at high efficiency, and is important for proper completion of reverse transcription.91
Removal of tRNA
In the next step, the primer tRNA at the 5′ end of the minus strand DNA is removed by RNase H. Its removal may occur in two stages: with an initial cleavage near the RNA–DNA junction and a second one within the tRNA. The cleavage need not occur exactly at the RNA–DNA junction, and a single ribonucleotide base (A) is normally left on the 5′ terminus of the HIV-1 minus strand without affecting subsequent processes. The posttranscriptional modifications present in natural tRNA are probably important for proper recognition by RT and for plus-strand strong-stop translocation.
The Second Translocation
The removal of tRNA exposes the 3′ end of the plus-strand strong-stop DNA to permit its pairing with the 3′ end of the msDNA. The sequences anneal via the shared pbs sequences. This annealing forms a circular intermediate, with both 3′ termini in a suitable structure for elongation.
Completion of Both Strands
Both strands are now elongated. The final extension of minus strand DNA is coupled to displacement of the plus-strand strong-stop DNA from the 5′ end of the minus strand; as minus-strand elongation occurs, the plus-strand strong-stop is peeled away and transferred to the 3′ end of the minus
strand. At the end of this elongation, the circle is opened up into a linear DNA. The plus strands are then extended. When multiple plus-strand initiation events have occurred, the completed plus strand will consist of adjacent fragments and contain nicks or discontinuities. Displacement synthesis by an upstream fragment can slowly displace downstream RNAs and DNAs, leading to longer plus strands. However, some nicks or gaps may persist in the final double-stranded product. These breaks may be at heterogeneous positions, though strong sites of plus-strand initiation, such as the one at the central ppt of lentiviruses, can lead to specific sites for such discontinuities. Sequences near the central ppt of the lentiviruses cause termination of synthesis during elongation from upstream primers, ensuring the maintenance of a discontinuity at this site.92 This site retains a partially displaced sequence or overlap of a few nucleotides: 99 nt in the case of HIV-1. The structure has been shown to persist even to the time of integration of the DNA into the cell. Host DNA repair processes ultimately correct all such discontinuities.
strand. At the end of this elongation, the circle is opened up into a linear DNA. The plus strands are then extended. When multiple plus-strand initiation events have occurred, the completed plus strand will consist of adjacent fragments and contain nicks or discontinuities. Displacement synthesis by an upstream fragment can slowly displace downstream RNAs and DNAs, leading to longer plus strands. However, some nicks or gaps may persist in the final double-stranded product. These breaks may be at heterogeneous positions, though strong sites of plus-strand initiation, such as the one at the central ppt of lentiviruses, can lead to specific sites for such discontinuities. Sequences near the central ppt of the lentiviruses cause termination of synthesis during elongation from upstream primers, ensuring the maintenance of a discontinuity at this site.92 This site retains a partially displaced sequence or overlap of a few nucleotides: 99 nt in the case of HIV-1. The structure has been shown to persist even to the time of integration of the DNA into the cell. Host DNA repair processes ultimately correct all such discontinuities.
Although most of the viral DNA is made in the cytoplasm, it may not always be completed in the cytoplasm. For some viruses, completion of the two DNA strands may occur only after entry into the nucleus. Specific mutants with alterations in the Cys-His residues of the NC protein show an interesting phenotype: the formation of linear DNA with heterogeneous and truncated ends.208 These experiments suggest that NC plays a role in the completion, or the stabilization of the ends, of the viral DNA.
A key consequence of the two translocation events that occur during reverse transcription is the duplication of sequences: duplication of U5 during minus-strand strong-stop DNA translocation and of U3 during plus-strand strong-stop DNA translocation. The resulting DNA thus contains two LTRs that have been assembled during reverse transcription. Each LTR consists of the sequence blocks U3-R-U5. The positions of the LTR edges—the left edge of U3, and the right edge of U5—are determined by the sites of initiation of DNA synthesis for the two DNA strands. Thus, the terminal sequences of the complete DNA molecule are also determined by these sites of initiation. These sequences for most viruses are perfect or imperfect inverted repeats, and serve an important role during integration of the DNA (see the Viral att sites section).
Biochemistry and Structure of Reverse Transcriptase
The enzyme that mediates the complex series of events outlined in the previous section is RT, one of the most famous of the viral polymerases (25; for review, see 553). All RTs contain two separate activities present in two separate domains: a DNA polymerase able to incorporate deoxyribonucleotides on either an RNA or a DNA template, and an RNase H activity able to degrade RNA only in duplex form. These two activities are responsible for the various steps of reverse transcription. Two distinct domains of the enzyme contain these two activities: an aminoterminal domain contains the DNA polymerase, and a carboxyterminal domain contains the RNase H activity.587 While isolated domains can be shown to exhibit either one of the two activities separately, an intact enzyme is required for full activity and specificity. However, the two functions can be provided by two mutant RT molecules so long as they are co-incorporated into a single virion.
DNA Polymerase
DNA polymerase activity is similar to that of all host and viral polymerases in requiring a primer, which can be either RNA or DNA, and a template, which can also be either RNA or DNA. RTs incorporate dXTPs to a growing 3′OH end with release of PPi, and require divalent cations, usually Mg++. The primer must contain a 3′OH end that is paired with the template. RTs cannot perform nick-translation reactions, but they can efficiently perform strand displacement synthesis. The only fundamental way in which RTs are unusual among the DNA polymerases is that they exhibit comparable specific activity on either DNA or RNA templates.
RTs are readily isolated from purified virion particles, and can be even more easily prepared as recombinant proteins expressed in bacteria. RTs are relatively slow DNA polymerases, under standard conditions only incorporating 1 to 100 nucleotides per second, depending on the template. Further, they exhibit poor processivity, and tend to release primer-template frequently in vitro. The enzyme must then rebind to the substrate to continue synthesis. Secondary structures in RNA templates can strongly enhance the pausing of RT and its tendency to release from the template.226 The enzyme also exhibits low fidelity, and though the values of the error rate vary widely with the primer, template and type of assay, the misincorporation rate of most RTs under physiologic conditions is on the order of 10−4 errors per base incorporated. This rate suggests that during replication there would be approximately one mutation per genome per reverse transcription cycle. The mutation rate observed in vivo is roughly consistent with this high error rate, though fidelity in vivo may be somewhat better than in vitro. Drug-resistant variants that do not incorporate chain-terminating analogs are often found to exhibit higher fidelity, perhaps because they require a more precise fit for the correct incoming triphosphate to allow for discrimination against the analog. A wide range of types of mutations are created by RT errors, and both the type and the frequency of appearance of each type of mutation exhibit a complex dependence on sequences and structures in the template.
RTs do not generally exhibit a proofreading nuclease activity,35 and misincorporated bases are not removed as efficiently by most RTs as they are by host DNA polymerases. However, mutants of the HIV-1 RT resistant to AZT have been shown to exhibit an enhanced ability to remove the incorporated AZT moiety at the 3′ end through a pyrophosphorolysis reaction.382 Thus, it is possible for RT to remove some such analogs and rescue a terminated chain for continued elongation.
RNase H
The RNase H activity of RT is an endonuclease that releases oligonucleotides with a 3′OH and a 5′PO4. This property allows the products of RNase H action to serve as primers for initiation of DNA synthesis by the DNA polymerase function of RT. There is an obligate requirement that the RNA be in duplex form, normally an RNA–DNA hybrid. However, retroviral RTs are also able to degrade RNA–RNA duplexes, an activity termed RNaseH*.243 The RNase H enzyme is capable of acting on the RNA of a template in concert with the polymerase as it moves along a nucleic acid, and as it does so its active site is located about 17 to 18 bp behind the growing 3′ end.206 RNase H can also act independently of polymerization. All RNase H activity requires a divalent cation.
Subunit Structures
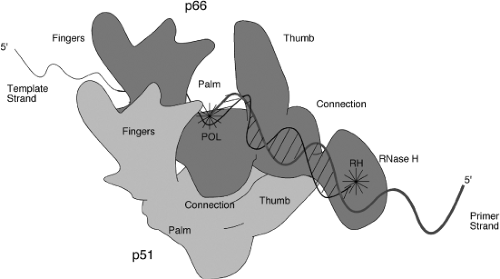
RT is incorporated into the virion particle during assembly in the form of a large Gag-Pol precursor (see below), and is released by proteolytic processing of the precursor during virion maturation. Different viruses make somewhat different cleavages in the precursor, and thus the RTs exhibit several different subunit structures (see below). In the gammaretroviruses, RT is a simple monomer in solution, corresponding only to the aminoterminal DNA polymerase and the carboxyterminal RNase H domains. These two domains can be expressed separately, and the isolated proteins exhibit their respective activities,587 though the specificity of the RNase H is affected by this separation. In the avian viruses, the RT is present as an αβ heterodimer, comprised of a smaller subunit containing the DNA polymerase and RNase H domains; and a larger β subunit containing these two domains but also retaining the integrase domain. In the lentiviruses, RT is again a heterodimer with a larger subunit (p66) containing the DNA polymerase and RNase H domains, and a smaller subunit (p51) lacking RNase H. The properties of the different enzymes as DNA polymerases are very similar in spite of these different subunit structures, and thus the significance of these various compositions for RT function is unclear. A curious observation was made that some RT inhibitors—the so-called nonnucleoside RT inhibitors—can potently enhance the association of p66 and p51, locking them into an inactive dimer.580
Crystal Structures
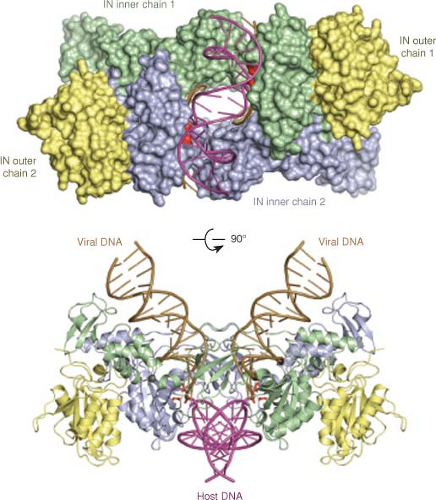
The three-dimensional structure of a number of RTs have been determined by X-ray crystallographic studies. Structures of the unliganded HIV-1 RT,246,515 RT bound to nonnucleoside RT inhibitors,127,135,300,504 RT bound to an RNA pseudoknot inhibitor,260 RT bound to a duplex oligonucleotide,17,248,258,259 and RT bound to a polypurine tract RNA:DNA hybrid,531 as well as the isolated RNase H domain,128 have all been reported. The two subunits are folded very differently so that the overall structure is highly asymmetric. The structure of the p66 is similar to that of a right hand, with fingers, palm, and thumb domains named on the basis of their position in the structure (Fig. 47.8). The nucleic acid lies in the grip of the hand, held by the fingers and thumb. The YXDD motif present at the active site for the DNA polymerase lies at the base of the palm. The RNase H domain is attached to the hand at the wrist. The p51 subunit, while made up of the same domains as the aminoterminal part of p66, is folded differently and lies under the hand, not making direct contact to the nucleic acid and thus not thought to participate in chemistry. The structure of p66 with and without a liganded nucleic acid is very different, with the thumb domain flexing to allow substrate binding. A surprising aspect of the structures is that the nucleic acid helix can be highly bent, perhaps accounting for the enzyme’s ability to sense conformationally strained substrates.531 Theoretical considerations suggest that the thumb may move during elongation.
Inhibitors
RT is a major target of antiviral drugs useful in the treatment of retroviral diseases such as AIDS. All such drugs used to date are inhibitors of the DNA polymerase activity of RT, and fall into two classes: nucleoside analog inhibitors (chain terminators), and nonnucleoside RT inhibitors (NNRTIs). The nucleoside analogs are typically prodrugs, and need to be activated by phosphorylation to the triphosphate form. These are then incorporated by RT into the growing chain, and serve to block further elongation. Examples include AZT, ddC, ddI, d4T, and 3TC. The NNRTIs are a group of compounds that are structurally diverse, but nevertheless interact with a common binding pocket in RT to prevent its normal activity.600 There are indications that the binding may inhibit the enyzme’s flexibility. For both classes of inhibitors, monotherapy with a single drug selects for drug-resistant variants that quickly predominate in the virus population, and for each drug, a pattern of mutations has been identified that serves to indicate the appearance of drug resistance.315 In many cases these mutations alter the binding side for the nucleoside or NNRTI such that the drug cannot bind and therefore cannot inhibit the enzyme. In the case of AZT, however, the mutations do
not prevent the binding and incorporation of AZTTP into the growing chain, but rather seem to activate a reverse reaction in which the AZT nucleotide is removed from the chain, subsequently permitting normal elongation.382 Combination therapy, typically involving the simultaneous treatment with three different drugs, can suppress virus replication to such an extent that variants resistant to all the drugs do not appear, at least for months or years.
not prevent the binding and incorporation of AZTTP into the growing chain, but rather seem to activate a reverse reaction in which the AZT nucleotide is removed from the chain, subsequently permitting normal elongation.382 Combination therapy, typically involving the simultaneous treatment with three different drugs, can suppress virus replication to such an extent that variants resistant to all the drugs do not appear, at least for months or years.
Recombination
The process of reverse transcription could in principle take place using a single template RNA molecule. In fact, however, retrovirions contain two copies of the RNA genome co-packaged into one particle, and the course of reverse transcription typically makes use of both RNAs.247,573 Recombination occurs between homologous sequences in the two RNAs, happening at surprisingly high frequencies, more than once per replication event per genome on average.511,667 Normally the two RNAs in a virion are identical, so that homologous recombination events are invisible and without consequence. When the two RNAs are distinct, however, as when they derive from two viruses or viral strains, the result is a very high frequency of recombination between them among the resulting proviral DNAs. Thus, physical markers and genetic markers recombine rapidly whenever the two genomes are co-packaged into one virion and thus are co-extant during a single round of reverse transcription. The frequency is highly dependent on the sequence and structure of the RNA in the region undergoing recombination. Similar recombination does not occur at high frequency when cells are co-infected simultaneously with two separate virus preparations, suggesting that each incoming virus particle performs its own reverse transcription reaction in the cytoplasm in cis, and does not freely exchange RNAs with other reactions happening in the same cell.
Models for Recombination
Two mechanisms provide for recombination between two genomes. In one, the copy choice model, recombination occurs during minus-strand synthesis. As RT proceeds along an RNA, it has the potential to carry out a template switch in which an incomplete DNA copied from one template serves to prime further elongation on the other RNA molecule.351,465 Pausing may enhance this transfer, and secondary structures in the RNA may act as hot spots for such recombination. Breaks in the RNA genome, which may be encountered often, cause a “forced copy choice”: transfer to the other RNA. This rescues an otherwise dead virus, and may represent the major evolutionary basis for high-frequency recombination in the viruses. The RNase H activity of RT may help release an incomplete DNA, promoting its serving as primer on the new template; NC also facilitates the reaction.413 This mechanism is likely the more important one of the two.666
In the other mechanism, strand-displacement assimilation, recombination occurs when at least portions of two minus strands have been synthesized in one virion. While multiple plus-strand fragments are elongating on one minus-strand template, strand displacement can expose the 5′ end of such fragments, which can then pair with the other minus-strand DNA to form a bridged “H” structure as intermediate. Further synthesis and repair of these structures leads to the transfer of sequences to the new DNA.274
When a recombination event occurs, there is a nonrandom increase in the probability that another recombination will occur nearby, a phenomenon called negative interference. This suggests that RT or the genomes may become recombination prone at specific times. When multiple recombination events occur, the resulting DNA is a patchwork of the sequences derived from the two input RNAs.
The translocation of the two strong-stop DNAs provides a special opportunity for recombination between the two viral genomes. When the minus-strand strong-stop DNA is formed, it has the potential to translocate from the 5′ end of its template to the 3′ end of either RNA molecule; though this event has been reported to occur strictly in cis, or strictly in trans, it most likely occurs randomly. Similarly, when plus-strand strong-stop DNA is formed, it too could in principle translocate to the 3′ of either minus strand. However, this translocation seems most often to occur in cis, perhaps simply because the frequency with which two long minus-strand DNAs are successfully formed, and thus are available to serve as acceptors, is low.
Recombination between two RNAs during reverse transcription can also occur between nonhomologous sites at lower frequency. Reconstructions suggest that these events are perhaps 100 to 1000 times less frequent than homologous recombination. These events can result in duplications or deletions in the DNA product of the reaction. Furthermore, if nonviral RNAs or chimeric RNAs containing viral and nonviral sequences are packaged into virions, such nonhomologous recombination events can create new joints and link a viral sequence to the nonviral sequences. These events are thought to play a central role in the process of transduction of cellular genes, most importantly during the formation of acute oncogenic retroviral genomes (see below).
Integration of Proviral DNA
The integration of linear retroviral DNA, like reverse transcription, is a crucial and defining feature of the retroviral life cycle. Integration is required for efficient replication of most retroviruses; mutants that are unable to integrate do not establish a spreading infection. The orderly and efficient integration of viral DNA is unique to the retroviruses. Although infection by some DNA viruses can result in the integration of viral DNA fragments into the host genome at low efficiency, these events are not the result of specific viral functions. Further, the establishment of the integrated provirus is responsible for much of retroviral biology. It accounts for the ability of the viruses to persist in the infected cell; for their ability to permanently enter the germ line; and for the mutagenic and oncogenic activities of the leukemia viruses. It also establishes a reservoir of latently infected cells in AIDS patients that resists antiviral drug therapy and that can be reactivated to induce virus replication.
Once the provirus is established, the DNA is permanently incorporated into the genome of the infected cell. There is no mechanism by which it can be efficiently eliminated. At very low frequencies, homologous recombination between the two LTRs can delete most of the provirus, but even here a single (“solo”) LTR remains.609 As the host cell divides, the provirus is transmitted to daughter cells as a new Mendelian locus. Thus,
it is likely to persist in the cell for its normal life span and to convert the cell permanently to a chronic producer of progeny virus.
it is likely to persist in the cell for its normal life span and to convert the cell permanently to a chronic producer of progeny virus.
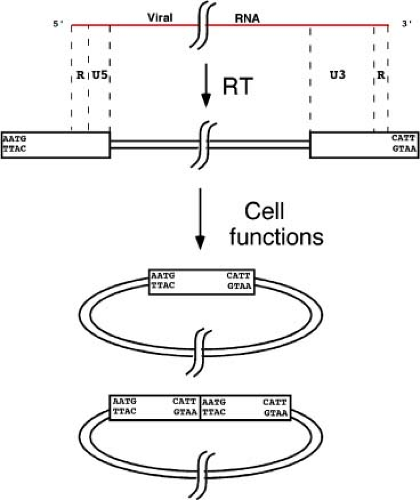
Unintegrated DNA Forms
The product of the reverse transcription reaction, as outlined in the previous section, is a full-length double-stranded linear DNA version of the genome, flanked at each end by copies of the LTR. The next step is the movement of the DNA into the nucleus, and the appearance two new DNA forms: closed circular molecules containing either one or two tandem copies of the LTR (Fig. 47.9). A small amount of the one-LTR circle may be formed during reverse transcription (see the Steps in reverse transcription of the retroviral genome section), but the bulk is thought to be formed by homologous recombination between the two LTRs of the linear DNA. The tandem two-LTR circles are apparently formed by the blunt-end ligation of the termini of the linear DNA. This event creates a unique sequence, termed the LTR–LTR junction, that is often used as a hallmark of nuclear entry of the viral DNA. The joints are often imperfect, with loss of nucleotides from one or both termini at the joint.556,626 There are also some circles that arise by autointegration of the ends of the linear DNA into internal sites, forming DNAs with deletions or inversions551; these circles are generally nonfunctional in terms of generating progeny virus.
Since three distinct unintegrated DNA form—one linear and two circular—coexist in the nucleus, it was uncertain for many years which form might serve as the precursor for establishment of the integrated provirus. In spite of prejudices based on such precedents as phage lambda, it is now clear that circles are not efficient substrates in the integration reaction and that the immediate precursor for the integration reaction is the linear duplex DNA. The circles are apparently dead-end products of a side reaction, formed by host enzymes acting on linear DNAs that have failed to integrate. There are settings and cell types in which unintegrated viral DNAs are observed to accumulate to high levels; various tissues in human HIV disease show considerable circular DNAs. While this DNA may reflect some unusual processing of the DNA, much of it is probably formed simply by massive infection occurring shortly before the DNA is harvested.
Unintegrated DNA is not a good substrate for forward transcription,527 perhaps because it is still retained in a complex that is poorly accessible to RNA polymerase. Mutant viruses that cannot integrate are unable to establish an efficient spreading infection, although low levels of virus can be produced.541 A very small subset of cells infected with such integration-defective mutants do integrate viral sequences through nonviral means, creating oligomeric tandem repeats similar to those formed after naked DNA–mediated transformation.222
Entry into the Nucleus
A key step that must take place before integration can occur is the entry of viral DNA into the nucleus. The mechanisms of nuclear entry are largely unknown, but there are probably at least two distinct routes used by different retroviruses. Simple retroviruses show a profound requirement for passage through mitosis for successful establishment of the integrated provirus,326,385,517,608 and the block in nondividing cells is at or close to the step of nuclear entry. Tests of the state of the viral DNA in nondividing cells are consistent with the notion that the preintegration complex must await the breakdown of the nuclear membrane in order to have access to the cellular DNA. Infection of nondividing cells results in the accumulation of linear dsDNA in the cytoplasm, and no further signs of infection. The viral DNA will persist in the cell for some time, and if the cell is stimulated to undergo division, the viral DNA will integrate and infection will proceed. However, the DNA loses its capacity to become activated in this way fairly rapidly.15,385 Some simple retroviruses are not strongly dependent on mitosis,228 and some postmitotic cell types may be susceptible to infection.338 For many viruses the restriction is quantitatively very significant, and profoundly limits the utility of simple retroviral vectors for gene therapy.
In contrast, lentiviruses and spumaviruses are able to successfully infect nondividing cells, suggesting that there must be an active transport of viral DNA through an intact nuclear membrane.77,325,326,409,623 This capability has made lentiviruses very attractive as gene delivery vectors for gene therapy applications. The molecular basis for this capability is a subject of great controversy. The lentiviral MA protein has been argued as essential for the infection of nondividing cells, and the phosphorylation of MA has been argued as necessary to promote
dissociation from the membrane and allow nuclear import, but these findings were discounted in later studies. Similarly, it has been shown that the Vpr protein is present in the preintegration complex, and can bind to nucleoporin components that may mediate nuclear import. DNA structures present at the second internal copy of the polypurine tract have also been suggested as important for infection of nondividing cells, but this notion has also been discounted. Another attractive model is that the IN protein might be involved in the nuclear import of the complex. IN itself contains nuclear localization signals that can function to target ectopically expressed IN to the nucleus, but these seem not to mediate PIC nuclear import or nuclear retention.
dissociation from the membrane and allow nuclear import, but these findings were discounted in later studies. Similarly, it has been shown that the Vpr protein is present in the preintegration complex, and can bind to nucleoporin components that may mediate nuclear import. DNA structures present at the second internal copy of the polypurine tract have also been suggested as important for infection of nondividing cells, but this notion has also been discounted. Another attractive model is that the IN protein might be involved in the nuclear import of the complex. IN itself contains nuclear localization signals that can function to target ectopically expressed IN to the nucleus, but these seem not to mediate PIC nuclear import or nuclear retention.
Recent experiments suggest that the CA protein of the incoming PIC may define competence for nuclear import.646 The lentiviral CA may serve to deliver the PIC to particular Nups, nuclear pore components, to initiate import. Studies of HIV-1 mutants with single changes in CA suggest that PICs can be imported via either of two alternative pathways, with wild-type virus virus using Nup153 and TNP03, and the N74D mutant using Nup155.99,306,320 Other studies have implicated Nup98 in HIV-1 PIC entry into the nucleus.151 Another study of import in vitro has suggested that a specific importer protein, importin 7, is required for PIC entry,167,661 though this has been disputed.672 Fractionation of extracts using similar in vitro import assays showed, remarkably, that tRNAs can promote uptake of PICS into nuclei.662 Whether tRNAs mediate import in vivo remains uncertain.
Structure of the Provirus
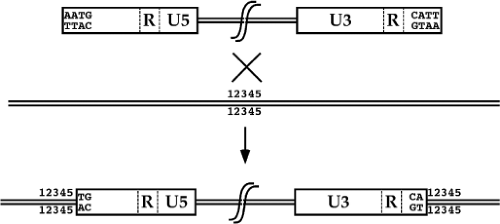
An important aspect of retroviral integration that distinguishes the process from nonviral or other viral mechanisms of DNA integration is the fact that the insertions create a consistent provirus structure. The integrated provirus is collinear with the product of reverse transcription, and consists of a 5′ LTR, the intervening viral sequences, and a 3′ LTR, inserted cleanly into host sequences. The joints between host and viral DNA are always at the same sites, very near the edges of the viral LTRs. As compared to the unintegrated linear DNA, there is a loss of a small number of base pairs, usually two, from each terminus of the viral DNA. There is also a duplication of a small number of base pairs of host DNA initially present once at the site of insertion that flank the provirus (Fig. 47.10). The number of base pairs duplicated is characteristic of each virus, and ranges from 4 to 6 bp.
Biochemistry of Integration
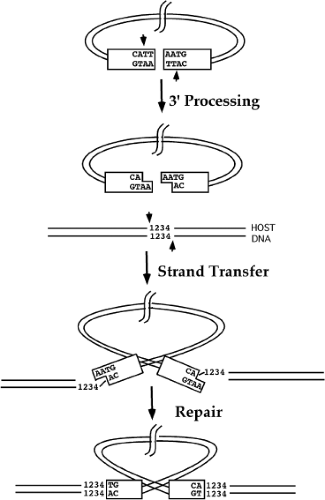
The actual integration of viral DNA into a target is mediated in vivo by the viral integrase protein IN,450,495,541 which is brought into the cell inside the virion, and acts to insert the linear DNA into the host chromosome. Some aspects of IN function have been studied by analysis of viral DNA formed in vivo.521 Most of our understanding of IN function, however, has been obtained through analysis of in vitro integration reactions, first using complexes extracted from infected cells,74,186 and later using recombinant IN protein. The reaction proceeds in two steps: 3′ end processing and strand transfer. A schematic view of these reactions is shown in Figure 47.11.
3′ End Processing
In the first step, the two terminal nucleotides at the 3′ ends of the blunt-ended linear DNA are removed by the integrase to produce recessed 3′ ends and correspondingly protruding 5′ ends. This cleavage occurs endonucleolytically at a highly conserved CA sequence, and releases a dinucleotide. For most viruses the terminal sequence is such that a TT dinucleotide is released, though this rule has exceptions. The ends do not remain covalently bound to protein, and the energy of the hydrolyzed phosphodiester bond is not retained.
Strand Transfer
In the second step, the 3′OH ends created by processing are used in a strand transfer reaction to attack the phosphodiester bonds of the target DNA.186 The attack occurs by an Sn2-type reaction, with inversion of the phosphorus center as detected with chiral labeling of the phosphate.158 The formation of the new phosphodiester bond between the viral end and host DNA displaces one of the phosphodiester bonds in the host DNA, leaving a nick. The protruding 5′ end of the viral DNA is not joined to the host DNA by IN. The reaction is a direct transesterification, and thus no ATP or other energy source is required. Mutational studies strongly suggest that the two activities—processing and joining—utilize the same active site residues. In fact, the two steps involve similar chemistry: 3′ end processing is an attack on DNA by a hydroxyl residue of water, while joining is an attack on DNA by a 3′ hydroxyl residue of another DNA. It should be noted that other hydroxyl residues can participate; alcohols such as glycerol can be utilized, and
the 3′OH of a DNA can even attack a phosphodiester bond on the same DNA, forming a cyclic product.158
the 3′OH of a DNA can even attack a phosphodiester bond on the same DNA, forming a cyclic product.158
Disintegration
The IN protein exhibits a third enzymatic activity in vitro: a reversal of the integration reaction known as disintegration.98 This activity releases DNA from a branched structure and seals the nick at the site of the branch. The significance of the activity in vivo is uncertain.
Target Site Duplication
In a wild-type virus, when the two ends are joined to the two strands of the target DNA, the two sites of attack are staggered by a few base pairs. After the joining, the resulting structure contains short gaps in the host DNA and unpaired bases from each 5′ end of the viral DNA. The 5′ ends of the viral DNA are not joined to host DNA by any known activities of IN. However, the 5′ ends are very quickly repaired in vivo, almost as quickly as the initial integration reaction.516 These discontinuities are presumed to be repaired by the host repair enzymes, though it is possible that the viral RT or IN could participate. The processing and filling in the gaps creates a short duplication of sequence that was present only once at the target site; these duplications flank the integrated provirus. The number of bases duplicated is characteristic of each virus. Thus, the murine and feline viruses cause a 4-bp duplication, HIV-1 causes a 5-bp duplication, and the avian viruses cause a 6-bp duplication.
Viral att Sites
The sequences at the termini of the viral DNA, the att sites, are recognized by the viral IN protein and are important for end processing and joining.79,104,109,610 These terminal sequences are imperfect inverted repeats. The most conserved residues are a CA dinucleotide pair that lies near the 3′ terminus and determines the site of 3′ processing. Sequences upstream from the CA for perhaps 10 to 12 bp are needed for efficient integration, but these sequences are different for different viruses, with no indication of broadly conserved sequence motifs. Since the two termini of any given virus are somewhat different, they usually show differential efficiency of utilization in various assays. The fact that two distinct ends are bound together in a complex may be important for the concerted integration of these ends into the target.615
The sequence-specific binding to the att site is probably performed by the core domain of IN. The nonspecific DNA binding activity of IN has made it difficult to detect sequence-specific binding to these regions, though under some conditions preferential binding to the authentic sequences can be demonstrated.161 The observation that an IN mutation can compensate for a mutation in the DNA termini provides evidence for the delicate interaction between IN and the DNA termini.
Both 3′ processing and strand transfer reactions are concerted reactions in vivo. The processing step occurs simultaneously at both termini of the viral DNA and requires the correct sequences at both termini. Thus, a mutation altering the sequence at one end of the viral DNA of MuLV blocks the processing reaction at both ends.405 This result suggests strongly that the reaction requires both termini to be loaded into a complex before hydrolysis proceeds. Similarly, the strand transfer reaction normally occurs so that both ends are joined to the target DNA, and at a fixed spacing between the two sites along the DNA helix. The 3′ processing and strand transfer reactions can both be carried out in vitro using native PICs, extracted from recently infected cells, and these reactions reconstruct the concerted nature of the in vivo reactions. Alternatively, integration can be performed using artificial DNA constructs and recombinant IN protein. However, these systems typically only mediate a half-reaction: that is, the uncoupled processing of one viral terminus and its joining into a single target DNA. Efforts have led to the identification of conditions and factors that mediate formation of a complex and that enhance concerted joining.7,175,616 Once such a protein–nucleic acid complex is formed, it is very stable.
Structure of the Integrase
The IN protein consists of three distinct domains: an N-terminal region containing an HHCC zinc-finger motif; a
central catalytic core containing the so-called D,D-35-E motif; and a less well-conserved C-terminal region. The IN protein is a multimer: it readily dimerizes, and at high concentration forms tetramers as well. All three regions may be involved in the multimerization of IN and in DNA binding. Many of the residues important for enzymatic activities have been identified by mutagenesis. The most crucial residues for catalysis are the acidic amino acids in the D,D-35-E motif, a highly conserved array of three residues in the core region of many integrases and transposases.308 Mutants indicate that both the N- and C-terminus are also important for function. Surprisingly, pairs of IN mutants with alterations in different regions of the molecule can often complement to restore normal function. The separate N-terminal domain can even complement a nonoverlapping fragment, suggesting that these domains can still co-assemble into a functional oligomeric complex.
central catalytic core containing the so-called D,D-35-E motif; and a less well-conserved C-terminal region. The IN protein is a multimer: it readily dimerizes, and at high concentration forms tetramers as well. All three regions may be involved in the multimerization of IN and in DNA binding. Many of the residues important for enzymatic activities have been identified by mutagenesis. The most crucial residues for catalysis are the acidic amino acids in the D,D-35-E motif, a highly conserved array of three residues in the core region of many integrases and transposases.308 Mutants indicate that both the N- and C-terminus are also important for function. Surprisingly, pairs of IN mutants with alterations in different regions of the molecule can often complement to restore normal function. The separate N-terminal domain can even complement a nonoverlapping fragment, suggesting that these domains can still co-assemble into a functional oligomeric complex.
Early X-ray crystallography work first defined the structures of the HIV-1 and avian virus IN core domains, and NMR methods defined the structures of the N- and C-terminal domains. Very recently a crystal structure of the complete integrase from a foamy virus in complex with a model target DNA oligonucleotide has been obtained.359 This structure reveals a tetramer, arranged as a dimer of dimers, holding the target DNA in a strongly bent conformation. The catalytic sites for strand transfer are nicely positioned to hold the viral DNA termini for attack of the target DNA (Fig. 47.12).
Preintegration Complex
Integrase does not normally act alone; a large complex of proteins and nucleic acid is responsible for mediating the formation of the provirus in vivo.66,94 The nature and components of the preintegration complex (PIC), or intasome, are not known in any detail for either the simple or the complex viruses. The PICs of the simple gammaretroviruses contain p12, CA, RT, and IN, but other viral proteins may be present.66,489 The PICs of the complex viruses contain only lower levels of CA, but contain MA, NC, Vpr, RT, and IN.388 Thus, the PICs of the complex viruses may be very different from those of the simple viruses, consistent with their distinctive ability to infect nondividing cells.165,166 Many of these proteins probably stay with the DNA even after entry into the nucleus. The PICs contain a large structure protecting the two ends of the DNA, and perhaps holding them in proximity. The formation of this structure, detected as a footprint in a modified nuclease sensitivity assay,621 requires both IN and the correct sequences at the termini of the DNA.622
Host Proteins and Integration
A number of host proteins have been identified as potentially involved in the establishment of the provirus. One such protein is BAF-1, a low-molecular-weight protein recovered from the MuLV PIC for its ability to inhibit autointegration of the LTR edges into internal sites in the viral DNA.321 By inhibiting this reaction, BAF-1 can enhance normal integration into target DNAs in trans. However, infection of BAF-1-deficient cells occurs normally, suggesting that BAF-1 is not an essential player in the early events of the viral life cycle. Another such partner is LEDGF (lens epithelial-derived growth factor, a misnomer), a nuclear protein of uncertain function, which binds directly to the HIV-1 IN and dramatically enhances its integration activity.96,339,358
The integration of retroviral DNA has been shown to activate an apoptotic program in cells deficient in DNA-stimulated protein kinase (DNA-PK), an enzyme implicated in the DNA damage response125; the related kinase ATR and other components of the nonhomologous end joining repair machinery may also be involved.123,124 While it is not clear whether these kinases play any direct role in integration, they are likely involved in sensing the products of active integrase and responding to the damage. Their absence leads to substantial cell death in cells taking up the PIC.
Distribution of Integration Sites
An important issue affecting the ability of the retroviruses to create mutations is the distribution of integration sites in the host genome. Proviruses are inserted at very approximately random locations in the genome, and thus have the opportunity to create mutations in any gene. Various studies, however, have uncovered significant deviations from a completely random distribution. At the sequence level, examination of large numbers of integration sites has revealed weak but statistically highly significant preferences for symmetrical target sequences.212,240,643 Large-scale surveys of thousands of integration sites cloned from pools of infected cells have allowed analysis of the frequency of insertions into the 5′ upstream regions of genes, into transcribed regions, and into nontranscribed regions. The results show that different viruses show distinct biases for their target sites.30,389,535,642 HIV-1 tends to insert into transcribed regions, more or less equally along such regions; MuLV tends to selectively insert its DNA in sequences upstream from the 5′ end of transcribed regions, near transcriptional start sites; ASLV shows only very weak preference for active genes and none for 5′ regions. Activation of transcription per se can apparently, in some circumstances, inhibit avian retroviral integration at specific genes.372 These studies collectively show that various retroviruses have evolved mechanisms to choose aspects of their integration sites, presumably in support of their chosen life styles during infection. The biases are presumably determined largely by their respective IN proteins, but could also involve other viral proteins.
Expression of Viral Rnas
The integration of the provirus signals a dramatic change in the life style of retroviruses; it marks the end of the early phase of the life cycle and the beginning of the late phase. The early phase is driven by viral enzymes performing abnormal events such as reverse transcription and DNA integration, while the late phase is mediated by host enzymes performing such relatively normal processes as transcription and translation. This late phase of gene expression may begin immediately with the synthesis of viral RNAs and proteins, and the assembly of progeny virions (see Fig. 47.13 for an overview). For many viruses, the transcriptional promoters that drive this expression are constitutively active and cause the production of virions in a relatively unregulated way. In other viruses the activity of the promoter may be regulated, either by viral or host factors. The basic phenomenology of proviral gene expression will be reviewed, and the regulation exhibited by the complex retroviruses will be mentioned briefly.
Overview of Viral RNA Synthesis
The synthesis of viral RNA from viral DNA leads to the formation of a long primary transcript, which is then processed and may be spliced to form a small number of stable transcripts. The U3 region of the LTR contains a promoter recognized by the RNA polymerase II system; these sequences direct the initiation of transcription starting at the U3-R border. Cellular machinery then caps the 5′ end of the RNA with m7G5′ppp5′Gmp. The first G residue after the cap is a templated base in the provirus. Transcription proceeds through the genome, and continues through the 3′ LTR and into the downstream flanking host DNA. Finally, the RNA is cleaved and polyadenylated at the R-U5 border of the 3′ LTR, generating a complete, unspliced viral genomic RNA suitable for incorporation into the virion particle. Most genomes contain an AAUAAA sequence acting as the signal for this 3′ processing. The sequence normally lies in the R region, but the complete sequence needed for recognition can be complex, lying upstream or downstream, and may even be discontinuous, brought together by RNA folding to create the functional signal. The exact site of polyadenylation is not critical for virus replication; mutants in which the polyadenylation signal is inactivated generate longer RNAs that extend into downstream flanking sequences.668 These RNAs very efficiently mediate normal replication.578 A subset of the RNA is spliced to give rise to one or more subgenomic RNAs. The patterns of spliced mRNAs can be simple or exceedingly complex. Both the unspliced and spliced RNAs are then exported from the nucleus for translation.
Initiation of Transcription
The efficiency of initiation of transcription at the 5′ LTR is the major determinant of the levels of viral RNA formed in the cell. The promoter in the LTR is typically a very potent one, and the levels of viral RNA are often constitutively high. However, the cell type, the physiologic state, and the integration site169 can all result in substantial variation in the efficiency of transcription. In some viruses, the promoter is not constitutively active but depends on the activity of specific transcription factors such as the glucocorticoid receptors.
Positive Regulatory Elements in U3
The transcriptional elements in the U3 region of the simple viruses contain both core promoter sequences and enhancers. The core promoters contain a TATA box, bound by TFIIB; a CCAAT box, bound by CEBP525; and sometimes an initiator sequence near the U3-R border. The U3 regions of even closely related retroviruses are very diverse, and can evolve rapidly during viral replication. The enhancers are similar to those found
in many host promoters in containing multiple short-sequence motifs, arranged in very close packing; often there are tandemly repeated copies of some of these motifs. These short sequences are the binding sites for a large number of host factors that regulate transcription (e.g., see 562). Different cells and cell types will make use of distinct arrays of these factors to mediate transcription from a given viral LTR.213 The factors are not simply additive but may interact in complex ways on particular viral sequences. A partial list of these factors used by various retrovirus LTRs includes: Sp1; USF-1; the Ets family of factors, which include more than 20 members in vertebrates; the core-binding factor (CBF), consisting of an a-b heterodimer; nuclear factor 1 (NF1); and a mammalian type C retrovirus enhancer factor (MCREF-1). Specific viruses may often contain recognition sites for other more specific positive regulatory factors. Major examples of such factors include the glucocorticoid receptors, driving expression of the MMTV genome, and to a much lesser extent, other MuLVs; NF-κB, important for expression from the HIV-1 LTR in certain cell types; the GATA factors for Cas-BR-E and other viruses; and the myb protein. Evidence has been obtained that the STAT factors, DNA binding proteins normally activated the Janus kinases (Jaks) may also be important for MMTV transcription.493
in many host promoters in containing multiple short-sequence motifs, arranged in very close packing; often there are tandemly repeated copies of some of these motifs. These short sequences are the binding sites for a large number of host factors that regulate transcription (e.g., see 562). Different cells and cell types will make use of distinct arrays of these factors to mediate transcription from a given viral LTR.213 The factors are not simply additive but may interact in complex ways on particular viral sequences. A partial list of these factors used by various retrovirus LTRs includes: Sp1; USF-1; the Ets family of factors, which include more than 20 members in vertebrates; the core-binding factor (CBF), consisting of an a-b heterodimer; nuclear factor 1 (NF1); and a mammalian type C retrovirus enhancer factor (MCREF-1). Specific viruses may often contain recognition sites for other more specific positive regulatory factors. Major examples of such factors include the glucocorticoid receptors, driving expression of the MMTV genome, and to a much lesser extent, other MuLVs; NF-κB, important for expression from the HIV-1 LTR in certain cell types; the GATA factors for Cas-BR-E and other viruses; and the myb protein. Evidence has been obtained that the STAT factors, DNA binding proteins normally activated the Janus kinases (Jaks) may also be important for MMTV transcription.493
Negative Regulatory Elements
A number of negative regulatory factors that reduce viral expression have been identified. Embryonic carcinoma cells, and true embryonic cells, are the best-characterized examples of cell types that strongly repress LTR-mediated transcription through expression of negative regulatory proteins. The MuLVs are silenced via a stem-cell specific repressor that binds to a site, curiously, overlapping almost perfectly with the proline tRNA primer binding site.289,473 The proteins responsible for this silencing in mouse embryonic stem cells have recently been identified as TRIM28 (Kap-1) and the zinc-finger protein ZFP809.637,638 Viruses that use an alternate primer tRNA and thus lack the pbs recognition site for these proteins can escape the repression.239 Other negative factors include one known variously as UCRBP, NF-E1, or YY1,176 and a cellular embryonal LTR-binding protein (ELP; 598).
trans-Acting Viral Regulatory Factors
The complex retroviruses encode an array of small regulatory proteins that can activate transcription from the viral LTR in trans. Examples of these transactivators include the HTLV-1 Tax protein132 and the HIV-1 Tat protein.120 The Tax protein acts in concert with a complex of host proteins, the activating transcription factor/CRE-binding protein (ATF/CREB), and binds to three cAMP response elements in the viral LTR. Tax thus sets up a positive feedback loop that results in high levels of viral transcripts. The Tat protein is unusual among transcriptional activators in that it binds to a structure in the 5′ end of nascent viral RNA, rather than to DNA.136,548 Tat binds to a bulged hairpin structure, the TAR element, and recruits a pair of host proteins, cyclinT/cdk9, to the RNA. These proteins enhance the ability of RNA polymerase to elongate beyond the LTR and down the genome with high processivity, probably by phosphorylation of the C-terminal repeat domain (CTD) of the polymerase. Again, the result is a strong positive feedback loop that results in high levels of viral RNA. (For more detailed discussion of tat function, see 120, and Chapter 49 of this book.)
Beginning and Ending the RNA
Because proviruses contain two identical LTRs, transcription can be initiated at both 5′ LTR and 3′ LTR. However, the 5′ LTR is generally much more efficiently utilized than the 3′ LTR.235 One possible mechanism is promoter interference, in which the upstream promoter being active suppresses the utilization of the downstream promoter. It is possible that elements near the 3′ LTR may restrict use of the downstream LTR, so that generally transcripts initiating at the 5′ LTR predominate. These restraints may be lost in tumors, in which transcription from the 3′ LTR can be significantly enhanced.59 Similarly, since there are two LTRs, transcripts might in principle be subject to 3′ end processing at either the 5′ LTR or the 3′ LTR, but most of the RNAs formed extend from the 5′ LTR to the 3′ LTR.
RNA Processing
The full-length transcript of the retroviral genome is directed into several pathways. A portion of the transcripts is exported directly from the nucleus and serves as the genome to be packaged into the progeny virion particle, assembling either at the plasma membrane or in the cytoplasm. Another portion with identical structure is also exported and used for translation to form the Gag and Gag-Pol polyproteins. It is not yet clear if these two subsets are truly distinct, whether there can be interchange between the pools, or whether there is a single pool of such molecules used for both purposes. A third portion is spliced to yield subgenomic mRNAs. For the simple retroviruses, there is a single spliced mRNA encoding the Env glycoprotein. For the complex viruses, there can be multiple alternatively spliced mRNAs, encoding both Env and an array of auxiliary proteins. Examples of the complicated array of mRNAs that are formed for both simple and complex viruses are shown in Figure 47.14. The protein products of these multiply spliced mRNAs will be discussed in Chapters 48–52.
The splicing and subsequent export from the nucleus of only a portion of an initially transcribed RNA is an extraordinary process; normally splicing of cellular mRNA precursors goes to completion, and only then is the mRNA exported. The export of a precursor mRNA is prevented until splicing is complete. At least three aspects of the retroviral genome may promote the export of unspliced mRNAs. First, the splice sites of the viral RNA may have poor overall efficiency of utilization by the splicing machinery in the cell.280 The sequences at the splice donor and acceptor regions are often poor matches to the consensus sequences for splice sites, and mutations that make the sites better matches increase splicing and are actually deleterious to virus replication. These mutations can be suppressed by secondary mutations that reduce splicing efficiency. The overall folding of the RNA may affect the efficiency of splicing; thus, sequences at some distance, as in the gag gene, may modulate splicing.565
Second, studies of ASLV have identified specific sequences that act as negative regulators of splicing (NRS) through their
interaction with host factors.11,378,379 These elements can be important for the expression of transduced genes in some viruses.558 Similar signals may exist in other viruses; mutations in the Gag region of MuLV can affect RNA processing in complex ways.
interaction with host factors.11,378,379 These elements can be important for the expression of transduced genes in some viruses.558 Similar signals may exist in other viruses; mutations in the Gag region of MuLV can affect RNA processing in complex ways.
In addition, unspliced mRNAs contain cis-acting elements that promote the export of the RNA out of the nucleus, the so-called constitutive transport elements (CTEs).68 These sequences are located near the 3′ end of the genomic RNA of MPMV, and possibly in similar regions of ASLV. The CTE is recognized by one or more host proteins that assemble a complex onto the RNA to mediate its export, including Tap and its cofactor Nxt. In the complex viruses, RNA export is regulated through complex interactions of the Rex or Rev gene products with cis-acting sites, the RRE elements that promote RNA export; and of various host factors with the CRS/INS elements that prevent it (see 121 for review). The key players include Crm1, a cellular nuclear export factor, and DDX3, an RNA helicase (see Chapter 49 for detailed discussion of the mechanism of Rev action).
Viral RNAs are subject to other modifications common to cellular mRNAs. Like cellular mRNAs, the N6 position on specific A residues can be methylated, and other sites can be modified by dsRNA adenosine deaminase. The significance of these modifications is uncertain.
Translation and Protein Processing
All retroviral genomes, at a minimum, contain ORFs designated the gag, pro, pol, and env genes. These genes are expressed by complex mechanisms to form precursor proteins, which are then processed during and after virion assembly to form the mature, infectious virus particle. The expression of the various proteins as large precursors that are subsequently cleaved provides several advantages: it allows for many proteins to be made from one ORF; it ensures that the proteins are made at proper ratios; and it allows for many proteins to be targeted to the virion during assembly as a single entity. The gag, pro, and pol genes are expressed in a complex way from the full-length unspliced mRNA. The arrangement of these genes, and especially the way pro is expressed, are different in different viruses. A summary of the arrangement of the ORFs of various viruses is shown in Figure 47.15.
Gag Gene Expression
The gag gene is present at the 5′ proximal position on all retroviral genomes. A full-length mRNA, identical in sequence to the genomic RNA, is translated in the cytoplasm to form a Gag precursor protein, in the 50 to 80 kDa range. Translation begins with an AUG initiator codon and proceeds to a terminator codon at the 3′ end of the ORF. The viral RNA typically contains a relatively long 5′ untranslated region, and there has been uncertainty regarding whether ribosomes could scan from the 5′ cap to the start codon for Gag translation. These 5′ RNA sequences are predicted to contain stable secondary structures that would inhibit scanning. Furthermore, the long 5′ UTRs often contain AUG codons in contexts that are favorable for translation, that are not in frame with the gag ORF, and presumably would inhibit successful translation of Gag. Experiments suggest that for the MuLVs and related endogenous RNAs, an internal ribosome entry site (IRES) is present near the start of the gag ORF and is used to initiate translation in a cap-independent mechanism.45,46,344 Thus, at least in these viruses, ribosomes can bind directly near the gag gene and do not need to scan the mRNA. Although the suggestion is not without controversy,383 it is likely that many other viruses, including HIV-1, also utilize IRES elements for translation of Gag.130,131,427 In the case of HIV-1, the IRES is remarkable in that critical sequences extend downstream of the AUG, lying within the Gag coding region.76
Some retroviruses encode an additional Gag protein besides the major product, termed gPr80gag or “glycoGag.” This Gag protein is longer than the major product and derives from translational initiation at a nonconventional CUG codon
upstream from the initiating AUG codon. Translation beginning at this codon first forms an N-terminal leader sequence and then proceeds in the same reading frame through the normal AUG and the rest of the Gag protein. Thus, where the proteins overlap their sequences are identical. The leader sequence contains a functional signal peptide directing the translation machinery to the endoplasmic reticulum, and specifying that the Gag protein be co-translationally inserted into the secretory pathway. The Gag become glycosylated at several sites, is transported via the golgi to the cell surface, and persists for some time as a membrane-bound glycoprotein, with the carboxyterminal domain exposed on the cell surface.476 The protein is processed into several fragments and has a relatively short half-life. It is not required for virus replication in some cells.542 However, the protein can facilitate release of virus at lipid rafts,420 apparently acting in concert with the host La protein,421 and can replace the function of the HIV-1 Nef protein in promoting virion release.479 Very recent work suggests that GlycoGag serves to inhibit the Apobec3 restriction factor.
upstream from the initiating AUG codon. Translation beginning at this codon first forms an N-terminal leader sequence and then proceeds in the same reading frame through the normal AUG and the rest of the Gag protein. Thus, where the proteins overlap their sequences are identical. The leader sequence contains a functional signal peptide directing the translation machinery to the endoplasmic reticulum, and specifying that the Gag protein be co-translationally inserted into the secretory pathway. The Gag become glycosylated at several sites, is transported via the golgi to the cell surface, and persists for some time as a membrane-bound glycoprotein, with the carboxyterminal domain exposed on the cell surface.476 The protein is processed into several fragments and has a relatively short half-life. It is not required for virus replication in some cells.542 However, the protein can facilitate release of virus at lipid rafts,420 apparently acting in concert with the host La protein,421 and can replace the function of the HIV-1 Nef protein in promoting virion release.479 Very recent work suggests that GlycoGag serves to inhibit the Apobec3 restriction factor.
The major Gag product is often modified by the addition of myristic acid, a relatively rare 14-carbon fatty acid, to the penultimate aminoterminal residue, a glycine.234 The addition is mediated by a myristyl CoA transferase that co-translationally transfers myristate from a myristyl CoA donor to the amino group of the glycine residue, forming an amide bond. The fatty acid is important for the membrane localization and binding of the Gag precursor, increasing the hydrophobicity of the aminoterminal domain. Mutant Gags in which the glycine is altered are not modified; these Gags do not associate with membrane properly and do not aggregate to form virions.75,210,500 It should be noted that although the myristate is important, it is not sufficient for membrane targeting; hydrophobic residues in the MA domain are also required. Furthermore, basic residues further downstream in the MA of some viruses form a patch of positive charge that interacts with negatively charged phospholipids in the membrane.
An aminoterminal myristate is not found on the Gags of BIV, EIAV, visna, or ASLV. For the avian retroviruses, the aminoterminus is not myristylated but rather acetylated. The Gag protein of these viruses is apparently sufficiently hydrophobic to be targeted to the membrane without the fatty acid in avian cells, though, curiously, not for ASLV in mammalian cells. Alteration of the avian Gag to allow its myristylation permits virion assembly in mammalian cells633 and does not block its function in avian cells.
pro Gene Expression
The relative position of the pro gene on retroviral genomes is always similar—in between gag and pol. However, the pro gene is expressed in very different ways in different viruses. Sometimes it is fused in frame onto the 3′ end of gag, sometimes it is fused to the 5′ end of pol, and sometimes it is present as a separate reading frame. These various patterns have led to considerable confusion in the literature; sometimes pro is considered a portion of gag, or sometimes of pol. Because of these different patterns of expression, it is best to consider this ORF as a separate gene.
The various arrangements of the pro gene and its mode of expression are as follows. For the alpharetroviruses, gag and pro are fused and expressed as a single protein; pol is in a different reading frame, and a frameshift is used to express the Gag-Pro-Pol polyprotein. For the betaretroviruses and deltaretroviruses, gag, pro, and pol are all in different frames and successive frameshifts are used to express Gag-Pro and Gag-Pro-Pol polyproteins. For the gammaretroviruses and epsilonretroviruses, gag and a pro-pol fusion are in the same reading frame and separated by a stop codon, and translational readthrough is used to make Gag-Pro-Pol. For the lentiviruses, gag and a pro-pol fusion are in different reading frames, and frameshifting is used to make Gag-Pro-Pol. Finally, for the spumaviruses, pro is fused to pol, and the Pro-Pol protein is expressed without Gag, from a spliced mRNA. More about these varied mechanisms of expression is presented in the following section.
pol Gene Expression
The pol gene encodes several proteins needed at lower levels for the replication of the virus, including the reverse transcriptase and integrase enzymes. The pol ORF is not expressed as a separate protein in most retroviruses, but rather is expressed as a part of a larger Gag-Pro-Pol fusion protein. The Gag-Pro-Pol protein must be made at the correct abundance, in proportion to the amount of Gag protein, for efficient assembly of infectious virus; expression of only Gag-Pro-Pol does not result in virion assembly.170,458 The formation of this protein is mediated by one of two mechanisms, depending on the virus.
Translational Readthrough
In the gammaretroviruses and epsilonretroviruses, the Gag and Pro-Pol ORFs are in the same reading frame, separated by a single UAG stop codon at the boundary between Gag and Pro-Pol. The translation of Gag-Pro-Pol in these viruses occurs by translational readthrough—that is, by suppression of termination—at the UAG stop codon.651 Most of the time, translation of the RNA results simply in the formation of the Gag protein. But approximately 5% to 10% of the time, ribosomes translating the RNA do not terminate at the UAG codon, and instead utilize a normal aminoacyl tRNA, usually a glutamine tRNA, to insert an amino acid at the position of the stop codon. Translation then continues, in frame, through the entire long pro-pol ORF, resulting in the formation of a long Gag-Pro-Pol precursor protein.
The high-level suppression of termination is specified by a specific structure in the RNA immediately downstream of the UAG stop codon.241,449 The precise features of this structure that are required for suppression are not completely known, but they include a purine-rich sequence immediately downstream of the stop codon, and a pseudoknot formed from the next 60 or so nucleotides.172 The structure may slow translation, and it may also in some other way alter the balance between termination, which requires binding of termination factors eRF1 and eRF3 by the ribosome, versus incorporation of an amino acid, which requires misreading of the codon by an aminoacyl tRNA. No changes in the tRNA pool occur during infection. The signals in the RNA can operate to mediate suppression of both UAA and UGA termination codons as well as UAG.
A screen for proteins interacting with the MuLV RT resulted in the identification of the eukaryotic termination factor eRF1, and subsequent studies showed that overexpression of RT could inhibit termination and promote translational readthrough of the Gag stop codon in vivo.434 Mutant viruses with point mutations in RT blocking the interaction with eRF1 were unable to express normal levels of Gag-Pol and failed to replicate. These results suggest that RT, likely in the context of the nascent
Gag-Pol protein, can bind and inhibit eRF1, increasing the level of readthrough to increase its own synthesis. The final level of Gag-Pol produced in this positive feedback loop presumably is ultimately limited by other factors.
Gag-Pol protein, can bind and inhibit eRF1, increasing the level of readthrough to increase its own synthesis. The final level of Gag-Pol produced in this positive feedback loop presumably is ultimately limited by other factors.
Translational Frameshifting
In the alpharetroviruses and lentiviruses, the gag and pol ORFs lie in different reading frames, and the formation of the Gag-Pro-Pol fusion is mediated by a translational frameshift mechanism.257 Most of the time, translation again results in the simple formation of the Gag protein. But approximately 10% of the time, as the translation approaches a specific site near the end of the gag ORF, the ribosome slips back one nucleotide (a –1 frameshift) and proceeds onward in the new reading frame. The ribosome passes through the stop codon out of frame and continues to synthesize protein using the codons of the pol ORF. As for readthrough, the determinants of frameshifting lie in the RNA sequence and structure near the site of the event. The requirements for frameshifting include a “slippery site,” a string of homopolymeric bases where the frameshift occurs; these are oligo U or oligo A in different viruses. In addition, the frameshifting requires either a very large and near-perfect hairpin or stem-loop structure (as for HIV-1 group M viruses); or a large pseudoknot structure (as for HIV-1 group O viruses), similar to those used in readthrough, though apparently containing a distinctive bend at the junction of the two paired sequences. As for readthrough, the proper frameshifting efficiency is crucial for normal virus replication.
In the betaretroviruses (e.g., MMTV) and deltaretroviruses (e.g., BLV, HTLV-1), the pro gene is present as a separate ORF, in a different reading frame from that of gag or pol. Two successive frameshifts are utilized to make the long Gag-Pro-Pol fusion protein. Near the 3′ end of the gag ORF, ribosomes carry out a first (–1) frameshift and continue into the pro ORF; near the 3′ end of the pro ORF, they perform a second (–1) frameshift and continue on into the pol ORF. These two frameshifts occur at extremely high frequencies—as much as 30% of the time that the ribosome transits through each site—so that the overall frequency of formation of the Gag-Pro-Pol protein is perhaps 10% that of formation of Gag.
Separate Pol Expression
The spumaviruses are unique among the retroviruses in that the synthesis of the Pol protein is not mediated by the formation of a Gag-Pol fusion protein. Instead, a subgenomic spliced mRNA is translated directly to form a separate Pro-Pol protein.159,340 This protein must be directed to the assembling virion by distinct domains rather than by the Gag portion of a Gag-Pol fusion.
env Gene Expression
In all retroviruses the env gene is expressed from a subgenomic mRNA. The env message is a singly spliced mRNA, in which a 5′ leader is joined to the coding region of env. Thus, the bulk of the gag and pol genes are removed as an intron from the mRNA. The resulting message is exported to the cytoplasm and translated from a conventional AUG initiator codon. In the alpharetroviruses, the AUG is actually the same one used for Gag translation; it lies in the leader, and the splicing brings this AUG and the first six codons into frame with the env coding region. The first translated amino acids constitute a hydrophobic signal peptide, and direct the nascent protein to the rough endoplasmic reticulum. The leader is removed by a cellular protease (the signal protease) in the ER, and the protein is heavily glycosylated by transfer of oligosaccharide from a dolichol carrier to asparagine residues on Env. These residues lie in the conventional Asn-X-Ser/Thr motifs recognized by the modification enzymes. Near the end of the co-translational insertion of Env into the ER, a highly hydrophobic sequence acts as a stop transfer signal to anchor the protein in the membrane. The remaining C-terminal portion of the protein stays on the cytoplasmic side of the membrane.
Before the Env proteins are transported to the cell surface, they are folded and oligomerized in the ER. The formation of oligomers is required for stable expression of the protein, and is sensitive to overall conformation; many mutants of Env show defects in oligomerization.599 Envelope proteins generally form trimers in the mature virus.347 The most studied envelope proteins (ASLV and HIV-1) may pass through dimeric or tetrameric intermediates, but the nature of these intermediates is not clear. The folding of the protein is presumably catalyzed by chaperone proteins in the ER and the formation of disulfide bonds between various pairs of cysteine residues by disulfide interchange enzymes.
The Env protein is then exported to the Golgi and cleaved by furin proteases to form the separate SU and TM subunits. This cleavage is essential for the normal function of the Env protein. The cleavage occurs at a dibasic pair of amino acids,139 producing a hydrophobic N-terminus for the TM protein that is required to mediate fusion of the viral and host membranes during virus entry. In the Golgi the sugar residues are modified by the sequential removal of mannose residues and addition of N-acetyl glucosamine and other sugars to many of the oligosaccharide. O-linked glycosylation and sulfation of Env glycoproteins have also been documented.478 The pathway by which Env is transported to the cell surface is not fully understood, but presumably host vesicular transport systems are utilized. There is evidence that clathrin adaptor complexes interact with the cytoplasmic tail of Env and direct its movement to the plasma membrane. The protein typically becomes a prominent cell-surface protein on the infected cell.
In polarized epithelial cells, Env proteins are often restricted to the basolateral surface of the cell.441 This localization is mediated by a tyrosine-based motif, Yxxf, present in the cytoplasmic tail of Env442 (x, any amino acid; f, hydrophobic residue). Remarkably, this targeting of Env can redirect the budding of Gag proteins to this surface.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree