Fig. 7.1
Pediatric glioblastoma exhibiting pseudopalisading necrosis. Hematoxylin and eosin staining of a Grade IV GBM (40× magnification). Arrows point to pseudopalisading necrosis
Anaplastic astrocytomas differ from anaplastic oligodendrogliomas by having hyperchromatic nuclei that are often elongated, irregular, or smudged. Mitotic figures may not be common, but can be found by immunohistochemical markers of proliferation such as the MIB-1 index. While the tumor cells are not as densely packed as in the oligodendroglioma, they are nonetheless, obviously more hypercellular than the white matter by a factor of three- to tenfold. Vascular changes are frequent in anaplastic astrocytoma, but the defining element of vascular proliferation as found in the Grade IV glioblastoma, is debated. While most brain tumor neuropathologists agree that endothelial duplication circumferentially ringing the vessel is a feature of glioblastomas, it is unclear where glomeruloid vessels and vascular garlands or loops of vessels fall in astrocytoma grading. Among astrocytic tumors, foci of necrosis are reserved for glioblastomas.
Glioblastoma multiforme, the most malignant and most common of the high-grade gliomas, is an astrocytic neoplasm that has been found to most commonly arise de novo without a preceding history of glioma, with only 5 % of glioblastomas occurring as a result of progression from a lower grade astrocytoma. Both de novo and progressive GBMs exhibit nuclear pleomorphism, mitotic activity, vascular proliferation, and/or necrosis. While densely cellular in some foci of the tumor, there is commonly an infiltrative component that can be found distant from the central mass. This infiltrative capability currently makes GBMs impossible to cure by surgery or local–regional radiation therapy.
Epigenetics
With the advent of next generation sequencing, our understanding of the genetic alterations in pHGGs has been turned upside down. This technological advance has led to the discovery of heterozygous K27M or G34R/V mutations in the tail of histone 3.3 or K27M mutations in the tail of histone 3.1 and loss of function mutations in the chromatin remodelers ATRX (α-thalassaemia/mental retardation syndrome X-linked) and DAXX (death-domain associated protein) in pHGGs (Fig. 7.2; [3, 4]). As a brief review, the fundamental repeating unit of chromatin is the nucleosome, which consists of approximately 147 bp of superhelical DNA wrapped around the radial surface of an octamer of highly conserved core histone proteins (two copies of each H2A, H2B, H3, and H4). Histone proteins are subject to a wide array of covalent modifications that occur primarily at amino (N−) and carboxy (C−) termini. The tail regions of core histones contain flexible and highly basic amino acid sequences that are highly conserved and serve as substrates for several posttranslational modifications such as acetylation, methylation, ADP-ribosylation, ubiquitylation, and phosphorylation. These modifications impact gene transcription, DNA replication, and chromatin assembly. The histone code states that distinct patterns of histone modifications act in concert with DNA methylation, noncoding RNAs, and transcription factors to generate “histone-epigenetic codes” that are read by effector proteins [5]. Lastly, another level of complexity is histone variants (e.g., H3.3 vs. H3.1), which are relevant to pHGGs. Although the difference between H3.3 and H3.1 is only four amino acids, H3.1 (also called canonical core H3) is only incorporated into nucleosomes during the S-phase of the cell cycle while H3.3 incorporation into nucleosomes is cell-cycle independent. Furthermore, ATRX and DAXX are both H3.3 chaperones and together they facilitate the deposition and remodeling of H3.3 containing nucleosomes [6].
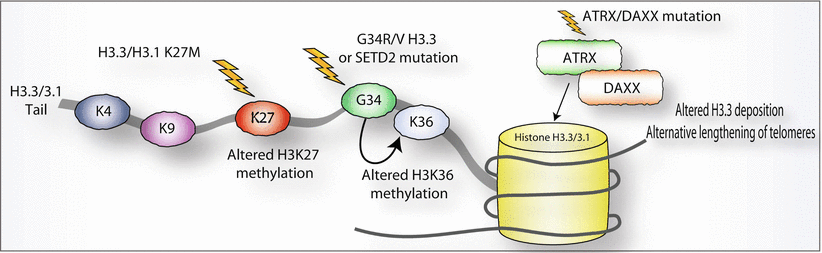

Fig. 7.2
Epigenetic alterations associated with pediatric high-grade gliomagenesis. This schematic of the H3.3/H3.1 tail illustrates the three types of epigenetic mutations seen in pHGG: (1) K27M mutations which impact H3K27 methylation, (2) G34R/V or SETD2 mutations, both of which impact H3K36 methylation, and (3) ATRX/DAXX mutations which likely impact H3.3 deposition and alternative lengthening of telomeres
The initial two manuscripts describing these mutations noted, in the brainstem there are K27M mutations in either H3.3 or H3.1 in up to 80 % of DIPGs, while the G34R/V mutations are found only in H3.3 and were primarily found in pHGGs located in the cerebral cortex. In addition, G34R/V mutations co-occur with loss of function ATRX or DAXX mutations and are associated with the ALT (alternative lengthening of telomeres) phenotype [3, 4, 7, 8]. In a follow-up paper, Sturm and colleagues [9] pursue an integrative approach based on epigenetic, copy number, expression, and genetic analyses on over 200 adult and pediatric GBMs to identify six distinct DNA methylation clusters which were labeled as “IDH,” “K27,” “G34,” “receptor tyrosine kinase (RTK) I (platelet-derived growth factor receptor A or PDGFR-A),” “mesenchymal,” and “RTK II (Classic).” A key finding of this analysis was that H3F3A K27 and H3F3A G34 mutations were exclusively distributed to the K27 and G34 clusters, respectively, and these were mutually exclusive of the isocitrate dehydrogenase (IDH1) mutations. The RTK I (PDGFR-A) group had a high frequency of PDGFR-A amplification and the RTK II Classic group had a very high frequency of whole chromosome seven gain, whole chromosome ten loss, frequent deletion of cyclin-dependent kinase inhibitor 2a (CDKN2A), and amplification of epidermal growth factor receptor (EGFR). This RTK II Classic subgroup is completely devoid of pediatric patients. Remarkably, the clusters are associated with patient age, with K27M patients being the youngest (median age 10.5; range 5–23) and G34 patients being the second youngest (median age 18 years; range 9–42). The RTK I “PDGFR-A” cluster (median age 36 years, range 8–74 years) and the IDH cluster (median age 40 years, range 13–71 years) mostly comprised young adults, while the oldest cluster, the RTK II “Classic” cluster, comprised older adults (median age 58, range 36–81 years). The other remarkable finding was that the epigenetic GBM subgroups showed region-specific predilection within the CNS whereby the K27-mutant tumors were predominantly seen in midline locations such as the thalamus, pons, and the spinal cord while the tumors in the other five subgroups almost exclusively arose in the cerebral hemispheres. This remarkable observation clearly suggests that the mechanism of gliomagenesis is distinct in different regions of the CNS. Lastly, these subgroups also correlated with survival with the K27 subgroup having the shortest survival, the IDH subgroup with the longest survival, and the other subgroups in between. Interestingly, both the IDH and H3F3A mutations co-occur with p53 mutations, suggesting that p53 mutations do not have independent prognostic significance [9].
How histone mutations contribute to pHGG pathogenesis is subject to current investigations, but Lewis and colleagues recently reported an initial glimpse into the mechanism. They reported that the K27M H3.3/H3.1 mutations inhibit polycomb repressive complex 2 (PRC2), the enzyme complex that adds methyl groups to H3 lysine 27. Under normal circumstances, this histone mark is repressive, and inhibition of PRC2 results in global loss of H3 lysine 27 trimethylation. The mechanism of the G34R/V histone mutations is less well understood, but results in a local decrease in H3 lysine 36 trimethylation [10]. In addition to the epigenetic mutations described above, pHGGs recently have also been reported to harbor loss of function mutations in SETD2, an H3K36 trimethyltransferase. Perhaps not surprisingly, SET2D mutations were mutually exclusive with H3F3A mutations, but they did overlap with IDH1 mutations [11]. High-grade gliomas with SET2D mutations were found exclusively in tumors arising in the cerebral hemispheres, reinforcing the notion that H3K36 is important for gliomagenesis in the cerebral hemispheres, while H3K27 is central in the etiology of tumors arising in midline structures of the CNS. Lastly, while IDH mutations are primarily found in adult gliomas, adolescents 13 years old and older can also harbor activating mutations in IDH1 in amino acid 132 [12, 13]. These IDH1 mutations have also been reported to impact histone marks, but through a completely different mechanism [14].
Cytogenetics
Several studies have investigated the spectrum of copy number aberrations in pHGGs [15–22]. Copy number changes in pHGGs are best subdivided between broad chromosomal gains and losses and focal gains and losses. Broad low-amplitude gains of chromosome 1q were identified as well as broad losses of 10q, 13q, and 14q. Focal gains have been reported in PDGFR-A, cyclin D1-3 (CCND1-3), cyclin-dependent kinase 4 (CDK4), cyclin-dependent kinase 6 (CDK6), MYC, v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog (MYCN), EGFR, V-Erb-A Avian Erythroblastic Leukemia Viral Oncogene Homolog-Like 4 (ERBB4), MET, hepatocyte growth factor (HGF), insulin-like growth factor-1 receptor (IGF1R), insulin-like growth factor 2 (IGF2), platelet-derived growth factor B (PDGFB), Neuregulin 1 (NRG1), phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA), PIK3C2B, PIK3C2G, PIK3R5, Kirsten rat sarcoma viral oncogene homolog (KRAS), v-akt murine thymoma viral oncogene homolog 1 (AKT1), AKT3, S6K1, and murine double minute 4 (MDM4). The most common homozygous focal loss was at CDKN2A/CDKN2B. Other homozygous focal losses included the following genes: CDKN2C, neurofibromin-1 (NF1), PTEN (phosphatase and tensin homolog), retinoblastoma (RB1), TP53, TP73, and protein tyrosine phosphatase receptor type D (PTPRD). Interestingly, copy number alterations are also age- and region specific. PDGFR-A is the most common amplified receptor tyrosine kinase (RTK) in pHGGs while EGFR amplification is the most common amplified RTK in adult high-grade glioma. Gains of chromosomes 2q, 8q, and 9q and losses of 16q, 17p, and 20p were significantly more frequent in DIPG than in non-brainstem pHGGs. Furthermore, focal deletions of CDKN2A are extremely rare in DIPGs and are found in 26 % of non-brainstem pHGGs [23]. Lastly, Barrow et al. described homozygous loss of ADAM3A in 16 % of pHGGs, although the function of this gene and how its loss contributes to pediatric gliomagenesis is not known [17]. Figure 7.3 includes a summary figure of the genetic alterations in pHGGs, as well as a table, which lists the genetic alterations in DIPG, non-brainstem pHGG, and adult HGG.
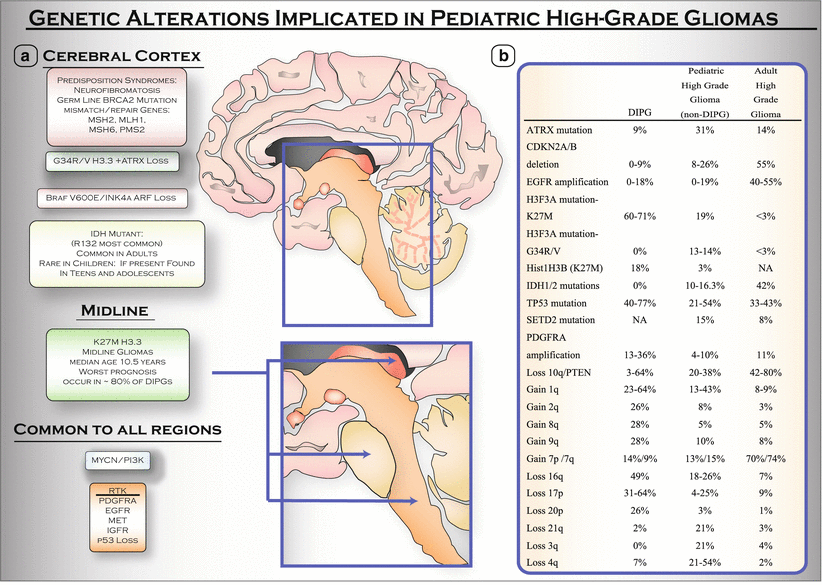

Fig. 7.3
Genetic alterations implicated in pHGGs. Genetic alterations vary by location and age. (a) Genetic alterations observed in pHGGs located in the cerebral cortex, or in midline areas of the CNS, or common to all pHGGs. Arrows pointing to midline areas from top to bottom: thalamus, pons, and spinal cord. (b) Distinct molecular genetics of DIPG as compared to pHGGs (non-DIPG) and adult high-grade gliomas. Table is adapted from: Kristin M Schroeder, Christine M Hoeman, and Oren J Becher, Children are not just little adults: recent advances in understanding of diffuse intrinsic pontine glioma biology, Pediatric Research, 2014 [57]
Gene Expression
Gene expression profiling, a method to analyze the mRNA expression of all genes in the tumor, is another useful technique to study the complex biology of cancer. In fact, mRNA analysis of a select number of genes is used to make clinical decisions in some types of breast cancer. In pHGGs, unsupervised hierarchical clustering identified three main tumor subgroups: HC1/proliferative, HC2/proneural, and HC3/mesenchymal [15]. Gene ontology analysis across the groups revealed that HC1 is most associated with cell-cycle genes; HC2 is most associated with neuronal differentiation, while HC3 is most associated with extracellular matrix–receptor interactions and cell adhesion. Interestingly, HC1 is most associated with amplifications targeting the PDGFR signaling cascade, which ties together PDGF signaling with cell growth. If one were to compare the expression profiling of pHGGs to adult high-grade gliomas, PDGFR-A mRNA is significantly overexpressed in pHGGs relative to adult high-grade gliomas while EGFR mRNA is significantly repressed in pHGGs relative to adult high-grade gliomas. With regard to the HC3/mesenchymal group, immune response genes were also enriched and more specifically associated with microglia/macrophages and monocytes [24]. Lastly, using principal component analysis (PCA), two independent groups noted that the expression profiling of DIPGs consists of a distinct cluster separate from non-brainstem gliomas, reinforcing the notion of region-specific differences in pediatric CNS gliomagenesis [23, 25].
Prognostic Stratification
Until recently, the concept that HGGs comprise several, biologically distinct diseases associated with age and location was not fully appreciated. As a consequence, current research efforts center on developing a better understanding of tumor biology and accordingly, devise appropriate classification schemes. Similar to the increased stratification of leukemias in children, molecular stratification of pHGG will continue to become increasingly refined, in parallel with advances in our understanding of disease biology. The ongoing challenge is how to best incorporate new molecular prognostic factors with well-established prognostic factors, such as extent of resection and tumor grade [2]. For over a decade, p53 overexpression has been recognized as a poor prognostic factor in pHGGs. Most importantly, this association was found to be independent of age, histologic features, the extent of resection, or tumor location [26]. Overexpression of p53 as determined by immunohistochemistry, however, is an imperfect proxy for oncogenic p53 mutations, and taken in context with our current knowledge of the molecular genetics of HGG, a clearer picture emerges. For example, as previously mentioned in the epigenetics section of this chapter, IDH mutations frequently co-occur with p53 mutations, and these tumors currently have the best prognosis. However, p53 mutations also overlap with K27M mutations, which appear to have the worst prognosis. According to retrospective studies, K27M is a poor prognostic factor in pediatric GBM, although it is unclear whether this is due to a different biology versus the midline location of these tumors, which limits surgical options [9, 27]. Within the K27M subgroup, DIPGs have the worst prognosis, with a median survival time of 9–12 months and greater than 90 % of children dying within 2 years [28]. Lastly, PTEN mutations or loss of PTEN expression by IHC have been reported to be significantly associated with decreased survival in pHGGs, but this has so far only been reported in small cohorts and will require further validation [29, 30].
Molecular Signaling Pathways
Three pathways that are most implicated in pHGG pathogenesis are the p53, retinoblastoma protein (Rb), and RTK/Ras/phosphoinositide 3-kinase (PI3K) signaling pathways. These pathways are dysregulated in both pediatric and adult high-grade gliomas, and the importance of these three pathways in adult gliomagenesis was recently underscored by the mutual exclusivity of alterations affecting these pathways [31]. Evolving knowledge of precisely how these pathways contribute to tumor initiation and growth is expected to lead to better-informed molecular targeted therapeutic approaches. With regard to activation of the RTK/Ras/PI3K pathway, 80 % of pHGGs activate this pathway through amplification of RTKs, and/or activating mutations in PI3K, and/or loss of PTEN either through deletion or promoter methylation [32]. Below is a brief summary of the key molecular signaling pathways.
RTK/Ras/PI3K Pathway
The axis of PI3K signaling in cancer begins with engagement of growth factors by RTKs such as PDGFR, MET, EGFR, and IGF-1R (Fig. 7.4). PI3K, a lipid kinase, is then recruited to plasma membrane-anchored receptors, is activated, and phosphorylates PIP2 to generate PIP3. Through its pleckstrin homology (PH) domain, the nodal kinase AKT (also known as PKB) binds to PIP3, where it is activated by two phosphorylation events, and triggers a complex cascade of signals that regulate growth, proliferation, survival, and motility. The lipid phosphatase, PTEN, antagonizes this process by dephosphorylating PIP3 to inhibit activation of AKT. PI3K is activated downstream of numerous RTKs that directly, or through adaptor proteins, bind and activate PI3K. PI3K activity is thus carefully regulated by growth factor–receptor interactions. In fact, the vast majority of PI3K remains inactive in the cytoplasm, removed from its plasma membrane-associated substrates, and only a small percentage of PI3K becomes activated upon growth factor stimulation. Therefore, even slight modulations in receptor activity can lead to many-fold increases in PI3K activity [33].
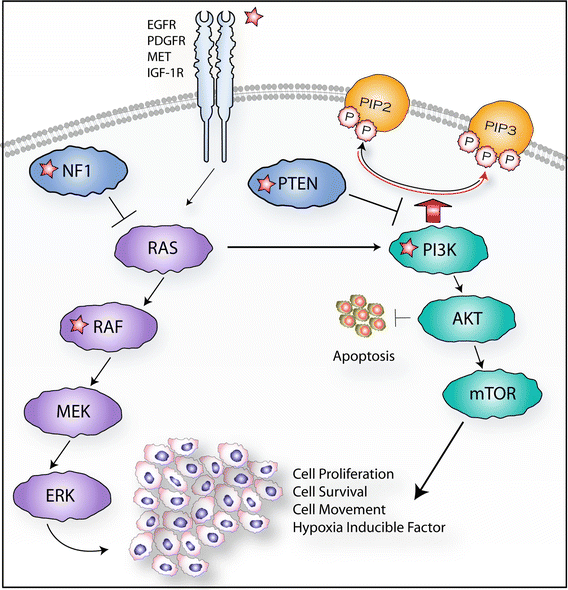

Fig. 7.4
RTK/Ras/PI3K pathway. Growth factors such as EGF, PDGF, HGF, and IGF2 engage with their respective RTKs leading to PI3K activation. PI3K activation phosphorylates PIP2 to generate PIP3. AKT binds to PIP3, becomes activated, and triggers a complex cascade of signals that regulate growth, proliferation, survival, and motility. PTEN, a naturally occurring antagonist of this pathway, dephosphorylates PIP3 inhibiting activation of AKT. Growth factors and RTK interactions also regulate cell proliferation and survival through activation of Ras followed by sequential activation of Raf, Mek, and Erk. Starred (asterisk) factors mutated in RTK/Ras/PI3K pathway are prevalent in pHGGs
In addition to copy number alterations, pHGGs can also harbor additional alterations in the RTK/Ras/PI3K pathways through somatic mutations or alternative splicing. The most commonly mutated RTK in pHGGs is PDGFR-A where mutations in the extracellular domain as well as in the tyrosine kinase domain have been described in approximately 10 % of these tumors [3, 25, 34]. By contrast, the most commonly mutated RTK in adult high-grade glioma is EGFR. EGFRvIII (an EGFR lacking exons 2–7 resulting in ligand-independent signaling) is an alternatively spliced EGFR variant found in 19 % of adult HGGs, but has also been reported in 17 % of pHGGs [31, 35]. Downstream of RTKs, activating mutations in PIK3CA have been described in a subset of pHGGs both inside and outside of the brainstem [36, 37]. While activating Ras mutations have rarely been described in pHGGs (G12V Kras reported by Schiffman et al. [16]), loss of function NF1 (neurofibromatosis type 1 which negatively regulates the Ras pathway) mutations has been identified in a subset of pHGGs outside of the brainstem [3]. Furthermore, activating V600E Braf mutations have also been described in pHGGs [11, 16, 38, 39]. V600E Braf mutations can also be found in low-grade gliomas, but usually in isolation, suggesting that cooperating mutations (such as CDKN2A) may be required for a high-grade phenotype in V600E Braf mutant tumors [16].
RB Pathway
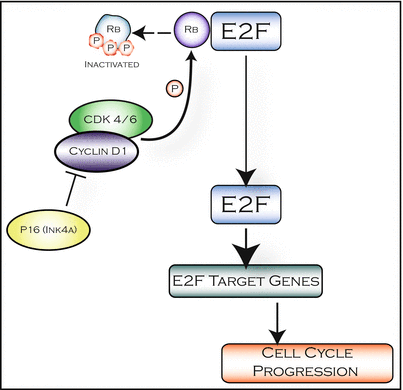
The retinoblastoma protein (RB) is a tumor suppressor protein and key regulator of cell-cycle control. The RB pathway consists of five families of proteins: CDKN (e.g., Ink4a), D-type cyclins, D-cyclin-dependent protein kinases (cdk4, cdk6), RB-family of pocket proteins (RB, p107, p130), and the E2F-family of transcription factors. Each Ink4-protein (p16Ink4a, p15Ink4b, and p18Ink4c) can bind to and inhibit the activity of cdk4 and cdk6. Each D-cyclin protein can associate with cdk4 or cdk6 to form the active kinase complex. Therefore, Ink4 proteins compete with the D-cyclins for cdk4/6 to prevent the formation of the active kinase complex [40]. Importantly, proteins in this pathway are commonly altered in pHGGs, primarily through copy number aberrations: cdkn2a deletions and/or amplification of cdk4, cdk6, cyclin d1, d2, and d3 [15, 19, 23].
The RB pathway regulates cell proliferation as the proteins in this pathway are activated and/or inhibited by growth-promoting, as well as growth-suppressing signals (Fig. 7.5). During regulated cell proliferation, as cells respond to mitogenic signals and commit to cell-cycle entry the complex of D-cyclin/cdk4/6 is activated. The primary cellular targets of the D-cyclin/cdk4/6 complex are the RB-family of pocket proteins (henceforward referred to as RB), which inhibit transcription by directly inhibiting the activity of E2F. Hyperphosphorylation of RB by activated D-cyclin/cdk4/6 complexes renders RB inactive and in turn allows for the release of E2F transcription factors, leading to the activation of E2F target genes involved in cell-cycle progression [40]. Recently, a preclinical trial using genetically engineered mouse models identified a population of pHGG patients that may be sensitive to treatment with a highly selective cdk4/6 inhibitor [41]. Inhibition of cdk4/6 in murine high-grade gliomas harboring CDKN2A loss provided a significant survival benefit and holds promise for translation into clinical trials.