Respiratory Syncytial Virus and Metapneumovirus
Peter L. Collins
Ruth A. Karron
History
Human respiratory syncytial virus (HRSV) was first isolated in 1955 from a laboratory chimpanzee with illness resembling the common cold.398 Shortly thereafter, the same virus was recovered from infants with respiratory illness, and serologic studies indicated that infection in infants and children was common.86,87 HRSV is now recognized as the most important viral agent of pediatric lower respiratory tract illness (LRI) worldwide. In many areas it outranks other microbial pathogens as a cause of pneumonia and bronchiolitis in infants. In addition, HRSV can infect and cause disease in individuals of all ages and severe disease in the elderly and in profoundly immunosuppressed individuals.146,163,233 Worldwide, acute respiratory infection (ARI) is the leading cause of mortality due to infectious disease, and HRSV remains one of the pathogens deemed most important for vaccine development. HRSV research is hampered by its poor growth in vitro and its physical instability. HRSV has a single serotype with two antigenic subgroups A and B.
Human metapneumovirus (HMPV) was first described in 2001 following its isolation from infants and children experiencing HRSV-like disease of unknown etiology.572 There is serologic evidence of extensive pediatric infection dating back more than 50 years, and thus HMPV is newly discovered rather than newly emerged.572,608,610 The virus had been overlooked because it grows slowly in vitro, has a delayed cytopathic effect, and usually requires added trypsin for activation of the fusion F protein. HMPV is recognized as an important agent of respiratory tract disease worldwide, especially in the pediatric and elderly populations, although its impact is less than that of HRSV.608 HMPV also has a single serotype with two subgroups A and B.515
Infectious Agent
Classification
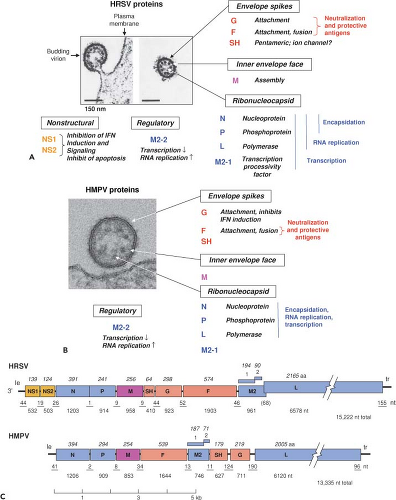
HRSV and HMPV are enveloped, cytoplasmic viruses with single-stranded nonsegmented negative-sense RNA genomes. They are members of the family Paramyxoviridae (Chapter 33) of the order Mononegavirales (Chapter 30). Paramyxoviridae has two subfamilies: Paramyxovirinae, which includes the human and animal parainfluenza viruses (PIVs, Chapter 34), mumps virus (Chapter 35), and measles virus (Chapter 36) among others, and Pneumovirinae, which consists of HRSV and HMPV and their animal relatives. There are two genera in Pneumovirinae: Genus Pneumovirus consists of HRSV, bovine RSV (BRSV), and pneumonia virus of mice (PVM), and genus Metapneumovirus consists of HMPV and avian metapneumovirus (AMPV, formerly called turkey rhinotracheitis virus [TRTV] or avian pneumovirus [APV]). Comparisons of
these viruses based on gene maps and nucleotide sequences are shown e-Figures 38.1 and 38.2.
these viruses based on gene maps and nucleotide sequences are shown e-Figures 38.1 and 38.2.
(Note that the descriptions in the following sections usually will begin with HRSV followed by HMPV.)
Virion
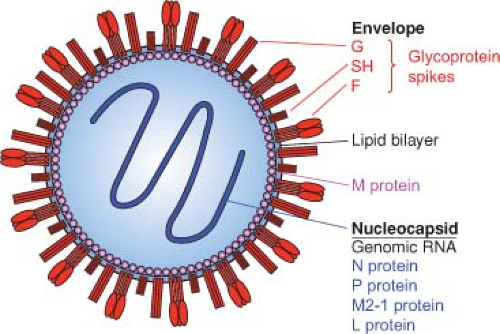
The HRSV virion consists of a nucleocapsid that is packaged in a lipid envelope derived from the host cell plasma membrane during budding (Fig. 38.1 and Fig. 38.2C). When visualized by electron microscopy (Fig. 38.2C and E), virions appear as irregular spherical particles of 100 to 350 nm in diameter and long filamentous forms that often predominate and are 60 to 200 nm in diameter and up to 10 μm in length. Despite the variability in size and shape, the ultraviolet (UV) inactivation kinetics of infectivity indicates that most particles contain a single functional genome.140 Both forms of the virion mostly remain cell associated. The filaments can be visualized as projections from the surface of infected cells by fluorescence photomicroscopy19,270,423,475 (Fig. 38.2B). The nucleocapsid appears in electron micrographs as a herringbone structure that is characteristic of Paramyxoviridae. However, the HRSV nucleocapsid is narrower than those of prototype members of Paramyxovirinae (12 to 16 nm compared to 17 to 20 nm) and has a steeper pitch.19,42,423
The HRSV envelope contains three virally encoded transmembrane surface glycoproteins: the major attachment protein G, the fusion protein F, and the small hydrophobic (SH) protein (Figs. 38.3A and 38.4). In addition, there is a nonglycosylated matrix M protein that is thought to form a layer on the inner face of the envelope. The viral glycoproteins are present as transmembrane homooligomers that are visualized as short (11 to 20 nm), closely spaced (intervals of 6 to 10 nm) surface projections or “spikes.” HRSV lacks a neuraminidase or a hemagglutinin; PVM alone agglutinates murine erythrocytes, via its G protein.339
The viral RNA is associated with four nucleocapsid/polymerase proteins: the nucleoprotein N, the phosphoprotein P, the transcription processivity factor M2-1, and the large polymerase subunit L (Fig. 38.3A). However, polymerase activity in purified virions, an activity found in prototypic members of Mononegavirales, has not been demonstrated for HRSV preparations.
HRSV virions can readily lose infectivity during handling, presumably reflecting particle instability. This can be partly overcome by agents such as sugars that reduce aggregation and improve thermal stability.17 When recombinant HRSV was engineered to contain a foreign attachment protein from baculovirus in place of the HRSV glycoproteins, the stability of infectivity was improved,491 suggesting that the lability of the particle may reside in one or more of the glycoproteins. Other data have implicated the F protein in thermo instability.469 The long filamentous shape of the particle may also contribute to fragility and loss of infectivity.
HMPV virions were visualized by electron microscopy as pleomorphic spheres and filaments that appear to have general similarity to those of HRSV443,572 (Fig. 38.2F). The spherical particles had a reported diameter of 150 to 600 nm with envelope spikes of 13 to 17 nm. The nucleocapsid diameter was reported as 17 nm, suggesting a possible difference compared to HRSV.443 HMPV appears to lack a hemagglutinin572; other virion-associated activities have not been reported. HMPV has the same array of structural proteins as HRSV (Fig. 38.3B). The infectivity of HMPV particles is markedly more stable than that of HRSV.557
RNA
The HRSV genome (Fig. 38.3C) is a single negative-sense strand of RNA ranging in length from 15,191 to 15,226 nucleotides for six sequenced strains including the subgroup A strains A2 (15,222 nucleotides; GenBank accession number M74568), Long (15,226 nucleotides; AY911262), S2 (15,191 nucleotides; NC_001803), and line 19 (15,191 nucleotides; FJ614813), and the subgroup B strains B1 (15,225 nucleotides; NC_001781) and 9320 (15,225 nucleotides; AY353550). More recently, complete sequences were reported and analyzed for 60 and four additional subgroup A and B strains, respectively.313a,491a,538a In addition, extensive sequence information and extensive inter-subgroup comparison is available for subgroup B strain 18537.274,276,278 The genome is neither capped nor polyadenylated. Both in virions and intracellularly, the genome is tightly and completely bound by N protein to create an RNase-resistant nucleocapsid, as is typical of Mononegavirales. This tight encapsidation likely protects the genome, which lacks stabilizing features of capping and polyadenylation, from degradation. It also likely shields the genome from recognition by host cell pattern recognition receptors, especially (a) the cytoplasmic helicases retinoic acid–inducible gene I (RIG-I) and melanoma differentiation–associated protein (MDA-5), which detect triphosphorylated RNA and double-stranded RNA (dsRNA) and activate the cellular transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor kappa B (NF-κB) to induce type I interferon (IFN) and proinflammatory cytokines, and (b) RNA-inducible protein kinase R (PKR), which also activates NF-κB and phosphorylates eukaryotic translation initiation factor 2a (eIF-2a) to inhibit translational initiation as part of antiviral defense.
The HRSV genome contains 10 genes in the order 3′ NS1-NS2-N-P-M-SH-G-F-M2-L (Fig. 38.3C) that are transcribed sequentially into 10 separate messenger RNAs (mRNAs).95,99,107,140,388 Each gene begins with a highly conserved nine-nucleotide gene-start (GS) transcription signal and ends with a moderately conserved 12- to 13-nucleotide
gene-end (GE) signal275,315 (e-Fig. 38.3A). The first nine genes are separated by intergenic regions that vary in length from 1 to 58 nucleotides for the strains sequenced to date.275 These lack any conserved motifs, are poorly conserved between strains, and appear to be unimportant spacers. The last two HRSV genes, M2 and L, overlap by 68 nucleotides106 (Fig. 38.3C and e-Fig. 38.3A). Specifically, the GS signal for the L gene is located upstream, rather than downstream, of the M2 GE signal. The same overlap occurs in BRSV; overlapping genes are not found in any other members of Paramyxoviridae, but sometimes are found in Rhabdoviridae and Filoviridae. The 3′ and 5′ ends of the genome consist of short extragenic leader and trailer regions (44 and 155 nucleotides long, respectively, in strain A2).
gene-end (GE) signal275,315 (e-Fig. 38.3A). The first nine genes are separated by intergenic regions that vary in length from 1 to 58 nucleotides for the strains sequenced to date.275 These lack any conserved motifs, are poorly conserved between strains, and appear to be unimportant spacers. The last two HRSV genes, M2 and L, overlap by 68 nucleotides106 (Fig. 38.3C and e-Fig. 38.3A). Specifically, the GS signal for the L gene is located upstream, rather than downstream, of the M2 GE signal. The same overlap occurs in BRSV; overlapping genes are not found in any other members of Paramyxoviridae, but sometimes are found in Rhabdoviridae and Filoviridae. The 3′ and 5′ ends of the genome consist of short extragenic leader and trailer regions (44 and 155 nucleotides long, respectively, in strain A2).
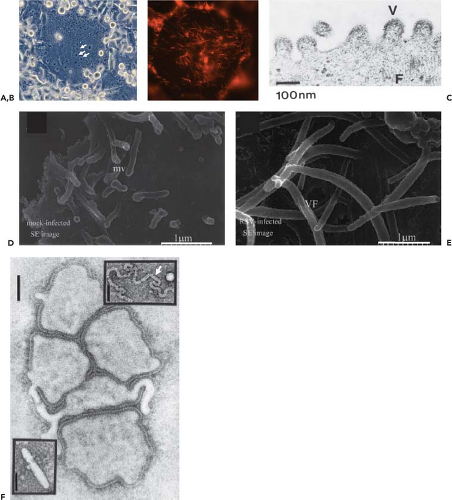 Figure 38.2. Photomicrographs (A and B) and electron micrographs (C–F) of human respiratory syncytial virus (HRSV)–infected cells (A–E) and human metapneumovirus (HMPV) virions (F). A: Photomicrograph of an HRSV-infected cell monolayer showing a virus-induced syncytium (several nuclei are indicated with arrows). (Courtesy of Dr. Alexander Bukreyev.) B: Fluorescence photomicrograph of an HRSV-infected syncytium (not the same one as in A) stained with an antibody specific to the F protein, showing filamentous viral projections. (Courtesy of Dr. Ursula J. Buchholz.) C: Negatively stained electron micrograph of budding HRSV virions: V indicates a budding virion and F indicates filamentous cytoplasmic structures that likely are nucleocapsids. (Courtesy of Dr. Robert M. Chanock284) D and E: Field emission scanning electron micrographs of the surface of uninfected (D) and HRSV-infected (E) cells, illustrating viral filamentous structures (VF in E) that are thought to form at sites of virus budding and may yield filamentous particles; also shown are microvilli (mv in D) that are found in uninfected cells. (D and E reprinted from Jeffree CE, Rixon HW, Brown G, et al. Distribution of the attachment (G) glycoprotein and GM1 within the envelope of mature respiratory syncytial virus filaments revealed using field emission scanning electron microscopy. Virology 2003;306:254–267; copyright 2003, with permission from Elsevier.) F: Negatively stained electron micrograph of HMPV (bar markers represent 100 nm): the main panel shows spherical-type particles, the upper insert shows a free nucleocapsid (arrow), and the lower insert shows a filamentous or rod-like particle. (Reprinted from Peret T C, Boivin G, Li Y, et al. Characterization of human metapneumoviruses isolated from patients in North America. J Infect Dis 2002;185:1660–1663, by permission of Oxford University Press.) |
The HRSV mRNAs contain a methylated 5′ cap structure m7G[5′]ppp[5′]Gp24 and are polyadenylated by reiterative copying on a U tract in the GE signal. Each HRSV mRNA encodes a single major protein except for M2, which has separate open reading frames (ORFs) for the M2-1 and M2-2 proteins. The M2-1 ORF is located in the upstream part of the mRNA, whereas the M2-2 ORF is located downstream and
overlaps by 32 nucleotides in strain A2. Translation of the downstream M2-2 ORF depends on reinitiation by ribosomes exiting the upstream M2-1 ORF, a process that appears to be influenced by the structure of a region of RNA located ∼150 nucleotides upstream of the M2-2 translational start site.202,203 Whereas the P genes of Paramyxovirinae encode additional accessory proteins by overlapping ORFs, alternative translational start sites, and RNA editing (Chapter 33), the P gene of HRSV (and HMPV) encodes only P.
overlaps by 32 nucleotides in strain A2. Translation of the downstream M2-2 ORF depends on reinitiation by ribosomes exiting the upstream M2-1 ORF, a process that appears to be influenced by the structure of a region of RNA located ∼150 nucleotides upstream of the M2-2 translational start site.202,203 Whereas the P genes of Paramyxovirinae encode additional accessory proteins by overlapping ORFs, alternative translational start sites, and RNA editing (Chapter 33), the P gene of HRSV (and HMPV) encodes only P.
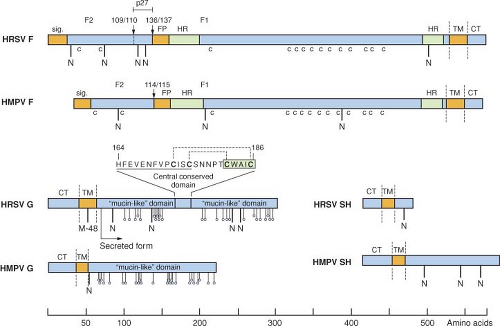 Figure 38.4. Primary structures (approximately to scale) of the F, G, and SH surface glycoproteins of human respiratory syncytial virus (HRSV) strain A2 and human metapneumovirus (HMPV) strain CAN97-83. Hydrophobic domains are brown bars: Sig., signal peptide; FP, fusion peptide; TM, transmembrane anchor; CT, cytoplasmic tail. Heptad repeats (HR) in the F protein are green and cysteine residues conserved between the F proteins of HRSV and HMPV are indicated underneath (indicated as c). Downward-facing arrows identify the cleavage-activation site(s) in the F protein. Potential acceptor sites for N-linked carbohydrate are indicated as downward facing stalks with N. For each G protein, the 25 potential acceptor sites for O-linked sugars predicted by NetOGlyc 2.0 to be the most likely to be utilized are indicated as downward facing stalks with small circles. The sequence excerpt above the HRSV G protein shows the conserved segment (underlined) and cystine noose; cysteine residues are bold; the disulfide bonding pattern is indicated by dotted lines201; and the fractalkine CX3C motif is boxed. M-48 in the HRSV G protein is the translational start site for the secreted form, and the mature secreted form is indicated.476 |
HRSV transcription follows the general Mononegavirales model111,140,316 (Chapters 31 and 33), involving initiation at a single 3′ promoter and a start-stop-restart sequential mechanism guided by the GS and GE signals. RNA sequences important in transcriptional initiation are shown in e-Fig. 38.4. Capping seems to be an essential step for efficient mRNA elongation: when capping was blocked using a novel HRSV-specific inhibitor, transcription produced uncapped abortive RNAs of ∼45 to 50 nt.343 Termination at the various GE signals typically is somewhat inefficient, resulting in readthrough transcription that creates mRNAs representing two or more adjacent genes and their intervening intergenic regions.107 These readthrough mRNAs account for approximately 10% of total mRNA.
The M2/L gene overlap raises two questions for the model of sequential transcription: (a) how do polymerases that exit the M2 gene find the upstream L GS signal (or does polymerase enter independently at that site), and (b) how do polymerases that initiate at the L GS signal avoid premature termination when they cross the M2 GE signal? Studies with a minireplicon system showed that when polymerase completes transcription of the M2 gene, it efficiently gains access to the L gene by retrograde scanning.170 The polymerase appeared to scan in both directions. This led to the realization that scanning may be a common function of the polymerase, and may occur at each gene junction as well as prior to the initiation of transcription and RNA replication. The M2 GE signal within the L gene indeed causes premature termination for 90% of L gene transcripts, producing a 68-nucleotide polyadenylated RNA that does not appear to encode a protein and is not known to have any further significance.106 The synthesis of full-length L mRNA depends on polymerase readthrough at
the M2 GE signal. Therefore, the “error” of reading through a GE signal is necessary for synthesis of this essential mRNA.106 Premature termination of 90% of L gene transcripts might be expected to severely downregulate the production of full-length mRNA, but the ability of the polymerase to recycle back to the L GS signal apparently relieves much of this effect, and the amount of L mRNA produced for HRSV relative to the other mRNAs appears to be about the same as for other members of Paramyxoviridae. The gene overlap does not seem to be of any particular benefit to the virus, and may be an accidental arrangement that can be tolerated due to the scanning function of the polymerase.
the M2 GE signal. Therefore, the “error” of reading through a GE signal is necessary for synthesis of this essential mRNA.106 Premature termination of 90% of L gene transcripts might be expected to severely downregulate the production of full-length mRNA, but the ability of the polymerase to recycle back to the L GS signal apparently relieves much of this effect, and the amount of L mRNA produced for HRSV relative to the other mRNAs appears to be about the same as for other members of Paramyxoviridae. The gene overlap does not seem to be of any particular benefit to the virus, and may be an accidental arrangement that can be tolerated due to the scanning function of the polymerase.
RNA replication by HRSV also follows the general Mononegavirales model (Chapters 31 and 33). RNA sequences important in the initiation of RNA replication are shown in e-Fig. 38.4. The replicating polymerase ignores the GS and GE signals and produces a complete positive-sense copy of the viral genome that is called the antigenome and that also is tightly encapsidated. The antigenome serves in turn as the template for producing progeny genomes. Chain elongation of nascent genomes and antigenomes depends on concurrent encapsidation.373 For viruses in the subfamily Paramyxovirinae, the nucleotide length of the genome must be an even multiple of six in order for efficient RNA replication to occur (“rule of six”; Chapter 33), reflecting a requirement for nucleocapsid organization, but there appears to be no comparable requirement for members of Pneumovirinae.303,488
The HMPV genome is approximately 13.2 kb—nearly 2 kb shorter than that of HRSV—and lacks the NS1 and NS2 genes (Fig. 38.3C). In addition, the order of the SH, G, F, and M2 genes differs between HRSV (SH-G-F-M2) and HMPV (F-M2-SH-G). Correcting for the lack of NS1 and NS2 for HMPV and the difference in gene order, the genomes of HRSV and HMPV share 50% nucleotide sequence identity. Strains for which complete sequences have been reported include the following: CAN97-83 (13,335 nt, GenBank accession AY297749) and NL/00/1 (13,350, AF371337) of subgroup A, and CAN98-75 (13,280, AY297748) and NL/1/99 (13,293, AY525843) of subgroup B.46,259
Features of the structure, encapsidation, transcription, and replication of the HMPV genome are generally similar to those described above for HRSV. The cis-acting signals of HMPV have considerable similarity to those of HRSV46,571 (e-Fig. 38.3B). The HMPV intergenic regions can be longer than those of HRSV, up to 190 nt in the case of CAN97-83 (Fig. 38.3C and e-Fig 38.3B). Unlike HRSV, the HMPV M2-2 protein appears to be translated independently of the upstream M2-1 ORF.66
Proteins
HRSV encodes 11 separate proteins (Fig. 38.3A). HMPV encodes nine proteins (Fig. 38.3B) that generally correspond to those of HRSV except for the lack of NS1 and NS2.46,571 Table 38.1 shows amino acid sequence relatedness between viruses within subfamily Pneumovirinae.
Fusion F Glycoprotein
The HRSV F and L proteins are the ones that most closely resemble their counterparts in Paramyxovirinae. As is typical for Paramyxoviridae, the HRSV F protein directs viral penetration by fusion between the virion envelope and the host cell plasma membrane. Later in infection, F protein expressed on the cell surface can mediate fusion with neighboring cells to form syncytia (Fig. 38.2A). Recent findings suggest that the HRSV F protein also plays a major role in viral attachment involving interaction with the cellular protein nucleolin.543
F is a type I transmembrane surface protein that has a cleaved signal peptide at the N-terminus and a membrane anchor near the C-terminus100 (Fig. 38.4). A predicted three-dimensional structure of the prefusion form of the HRSV F protein has been described based on homology modeling with the crystal structure of the Newcastle disease virus F protein,517 and crystal structures representing the postfusion form have recently been published.378,536 The structure of the Paramyxoviridae F protein is described in detail in Chapter 33.
Table 38.1 Percent Amino Acid Sequence Identity between the Proteins of HRSV Subgroup A (HRSV-A) or HMPV Subgroup A (HMPV-A) and the Indicated Virusesa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
As is typical for Paramyxoviridae, F is synthesized as an inactive F0 precursor that assembles into a homotrimer.74 HRSV F is activated by cleavage in the trans-Golgi complex by furin or a furin-like cellular endoprotease to yield two
disulfide-linked subunits: NH2-F2–F1-COOH.103,531 The N-terminus of the F1 subunit that is created by this cleavage contains a hydrophobic domain (the fusion peptide) that inserts directly into the target membrane to initiate fusion. The F1 subunit also contains two areas of heptad repeats that associate during fusion, driving a conformational shift that brings the viral and cellular membranes into proximity100,633 (Chapter 33). HRSV F protein can direct efficient fusion independent of other viral proteins.283 This differs from the situation for most members of Paramyxovirinae, for which efficient fusion requires interaction with the homologous attachment protein (Chapter 33).
disulfide-linked subunits: NH2-F2–F1-COOH.103,531 The N-terminus of the F1 subunit that is created by this cleavage contains a hydrophobic domain (the fusion peptide) that inserts directly into the target membrane to initiate fusion. The F1 subunit also contains two areas of heptad repeats that associate during fusion, driving a conformational shift that brings the viral and cellular membranes into proximity100,633 (Chapter 33). HRSV F protein can direct efficient fusion independent of other viral proteins.283 This differs from the situation for most members of Paramyxovirinae, for which efficient fusion requires interaction with the homologous attachment protein (Chapter 33).
The F proteins of HRSV and BRSV are unique in Paramyxoviridae in having two cleavage sites, rather than one cleavage site200,636 (Fig. 38.4). The site that is immediately upstream of the fusion peptide (with cleavage between residues 136/137 in HRSV strain A2) is the one that corresponds to other Paramyxoviridae. This site in HRSV contains six tandem Arg and Lys residues (Lys-Lys-Arg-Lys-Arg-Arg↓). The second, novel site is located 27 amino acids upstream (with cleavage between residues 109/110) and has the sequence Arg-Ala-Arg-Arg↓. Therefore, both sites contain the preferred furin cleavage motif (Arg-X-Arg/Lys-Arg↓). HRSV F appears to be readily cleaved during intracellular processing, although some of the F protein packaged in virions remained uncleaved at site 109/110. There is indirect evidence that the double-cleavage site in the F protein is associated with a hyperfusogenic character that is linked to reduced thermostabilty and the ability to fuse independent of a cognate attachment protein.469
Cleavage at the two sites in F0 releases a short peptide of 27 amino acids (p27) that contains two (strain A2) or three (strain Long) potential acceptor sites for N-linked sugars, of which at least two are utilized in strain Long.636 (Note that the remainder of the F protein has three acceptor sites for N-linked sugars, two in F2 and one in F1, all of which are utilized.639) For BRSV, this peptide contains a tachykinin sequence motif and is processed and released from BRSV-infected cells to yield a virokinin that induced smooth muscle contraction in vitro, which is a property of tachykinins.638 Tachykinins also have proinflammatory and immunomodulatory activities. Recombinant BRSV in which most of p27 was deleted, or in which the upstream cleavage site was mutated, replicated as efficiently as the wild-type parent in calves, but induced less pulmonary inflammation and a somewhat lower titer of serum neutralizing antibody.567,637 Therefore, p27 of BRSV has the potential to augment viral disease by effects on smooth muscle contraction and pulmonary inflammation, and might also stimulate the host immune response. In contrast, the sequence of p27 of HRSV does not resemble a tachykinin, and HRSV p27 did not have tachykinin-like properties in vitro.638
The HMPV F protein shares a moderate level of amino acid sequence identity with HRSV (33%)46,571 and is similar in general organization (Fig. 38.4). As with HRSV, HMPV F directs fusion involved in penetration and syncytium formation, and does so efficiently without need of the G and SH proteins.48 HMPV F also appears to play a role in attachment via interaction with cellular αvβ1 integrin.118 This integrin characteristically binds to a specific recognition sequence, Arg-Gly-Asp, and this motif is present in the HMPV F protein and mediates its binding to cells.
The HMPV F0 precursor contains only a single cleavage activation site (Fig. 38.4). The sequence at this site, Arg-Gln-Ser-Arg↓, does not conform to the consensus furin motif and, consistent with this, clinical isolates typically require exogenous trypsin for growth in vitro. In vivo, cleavage of the HMPV F protein presumably depends on secreted protease present in the lumen of the respiratory tract. Several observations indicate that this is not a limiting factor in pathogenesis. For example, in some cases, serial passage of HMPV clinical isolates in cell culture resulted in the emergence of mutants that no longer required added trypsin, but these did not exhibit increased replication in hamsters.492 These mutants contained the substitution of Pro in place of Ser in the –2 position of the cleavage activation site (Arg-Gln-Pro-Arg↓), which conferred intracellular cleavability even though it did not create a furin motif. Similarly, when the F cleavage site of recombinant HMPV was engineered so that it was multibasic and cleaved intracellularly, this did not increase its replication in African green monkeys (AGMs).45
Glycoprotein G
The HRSV G protein was originally thought to be the sole viral attachment protein,334 but it now appears that the F protein also plays a role, as noted. The HRSV G protein has no apparent relatedness by sequence or structure to the attachment HN, H, and G proteins of Paramyxovirinae and has only half the amino acid length.278,597 HRSV G is a type II transmembrane glycoprotein, with a hydrophobic signal/anchor located near the N-terminus (amino acids 38–63 in strain A2) and the C-terminal two thirds of the molecular oriented extracellularly (Fig. 38.4). A secreted version of G is produced by translational initiation at the second ATG in the ORF (codon 48), which lies within the signal/anchor sequence.255,476 This truncated form is then trimmed by proteolysis, removing the remainder of the signal/anchor and creating a new N-terminus at Asn-66 for the final secreted form. The secreted form constituted approximately 20% of the total G protein expressed in HRSV-infected cells in vitro, but because of its rapid secretion it accounted for 80% of the G protein released in cell culture by 24 h post infection, the remainder being virion associated.255
The polypeptide backbone of HRSV G has an Mr of approximately 32,000. In the case of strain A2, an estimated four N-linked sugar side chains are added co-translationally, increasing the Mr to 45,000.102,598 G assembles in the endoplasmic reticulum into oligomers that probably are trimers or tetramers, and O-linked sugars are added subsequently in the trans-Golgi compartment or network.102,598 Mature G migrates in gel electrophoresis as a broad, seemingly heterogeneous band of Mr 80,000–90,000. Analysis of an F/G fusion protein (created as a potential vaccine) expressed in insect cells by a recombinant baculovirus indicated that G contains approximately 24–25 O-linked side chains.592 However, when HRSV was grown in an in vitro model of human airway epithelium (HAE)—specifically, a differentiated pseudostratified mucociliary tissue that closely resembles the authentic airway epithelium631—the Mr observed for the G protein was 180,000.319 Because these cells would be considered to be a more authentic substrate than typical immortalized monolayer cultures, this suggests that the amount of carbohydrate present on G is substantially greater than previously thought. The presence of a sheath of host-specified sugars might shield the G protein from immune recognition, and might interfere with antigen processing and presentation. The secreted form of G appears to be
processed in a similar fashion to the membrane-anchored form except that it is secreted as a monomer, and no antigenic differences were detected when the secreted and membrane-bound forms were analyzed with an extensive panel of monoclonal antibodies (MAbs).155
processed in a similar fashion to the membrane-anchored form except that it is secreted as a monomer, and no antigenic differences were detected when the secreted and membrane-bound forms were analyzed with an extensive panel of monoclonal antibodies (MAbs).155
Most of the ectodomain of HRSV G consists of two large domains that are highly divergent between strains and have a “mucin-like” structure. Like mucin, these domains have a high content of serine, threonine, and proline residues; are heavily glycosylated; and are thought to have an extended, nonglobular secondary structure. Because there are more than 75 serine and threonine residues in the mucin-like domains as potential acceptor sites for O-linked sugars, there may be heterogeneity in site usage that could provide additional antigenic heterogeneity. The significance of this mucin-like character remains unknown. One possibility is that it somehow helps the virus penetrate the protective mucous layer overlying the respiratory epithelium.
The two mucin-like domains flank a central conserved domain that includes a 13-residue segment (positions 164–176 in strain A2) that is completely conserved among HRSV strains, as well as an overlapping segment containing four closely spaced invariant cysteine residues (positions 173, 176, 182, and 186)278 (Fig. 38.4). Disulfide linkages occur between Cys-173 and Cys-186, and between Cys-176 and Cys-182, to create a cystine noose.201 The cysteine-rich segment contains a Cys-X3-Cys (CX3C) motif involving Cys-182 and Cys-186 embedded in a region of limited sequence relatedness with the CX3C chemokine called fractalkine.561 A peptide containing this sequence mimicked the leukocyte chemoattractant activity of fractalkine in an in vitro assay.
The HRSV G protein participates in viral attachment in vitro by binding to glycosaminoglycans (GAGs), which are long unbranched chains of repeating disaccharide subunits that are part of the glycocalyx present on the outer surface of the cell. The conserved central domain of G seemed like an obvious candidate to be involved in attachment. However, this domain can be deleted from recombinant HRSV with little effect on virus replication in HEp-2 cells or in mice.548 A study that probed for possible GAG-binding domains by evaluating interaction between peptides spanning the ectodomain of G and the GAG heparin identified a potential binding site shortly downstream from the central conserved domain,175 but this also could be deleted with little effect on HRSV infection and replication in vitro or in mice.548 More recently, cleavage of the C-terminal domain of G was found to result in virus that bound less efficiently to GAGs and had reduced infectivity in HAE cultures, suggesting that this domain is important for the attachment function of G.319
Extensive passage of a subgroup B strain of HRSV in AGM Vero cells—in an attempt to attenuate the virus—resulted in mutants with various spontaneous deletions involving most of the G and SH genes.288 One such mutant virus that was evaluated as a potential vaccine replicated efficiently in Vero cells but was highly restricted in humans. Recombinant HRSV strain A2 from which the G gene was deleted (ΔG) also replicated efficiently in Vero cells. However, the ΔG virus was restricted in HEp-2 cells, with defects at the levels of virus binding, fusion, and assembly.544,545,547,550 In addition, the ΔG virus was highly restricted in mice and might not have replicated at all.550 Therefore, HRSV G is dispensable for replication in Vero cells but it is essential for efficient replication in HEp-2 cells and in vivo.
The HMPV G protein (Fig. 38.4) has a number of similarities with HRSV G, although it lacks significant sequence identity46,571 (Table 38.1). HMPV G has a comparable N-terminal proximal signal/anchor domain and a high content of serine/threonine/proline residues concentrated in the ectodomain.46,571 In addition it is modified by the addition of N-linked and O-linked sugars to yield a mature form that migrates in gel electrophoresis as a diffuse band of Mr 97,000.341 Its amino acid sequence also is highly divergent between the HMPV subgroups. However, HMPV G lacks the conserved central domain and cystine noose mentioned above for HRSV and is correspondingly shorter in length. A secreted form of HMPV G has not been described, and the amino acid sequence gives no suggestion that such a species exists. As with HRSV, the HMPV G protein binds to cell surface GAGs, which may reflect its role in attachment.551 Peptide binding studies suggested that two closely spaced clusters of basic amino acids at positions 149–155 and 159–166 may mediate binding to GAGs.551
Deletion of G from recombinant HMPV was less attenuating than for HRSV. Deletion of G had little effect on HMPV replication in vitro, and the HMPV ΔG mutant had a low-to-moderate level of replication in hamsters and AGMs.44,48 Interestingly, infection of epithelial cells in vitro with the ΔG mutant resulted in increased activation of the transcription factors IRF3 and NF-κB and increased expression of type I IFN and proinflammatory cytokines compared to wild-type HMPV, implying that the G protein otherwise inhibits these responses.21 The HMPV G protein was found to bind to the RIG-I cytoplasmic RNA recognition receptor, which explains its inhibitory effect.
Small Hydrophobic SH Protein
The HRSV SH protein (Fig. 38.4) is a short (64 amino acids for strain A2) transmembrane protein that is anchored by a hydrophobic signal-anchor sequence near the N-terminus, with the C-terminus oriented extracellularly.101,427 Most of the SH protein remains unglycosylated (Mr ∼7,500), but there are also forms that contain a single N-linked side chain (resulting in Mr ∼13,000 to 15,000), as well as the further addition of polylactosaminoglycan (Mr ∼21,000 to 60,000 or more), as well as N-terminally truncated forms that arise from translational initiation at the second methionine codon in the ORF.9,427 However, the significance of these multiple forms is not known.
Chemical cross-linking studies indicated that the HRSV SH protein associates into pentamers.101 When SH was expressed in bacteria, it was incorporated into the membrane, resulting in increased membrane permeability.446 Incorporation of partial or full-length SH into artificial membranes in vitro resulted in the formation of pentameric and hexameric pore-like structures and the acquisition of cation-selective channel-like activity.81,183 Therefore, the SH protein may have properties of a viroporin, which typically are small proteins that modify membrane permeability and can have roles in budding and apoptosis.
Recombinant HRSV from which SH was deleted replicates with wild-type efficiency in vitro and appears to be fully fusogenic.69,544 The SH protein was reported to reduce apoptosis, but the effect was small.181 SH also appeared to inhibit signaling from tumor necrosis factor (TNF)-α.181 The ΔSH virus was slightly attenuated in mice and chimpanzees,69,603 but deletion of the SH gene in an experimental live vaccine strain did not increase its level of attenuation in seronegative children.290 Therefore, the function(s) and impact of the HRSV SH protein seem unclear.
The SH protein of HMPV is nearly three times longer (177–183 amino acids) than its HRSV counterpart46,571 (Fig. 38.4). It has the same membrane orientation and a similar array of nonglycosylated and glycosylated forms. Passage of HMPV frequently results in mutations that ablate SH expression,43 suggesting that the protein may be somewhat toxic in cell culture. Infection of epithelial cells and mice with the ΔSH virus resulted in a modest increase in NF-κB activation and expression of proinflammatory proteins compared to wild-type virus, suggesting that the SH protein downregulates the innate response, but the effect may be small.20 In vivo, deletion of SH had little or no effect on HMPV replication in hamsters or in AGMs.44,48
Matrix M Protein
The HRSV M protein is a nonglycosylated internal virion component that is smaller than its Paramyxovirinae counterparts (256 amino acids versus 335–375 amino acids), with little apparent amino acid sequence relatedness. HRSV M appears to play two roles typical for Mononegavirales: it helps organize virion components at the plasma membrane for budding,254,547 and it may silence viral RNA synthesis in preparation for packaging into the virus particle.191 A crystal structure determined for the HRSV M protein revealed a monomer that is organized into compact N-terminal (residues 1 to 126) and C-terminal (residues 140–255) domains joined by a 13-residue linker.394 The surface of M was found to contain a large positively charged area that extends across the two domains and the linker and may mediate association with the negatively charged membrane as well as with nucleocapsids.394 The HMPV M protein (254 amino acids) likely is a close functional counterpart.
Nucleocapsid/Polymerase Proteins N, P, L, and M2-1
The N, P, and L proteins of HRSV appear to be close functional analogs of their counterparts in other members of Paramyxoviridae. The N protein binds tightly along the entire length of genomic RNA and antigenomic RNA to form separate RNase-resistant nucleocapsids that are templates for RNA synthesis. The P protein is a multifunctional adapter that helps mediate interactions between components of the nucleocapsid/polymerase complex. The L protein contains the polymerase catalytic domains. Studies with HRSV minireplicons showed that N, P, and L are necessary and sufficient to direct RNA replication.98,215,626 N, P, and L alone also have transcriptase activity, but fully processive transcription requires in addition the M2-1 protein.98,171
The 391 amino acid HRSV N protein is shorter than its counterparts in Paramyxovirinae (approximately 490–555 amino acids), and sequence relatedness is limited to several conserved segments located towards the C-terminus.27,542 HRSV N protein produced in bacteria was recovered in decamer rings bound to bacterial RNA, and a crystal structure was obtained542 that is described in e-Figure 38.5.
The HRSV P protein (241 amino acids) is shorter than its Paramyxovirinae counterparts (approximately 390–605 amino acids) and lacks evident sequence relatedness. However, it is thought to have the same general array of functions. Like its counterparts in Paramyxovirinae, HRSV P operates as a stable homotetramer formed through a multimerization domain in the middle of the molecule.15,82,344 The C-terminal region of the P tetramer interacts with N protein in the nucleocapsid by binding to a hydrophobic pocket surrounded by positively charged residues made up from discontinuous segments within amino acids 46–151 of the N protein, a structure distinct from that of other members of Mononegavirales182a (e-Fig. 38.5). Soluble P also binds to free N protein monomers—probably through the N-terminal domain of P—and delivers N to nascent genomes/antigenomes during RNA replication. P thus prevents N from self-aggregating or binding to nonviral RNA.82 In addition, P binds to the L294 and M2-112,559 proteins and helps mediate their interactions with the nucleocapsid. P is an essential polymerase co-factor. It may contribute to conformational changes that help the polymerase access the RNA template82 and appears to be necessary for promoter clearance and chain elongation by the viral polymerase.147 It also appears to have a role in dissociating the M protein from the nucleocapsid during uncoating to initiate infection.13
P is the major phosphorylated HRSV protein and contains phosphate 10 to 12 or more sites, with different sites exhibiting differing rates of turnover: the C-terminal domain contains low-turnover phosphates and accounts for most of the total phosphate, the middle domain contains intermediate-turnover phosphates, and the N-terminal domain contains high-turnover phosphates.14,418 Many of the activities of P described above appear to be directed by dynamic phosphorylation and dephosphorylation of P at a subset of these sites, usually involving a small percentage of the total phosphate content.12,13,14,15,580 Most of the constitutive phosphorylation, involving five sites, could be ablated in recombinant HRSV with only modest effects on virus growth.350
The 194 amino acid HRSV M2-1 protein is an essential transcription processivity factor; in its absence, the viral polymerase terminates prematurely and nonspecifically within several hundred nucleotides of the 3′ end of the genome, and downstream genes are not significantly transcribed.94,97,98,171 M2-1 also decreases the efficiency of termination at the GE transcription signals—possibly a reflection of the same processivity activity—resulting in increased production of readthrough mRNAs.245 HRSV M2-1 accumulates in phosphorylated and nonphosphorylated forms244 and forms a homotetramer via an oligomerization domain at residues 32 to 63.559 M2-1 contains a cysteine–histidine zinc finger motif (C-X7-C-X5-C-X3-H) near its N-terminus (residues 7 to 25) that is essential for its activity244; this motif is conserved in Pneumovirinae.571 The HRSV M2-1 protein binds RNA, and may be delivered to the RNA template by the P protein.80,120,559 M2-1 is unique to Pneumovirinae, although VP30 of Family Filoviridae has some similarities.
The 2,165 amino acid HRSV L protein is similar in length to its Paramyxovirinae counterparts and has low but unambiguous sequence relatedness along nearly its entire length.527 Specific segments of L are conserved within and beyond Mononegavirales and appear to include polymerase motifs.451 A putative nucleotide-binding domain involved in capping mRNA was identified in the central region of the L protein based on sequence analysis of HRSV mutants selected for resistance to novel capping inhibitors, in which resistance was associated with an amino acid substitution at position 1381, 1269, or 1421.343 In separate work, an amino acid substitution at position 1049 or 1169 in the L protein was associated with reduced efficiency of termination at the GE signals, resulting in increased synthesis of polycistronic mRNAs and reduced growth efficiency.79,282
The HMPV N, P, L, and M2-1 proteins are similar in size to their HRSV counterparts and share significant amino acid
sequence identity46,571 (Table 38.1). One notable difference is that, whereas M2-1 appears to be essential for HRSV, recombinant HMPV in which the M2-1 ORF has been silenced is viable and replicates in vitro with an efficiency that is only marginally reduced.66 HMPV lacking M2-1 appeared to execute processive transcription efficiently, although the level of RNA accumulation was somewhat reduced. Therefore, in contrast to HRSV, HMPV M2-1 appears to be a nonessential accessory protein of unknown function.66 However, in the hamster model, replication of HMPV lacking M2-1 could not be detected, indicating that M2-1 is important for replication in vivo.
sequence identity46,571 (Table 38.1). One notable difference is that, whereas M2-1 appears to be essential for HRSV, recombinant HMPV in which the M2-1 ORF has been silenced is viable and replicates in vitro with an efficiency that is only marginally reduced.66 HMPV lacking M2-1 appeared to execute processive transcription efficiently, although the level of RNA accumulation was somewhat reduced. Therefore, in contrast to HRSV, HMPV M2-1 appears to be a nonessential accessory protein of unknown function.66 However, in the hamster model, replication of HMPV lacking M2-1 could not be detected, indicating that M2-1 is important for replication in vivo.
M2-2 Protein
HRSV M2-2 is a small (90 amino acids for strain A2) protein that is expressed at a low level that may reflect inefficiency of the stop–restart mechanism of translation noted previously.5 It is not known whether M2-2 is packaged in the virion. Recombinant HRSV in which the M2-2 ORF has been silenced grows more slowly in vitro than wild-type HRSV, although it eventually achieves a similar titer.41,271 In ΔM2-2 virus–infected cells, the accumulation of mRNA was increased, whereas that of the genome and antigenome was decreased compared to wild-type virus. Conversely, when M2-2 was overexpressed during HRSV infection—using a co-transfected plasmid or a recombinant HRSV in which expression of M2-2 was upregulated by engineering it to be a separate gene89—HRSV replication was inhibited. Expression of M2-2 also was inhibitory to HRSV RNA synthesis by a minireplicon.98 These observations indicate that M2-2 plays a role in shifting RNA synthesis from transcription to RNA replication, that M2-2 can be inhibitory to RNA synthesis, and that the inhibitory activity occurs with increased M2-2 expression. The ΔM2-2 virus retained the ability to replicate in mice and chimpanzees, but it was attenuated approximately 500- to 1,000-fold compared to wild-type HRSV.41,271,549
HMPV encodes a 71 amino acid M2-2 protein from a comparable internal ORF in its M2 mRNA46,571 (Fig. 38.3C). The HMPV M2-2 protein co-immunoprecipitated with the L protein and inhibited RNA synthesis by a mini-replicon, an activity that was lost by short deletions at the N- or C-terminus of M2-2.301 Deletion of HMPV M2-2 from recombinant virus resulted in an attenuated virus in which transcription is increased and RNA replication decreased.66
Nonstructural Proteins NS1 and NS2
The NS1 and NS2 proteins (139 and 124 amino acids, respectively) are unique to the genus Pneumovirus. NS1 and NS2 can be co-immunoprecipitated537 and may occur in complexes of various stoichiometries.156 Monomeric NS2 was unstable with a half-life of 30 minutes.156
NS1 and NS2 strongly interfere with the induction and signaling of type I IFN and type III IFN (the latter comprising λ1, λ2, and λ3; also known as IL-29, IL-28A, and IL-28B, respectively) in human epithelial cells, macrophages, and dendritic cells. This suppresses a major component of host innate defense. The steps at which NS1 and NS2 act are summarized in e-Figure 38.6.
Deletion of NS2 from recombinant HRSV decreased its ability to induce activation of transcription factor NF-κB,524 and small interfering RNA (siRNA)–mediated knockdown of the expression of either NS1 or NS2 in HRSV-infected cells reduced activation of NF-κB and the serine-threonine kinase AKT (also known as protein kinase B), with the effect of speeding the onset of apoptosis.50 Therefore, the two NS proteins appear to activate prosurvival pathways, thereby prolonging the life of the cell and increasing the viral yield.50
In a minireplicon system, coexpression of NS1—and, to a lesser extent, NS2—inhibited transcription and RNA replication, affecting both the genomic and antigenomic promoters.16 These effects remain to be defined. The V and C accessory proteins of some members of Paramyxovirinae also have been shown to downregulate viral RNA synthesis (Chapters 33 and 34). This may be a general viral mechanism to prevent excessive RNA synthesis and avoid the accumulation of naked genomic/antigenomic RNA and the formation of dsRNA, which otherwise would activate the RIG-I/MDA5/PKR pattern recognition molecules.
Replicative Cycle
Efficient infection of cell lines in vitro by HRSV involves binding to cellular GAGs, especially heparan sulfate and chondroitin sulfate B.238 The G and F proteins each appear able to bind to GAGs.174,175,239,545 A number of additional potential receptor molecules for HRSV have been tentatively identified, including intracellular adhesion molecule (ICAM)-1,33 RhoA,438 the CX3CR1 fractalkine receptor,561 and annexin II.358 More recently, efficient HRSV infection in vitro and in the mouse model was shown to depend on binding to the cellular protein nucleolin,543 which also has been identified as a co-receptor for human parainfluenza virus 3 (HPIV3).58 Somewhat unexpectedly, binding to nucleolin is mediated by the F protein rather than by the G protein. This suggests that efficient attachment and infection by HRSV depends on two different binding events mediated by G and F, but the details of this process remain unclear.
HRSV entry occurs by fusion of the viral envelope with the cell plasma membrane283,526 or with endosomal membranes following clathrin-mediated endocytosis.304 However, HRSV entry does not require endosomal acidification, suggesting that there is no mechanistic difference between entry at the plasma versus endosomal membrane.304,526 When infection was monitored by video microscopy, the initiation of fusion by attached virions appeared to be a slow step, but once started the process was rapid.18 Genome transcription and replication occur in the cytoplasm and the virus can grow in enucleated cells and in the presence of actinomycin D, indicating a lack of essential nuclear involvement.
HRSV mRNAs and proteins can be detected intracellularly at 4 to 6 h after infection and reach a peak accumulation by 15 to 20 hours. The release of progeny virus begins by 10 to 12 h postinfection, reaches a peak after 24 hours, and continues until the cells deteriorate by 30 to 48 hours. Transcription and RNA replication occur concurrently but, as noted, the low-abundance M2-2 protein accumulates during infection and shifts the balance of RNA synthesis from transcription to RNA replication.41 In minireplicon experiments, increasing the level of accumulation of the N and P proteins did not shift the balance between RNA replication and transcription, indicating
that the availability of protein to encapsidate replicative RNA does not control this balance.172
that the availability of protein to encapsidate replicative RNA does not control this balance.172
Several factors influence the relative levels of expression of the various HRSV genes. Like other Mononegavirales, sequential transcription has a polar gradient due to polymerase fall-off, and thus promoter-proximal genes are expressed more efficiently. This gradient of expression is not very steep, with the exception of the L mRNA.107,307 The greatly reduced accumulation of L mRNA compared to the other mRNAs appears to be due to a posttranscriptional effect rather than polymerase fall-off, with one possibility being mRNA stability.318 Differences in the termination efficiency of the various GE signals may also have effects on the relative levels of gene expression.246,399 For example, if a GE signal is particularly inefficient, a greater fraction of the polymerase continues synthesis into the next downstream gene to produce a readthrough transcript, rather than terminating and reinitiating to produce an individual transcript of the next gene. Because ORFs in internal positions in eukaryotic mRNAs generally are not efficiently translated,306 this would have the effect of downregulating expression of the protein from the downstream gene. HMPV in particular has considerable variation in the efficiency of its GE signals, and in particular this appears to sharply downregulate the production of monocistronic SH mRNA, contributing to the observed low level of expression of SH protein.
HRSV assembly and budding occur at the plasma membrane. In polarized cells, this occurs at the apical surface.475,631 Video microscopy showed that budding occurs within circumscribed regions on the cell surface and appeared visually to be the reverse of fusion.18 These regions contain localized virus-modified lipid rafts involving all three viral surface proteins and the M protein.254,270,371,372,623 The minimum viral protein requirements for the formation of virus-like particles capable of delivering the viral genome to target cells are the F, M, N, and P proteins,547 and expression of these proteins induced the formation of viral filaments.564
As has been observed for other members of Paramyxoviridae, HRSV utilizes the host cytoskeleton in its replicative cycle. Viral substructures are associated with polymerized actin throughout the infectious cycle.269 Actin is packaged in the virion and was required, in combination with profilin (an actin-binding protein involved in restructuring actin polymers), for efficient RNA synthesis by intracellular nucleocapsids in a cell-free reaction.72 HRSV appears to hijack cellular apical recycling endosomes (ACEs) for budding, a pathway that is distinct from that described for a number of other enveloped RNA viruses.65,564
Efficient infection by HMPV in vitro depends on binding to cell surface GAGs and αvβ1 integrin.118,551 The F proteins of some strains of HMPV depend on acidification for activation, indicating a dependence on endosomal uptake rather than fusion at the plasma membrane: this requirement mapped to the presence of glycine at position 294.260,496,497 However, this was observed for only 6% of subgroup A strains and not for subgroup B, indicating an unexpected diversity in entry mechanisms. Finally, one notable difference between HRSV and HMPV is that the kinetics of infection of the latter in cell culture are slower, with the peak of intracellular protein expression occurring at 48 to 72 hours postinfection.
Propagation In Vitro
HRSV replicates most efficiently in vitro in immortalized cell lines derived from human epithelial cells, but can infect and replicate to a significant extent in a wide variety of cell lines representing various tissues from various hosts, including humans, monkeys, bovines, hamsters, and mice. The most commonly used cell line is human HEp-2, now thought to have been contaminated historically with, and outgrown by, the HeLa cell line.368 Virus is usually quantified by plaque titration; immunostaining is often used to enhance visualization of plaques. Fresh clinical isolates of HRSV may undergo some sort of adaptation to cell culture,361 but this is poorly understood. However, passaged laboratory strains retain their virulence for chimpanzees and humans.291,606
In cell culture, 90% of progeny HRSV virions remain associated with the cells, attached in a manner that suggests a failure to complete the budding process. To make virus stocks, cell-associated virus is dislodged by freeze-thawing, sonication, or vortexing. The yield is low, typically ten plaque-forming units (pfu) per cell. Virion preparations commonly have substantial contamination by cellular debris. A large fraction of released virions appeared to be empty and presumably noninfectious.19 Less than 5% of the infectivity of a preparation of HRSV passed through a 0.45-mm filter, whereas more than 85% passed through a 3-μm filter, consistent with the infectious particles being filaments475; similar findings had been reported for BRSV.433
HMPV is more restricted in its in vitro host range than HRSV, but is readily propagated in rhesus LLC-MK2 or AGM Vero cells.134,557 HMPV typically requires added trypsin to support cleavage of the F protein, although some strains mutate during passage to become trypsin independent, as noted.492 HMPV replicates more slowly than HRSV, its cytopathic effect is less prominent, and the yield of infectious virus is similarly low. Like HRSV, HMPV tends to remain cell-associated but is more stable.
Genetics and Reverse Genetics
HRSV (and presumably HMPV) has a high rate of nucleotide substitution, as is typical for RNA viruses. The rate of nucleotide substitution in RSV was estimated to be 6.47 × 10−4 substitutions/site/year, and the G gene had the elevated rate of 10−2.7 substitutions/site/year, reflecting relaxed selective constraints, as already noted.538a,641 HRSV and HMPV, like other Mononegavirales, also engage in nonhomologous recombination caused when the polymerase jumps from one template to another during synthesis. This can create defective interfering (DI) genomes,569 or deletion of parts of genes,288 or sequence duplications.560 Foreign sequence also can be acquired, as evidenced by a 1,015-nucleotide insert of unknown origin in the G gene of certain AMPV-C isolates.38 Recombination between co-infecting viruses appears to be rare, although potential mosaic genomes within a viral species are identified occasionally by sequence analysis.634 One study detected a single case of recombination between two co-infecting HRSV mutants in cell culture, involving both homologous (i.e., guided by sequence relatedness) and nonhomologous recombination522: this is the only such report for Mononegavirales. In addition, the difference in the gene orders of HRSV and HMPV may be evidence of past recombination, within one or the other species, resulting in gene rearrangement.
As with other Mononegavirales, HRSV and HMPV can be produced entirely from cloned cDNA by reverse genetics, involving co-transfection of plasmids encoding a copy of the genome or antigenome as well as the proteins of the nucleocapsid/polymerase complex. The viral protein requirements for recovering HRSV are N, P, L, and M2-1, but M2-1 is not
required for HMPV.47,66,97,259 A second type of reverse genetics system that has been used extensively for HRSV in particular is based on cDNA-encoded minireplicons in which the viral genes in the genome or antigenome cDNA have been replaced by one or more foreign marker genes, with the system driven by viral proteins supplied from co-transfecting plasmids. Depending on the supplied proteins, this system can recreate genome encapsidation, transcription, RNA replication, and particle morphogenesis.98,170,373,424,547 This system is ideal for detailed structure-function studies. In addition, mutations that might have drastic or lethal effects on infectious virus can readily be studied in the transient context.
required for HMPV.47,66,97,259 A second type of reverse genetics system that has been used extensively for HRSV in particular is based on cDNA-encoded minireplicons in which the viral genes in the genome or antigenome cDNA have been replaced by one or more foreign marker genes, with the system driven by viral proteins supplied from co-transfecting plasmids. Depending on the supplied proteins, this system can recreate genome encapsidation, transcription, RNA replication, and particle morphogenesis.98,170,373,424,547 This system is ideal for detailed structure-function studies. In addition, mutations that might have drastic or lethal effects on infectious virus can readily be studied in the transient context.
Infection of Experimental Animals
A number of animal species can be infected in the respiratory tract by intranasal administration of HRSV, including cotton rats, mice, ferrets, guinea pigs, hamsters, marmosets, lambs, and various nonhuman primates.35,73,210,460,521,606 Among experimental animals, only the chimpanzee approaches the human in being highly susceptible to infection by contact, in supporting moderate to high levels of virus replication, and in exhibiting rhinorrhea and cough resembling that of humans. The most widely used experimental animals, cotton rats and mice, support a low to moderate level of virus replication that peaks on day 4 and is cleared quickly. In situ hybridization of lung tissue from cotton rats at the peak of virus replication showed that only scattered cells were infected.413 Inbred strains of mice can vary 100-fold in permissiveness for replication.460 The BALB/c mouse is one of the more permissive strains, but it is less permissive than cotton rats. Rodents do not exhibit overt HRSV respiratory tract disease, although pulmonary histopathologic changes are evident and disease can be monitored by weight loss and changes in pulmonary function. More recently, newborn lambs have been used as models in detailed studies of neonatal HRSV disease.521
Intranasal administration of HMPV has been reported to infect guinea pigs, ferrets, mice, cotton rats, and hamsters, and several species of nonhuman primates.6,241,353 BALB/c mice and hamsters may be more permissive for HMPV than for HRSV. Among nonhuman primates, cynomolgus macaques and AGMs support moderate levels of virus replication that peak on days 5 to 6 and are mostly resolved by day 10.311,515 Captive cynomolgus macaques, AGMs, and chimpanzees have a high seroprevalence for HMPV, suggesting that they can be infected readily from their handlers and possibly by animal-to-animal transmission.
Antigenic Subgroups and Diversity
HRSV has a single serotype with two antigenic subgroups, A and B. These exhibit a three- to fourfold reciprocal difference in neutralization by polyclonal convalescent serum.92 Studies with MAbs have demonstrated extensive antigenic difference in the G protein with substantially less difference in the other viral proteins.10,400 Analysis of glycoprotein-specific responses in cotton rats or human infants by enzyme-linked immunosorbent assay (ELISA) with purified F and G glycoproteins (the HRSV neutralization antigens) showed that F has 50% antigenic relatedness between subgroups compared to 1% to 7% relatedness for G.256 Consistent with this, F protein expressed from a recombinant vaccinia virus was equally protective in cotton rats against infection with either subgroup, whereas the G protein was 13-fold less effective against the heterologous subgroup virus.277 Therefore, the F protein is responsible for most of the observed HRSV cross-subgroup neutralization and protection.
The genomes of the two HRSV subgroups share 81% nucleotide identity. The various proteins vary considerably in their level of subgroup divergence, with M2-2, SH, and G being the most divergent (Table 38.1). The divergence is greatest for the ectodomains of SH and G, which exhibit only 50% and 44% sequence identity between subgroups, respectively.105,278 The genome-wide nature of the sequence differences indicates that the two subgroups represent two lines of divergent evolution, rather than being variants that differ only at a few major antigenic sites. The divergence of the two HRSV subgroups has been estimated to have occurred approximately 350 years ago.640 In addition, there is considerable variation within each subgroup: for example, the G glycoprotein can have 20% difference between strains from the same subgroup.
The HRSV F protein is relatively stable antigenically, consistent with its high level of sequence conservation between the two HRSV subgroups. For example, analysis of 18 subgroup A strains and five subgroup B strains recovered from geographically diverse regions over 30 years using F-specific murine MAbs representing 16 separate neutralization epitopes in four antigenic sites showed that seven epitopes were conserved in all but one of the strains.32 Similarly, a major epitope in the N protein for CD8+ cytotoxic T lymphocytes (CTLs) was highly conserved in field strains.578
The G protein appears to be subject to greater change, although this occurs incrementally over a period of years. In one study, analysis of HRSV isolates collected over 47 years suggested that positive selection occurred at 13 codons in the G ectodomain, a number of which involved known epitopes.641 A second study of isolates collected over 38 years provided evidence of progressive amino acid changes at an average rate of 0.25% per year that were paralleled by changes in reactivity with MAbs.76 The mucin-like domains in G are unusual in that they are more divergent at the amino acid level than at the nucleotide level,278 which is the converse of what is observed for most proteins. This suggests that there is a selective pressure for amino acid substitutions, presumably driven by immune pressure. These regions in G may be relatively tolerant of amino acid change because of their proposed extended, nonglobular structure. In contrast, although F would be subjected to the same selective immune pressure, it likely is less tolerant of amino acid substitutions due to constraints from its folded structure and functional requirements. Analysis of BRSV isolates, including those from countries in which vaccination was in wide use, provided evidence of modest progressive changes not only in G, but also in N and F.568
HMPV also has a single serotype with two antigenetic subgroups, A and B, which have extensive cross-reactivity and cross-protection.55,259,442,572,574 The level of genome nucleotide sequence identity (80%) and the relatedness between the various proteins is similar to that described for HRSV (Table 38.1). The HMPV F protein is somewhat more conserved than that of HRSV and plays a major role in the high level of cross-neutralization and protection between the two subgroups.515 Vaccination against AMPV appears to drive antigenic drift in the field, especially in the G and SH genes, resulting in virus
that is sufficiently divergent from the vaccine strain that it is less restricted by vaccination.84
that is sufficiently divergent from the vaccine strain that it is less restricted by vaccination.84
Animal Counterparts
The BRSV gene map is very similar to that of HRSV.67,627 The genome of BRSV strain A51908 (NC_001989 and AF295543) contains 15,140 nucleotides and has 73% nucleotide identity with HRSV. Amino acid sequence identity ranges from 30% (G protein) to 93% (N protein) (Table 38.1), and there is extensive antigenic cross-reactivity. BRSV and HRSV have broadly overlapping host ranges in cell culture; however, in vivo, BRSV is highly restricted in nonhuman primates.68 Ovine and caprine RSV also have been described. Sequence analysis suggests that ovine RSV and BRSV represent two branches of ruminant RSV with a degree of divergence similar to that between the HRSV antigenic subgroups. Caprine RSV appears to be more closely related to BRSV than to ovine RSV.112
PVM was first identified during experiments in which clinical material was passaged in mice in an effort to identify new human pathogens, and an apparent mouse virus present in the animals became evident.264 PVM is a respiratory pathogen that readily infects mice and other rodents and can be highly virulent. However, the natural host of PVM is unclear: the virus occasionally appears in colonies of laboratory mice, but serologic studies usually find little evidence of PVM-specific antibodies in wild rodents under conditions where there is extensive seropositivity for other common rodent viruses.31,516 Serologic studies suggested that a substantial proportion of the human population has antibodies that react with PVM.464 However, further analysis suggested that infection of humans by PVM or a closely related virus is unlikely.64a A virus that was recently isolated from dogs with acute respiratory disease appears to be a strain of PVM (e.g., the percent amino acid sequence identity between the isolate and PVM strain 15 ranged from a low of 90.2% [SH] to a high of 98.1% [M]).473,474 Whether PVM commonly infects and causes respiratory tract disease in dogs remains unknown. The PVM gene map308 is essentially the same as that of HRSV and BRSV except that (a) the M2 and L genes of PVM do not overlap, and (b) the PVM P gene contains a second ORF that encodes a 137-amino acid product that represents a novel, 12th PVM protein.26 The complete nucleotide sequence of PVM strain 15 (AY729016) is 14,886 nucleotides in length and has 52% identity with that of HRSV.308 The level of amino acid sequence identity ranges from 10% (M2-2 protein) to 60% (N protein) (Table 38.1).
BRSV and PVM in their respective hosts are used as models for HRSV,190,285,441,481,614 although a review of these studies is beyond the scope of this chapter. The BRSV model is limited by the inconvenient nature of the large host. PVM differs from HRSV in that lethal infections are typical rather than an infrequent outcome, but it can serve as a model for severe pneumovirus disease and has the advantage of the many available murine immunologic reagents and inbred mouse strains.
AMPV causes respiratory tract disease of economic importance in turkeys and also infects chickens and other birds.421 Four AMPV antigenic subgroups have been described: subgroups A and B have been found in South Africa, Europe, Israel and Asia; subgroup C in North America; and subgroup D in France.421 Complete genomic sequences have been determined for strains of subgroup A (AY640317) and subgroup C (AY590688 and AY579780). These range in length from 13,134 to 14,150 nucleotides, have the same general gene map as HMPV, and share 61% to 68% nucleotide sequence identity with HMPV subgroup A. Subgroups A, B, and D are more closely related to each other than to C205,504 (e-Fig. 38.2B). Surprisingly, AMPV-C is more closely related to HMPV than to the other AMPV subgroups571,572 (e-Fig. 38.2B). Sequence analysis of isolates collected over a 25-year period suggested that the most recent common ancestor for AMPV-C and HMPV existed approximately 200 years ago, implying a cross-species jump.127 There is substantial host range restriction between human and avian MPV. AMPV-C (the subgroup most closely related to HMPV) is highly restricted in nonhuman primates.447 HMPV has been reported to be unable to infect chickens and turkeys,572 or to transiently infect turkeys.577
Pathogenesis and Pathology
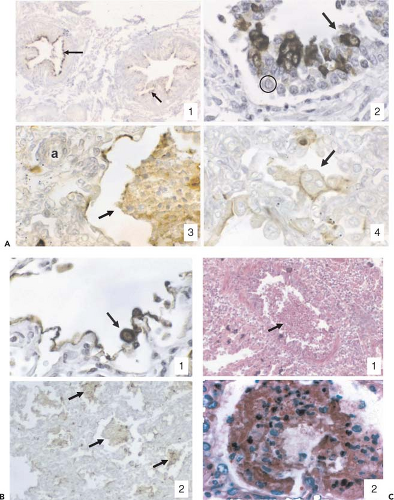
In typical monolayer cultures of immortalized cells, infection with HRSV or HMPV results in long surface filaments that bear viral antigen and probably give rise to filamentous virus particles489 (Fig. 38.2B and E). The formation of HRSV filaments and filamentous virus depends on activation of RhoA and actin rearrangement: when activation is blocked, the production of infectious virus shifted to the nonfilamentous form.71,206,371 HRSV-infected cells develop large electron-dense cytoplasmic inclusion bodies of up to several microns in diameter that include the N, P, M2-1, and L proteins and presumably contain active nucleocapsids78,185; HMPV forms similar inclusions.136 Recently, the HRSV inclusion bodies were found to sequester a number of cellular proteins involved in intracellular signaling pathways, with the effect of inhibiting innate immunity and stress responses.179a HRSV infection has slight inhibitory effects on cellular DNA and RNA synthesis and little effect on gross protein synthesis.335 As one factor in the lack of inhibition of protein synthesis, the HRSV N protein binds to PKR and prevents it from phosphorylating eIF-2a and inhibiting protein synthesis.216 Apoptosis of HRSV-infected cells occurs slowly and is inhibited by the NS1, NS2, and SH proteins, as already noted. HRSV blocks the formation of stress granules, which may otherwise restrict HRSV replication.179a,242 The formation of syncytia (Fig. 38.2A) is a major factor in cell death in typical nonpolarized monolayer cell cultures, but usually is not a prominent histopathologic finding in vivo (below).
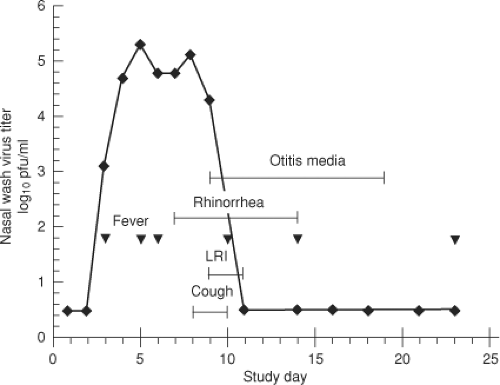
HRSV and HMPV are highly infectious viruses. Humans are their only natural host, although HRSV and HMPV can readily spread to nonhuman primates and are pathogenic in some situations.305 The major mode of spread by HRSV is by large droplets or through contaminated objects and depends on close contact with infected individuals or contact of contaminated hands to nasal or conjunctival mucosa (self-inoculation). It is generally thought that small-particle aerosols (which can remain airborne for extended periods and also can be inhaled deeply into the respiratory tract) are not an important mode of HRSV transmission,222,224 although there are contrary data.338 The incubation period from time of infection to onset of illness for HRSV is about 3 to 5 days137,286,291 (Fig. 38.5). The incubation period for HMPV is not defined, but is thought to be similar.
HRSV replicates initially in the nasopharynx. Signs of LRI, when it occurs, usually appear 1 to 3 days following the onset of rhinorrhea (Fig. 38.5). Viral spread to the lower respiratory tract likely involves aspiration of secretions. Infants hospitalized
for HRSV disease shed infectious virus in nasal secretions with peak titers of 104 to 106 infectious units per ml.138,139,225,615 Many infants continue to shed virus at hospital discharge and a few can continue to shed virus for weeks following clinical recovery. With the use of quantitative reverse transcription polymerase chain reaction (RT-PCR), the period of detection of HRSV shedding is prolonged, suggesting that virus is present but is neutralized by secretory antibodies made in response to the infection.137,351 Limited data suggest that the titer of virus in the lower respiratory tract, sampled in bronchoalveolar lavage fluids of ventilated patients, is similar to that in the upper respiratory tract. Infectious virus has been difficult to recover from adults during symptomatic infection despite positive serologic or RT-PCR tests, probably due to the presence of neutralizing antibodies.
for HRSV disease shed infectious virus in nasal secretions with peak titers of 104 to 106 infectious units per ml.138,139,225,615 Many infants continue to shed virus at hospital discharge and a few can continue to shed virus for weeks following clinical recovery. With the use of quantitative reverse transcription polymerase chain reaction (RT-PCR), the period of detection of HRSV shedding is prolonged, suggesting that virus is present but is neutralized by secretory antibodies made in response to the infection.137,351 Limited data suggest that the titer of virus in the lower respiratory tract, sampled in bronchoalveolar lavage fluids of ventilated patients, is similar to that in the upper respiratory tract. Infectious virus has been difficult to recover from adults during symptomatic infection despite positive serologic or RT-PCR tests, probably due to the presence of neutralizing antibodies.
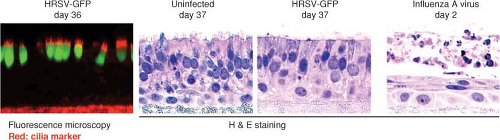 Figure 38.6. Infection of an in vitro model of human airway epithelium (HAE) with human respiratory syncytial virus (HRSV) and influenza A virus, shown in cross-section. The left hand panel is a fluorescence photomicrograph of cells 36 days following infection with HRSV expressing green fluorescent protein (GFP), and also stained with antibody specific to cilia (red; note that this antibody also stained the filter support underlying the cells). The three panels to the right show mock-infected, HRSV-infected, and influenza A virus–infected cultures visualized 37 (mock and HRSV) or 2 (influenza) days postinoculation.631 In the three right-hand panels, the cells were stained with hematoxylin and eosin. |
HRSV principally exhibits tropism for the airways and lung tissues. Studies in human adenoid or HAE epithelium models in vitro showed that HRSV preferentially infected ciliated cells. Infection was limited to the apical surface and did not spread to underlying cells, and virus was released from the apical surface562,616,631 (Fig. 38.6). In experimental HAE infections lasting several weeks, infected cells were shed and replaced with little visible alteration of the microscopic appearance of the tissue, although ciliary beating usually was impaired.616,631 Syncytia were not observed, probably because the F glycoprotein was expressed on the apical surface and did not contact adjacent cells. HRSV contrasted sharply with influenza A virus examined in parallel, which was rapidly destructive and quickly spread to underlying cells.616,631 HRSV infection of epithelial cells induces a rapid inhibition of Na+ transport, resulting in apical fluid accumulation.314 This might be a host mechanism to dilute and remove irritants, but also might contribute to excess fluid and virus spread. This effect was not unique to HRSV, and was observed with parainfluenza and influenza viruses as well as with bacterial pathogens.
Histopathology of fatal HRSV infections indicates that infection is limited to the superficial cells of the respiratory tract, consistent with the findings in HAE cultures, although it is likely that nonciliated cells also are infected4,187,273,596 (Fig. 38.7). There is necrosis and destruction of ciliated cells and occasional proliferation of the bronchiolar epithelium, but basal cells are spared. Ciliated epithelial cells rarely reappear before 2 weeks postinfection, and complete restoration requires 4 to 8 weeks,220 similar to the duration of postinfection altered lung function and airway reactivity in adults.237 There is an early influx of polymorphonuclear cells, and a peribronchiolar infiltrate of lymphocytes, plasma cells, and macrophages develops, with migration of the lymphocytes among the mucosal epithelial cells. Submucosal and adventitial tissues become edematous, and secretion of mucus is excessive, which combines with cell debris and inflammatory cells to obstruct the bronchioles and alveoli, causing either collapse or emphysema of distal portions of the airway. HRSV infects both type I and type II alveolar cells.273 In those instances in which pneumonia occurs, the interalveolar walls thicken as a result of mononuclear cell infiltration, and the alveolar spaces may fill with fluid. There is usually a patchy appearance of these pathologic changes, even though disease may be widespread. Immunostaining identified virus-infected cells in the bronchial, bronchiolar, and alveolar epithelium.273,419 Syncytia are sometimes observed, but are not prominent. However, syncytium formation and giant cell pneumonia are hallmarks of infection in individuals with extreme T-cell deficiency. HRSV antigen can be relatively abundant in lower respiratory tract infection, although the staining is usually focal, and in some cases of fatal HRSV bronchiolitis, antigen was present only in small amounts.187,419 The histopathology of HMPV infection is not well known but has been reported to involve a characteristic enlarged, darkly staining pneumocyte or “smudge cell” that is not seen with other respiratory paramyxoviruses.532 Studies in nonhuman primates indicate that HMPV has a similar tropism for the superficial cells of the respiratory epithelium.311
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree