Parainfluenza Viruses
Ruth A. Karron
Peter L. Collins
History
The four serotypes of human parainfluenza virus types 1 to 4 (HPIV1 to HPIV4) were first recovered between 1956 and 1960, following the application of cell culture and hemadsorption techniques to the study of pediatric respiratory tract disease.48,49,175 HPIV1, HPIV2, and HPIV3 were initially isolated from infants and children with lower respiratory tract illness (LRI), and HPIV4 was recovered from children and young adults with mild upper respiratory tract illness (URI). Soon after their discovery, these viruses were shown to be a major cause of croup (HPIV1, 2, and 3) as well as pneumonia and bronchiolitis (HPIV3).45,115,283 As a group, HPIV1, HPIV2, and HPIV3 are second only to human respiratory syncytial virus (HRSV) as a cause of serious viral respiratory tract disease in infants and children, whereas disease due to HPIV4 is less frequent and less serious.
There also are a number of parainfluenza viruses (PIVs) that infect animals. Indeed, the first PIV to be identified was the avian pathogen Newcastle disease virus (NDV). This virus was isolated following outbreaks in 1926 in Java, Indonesia, and Newcastle-upon-Tyne, England, of a seemingly new poultry disease with high mortality.91 The origin of this then-emerging pathogen remains obscure. This virus, or a close progenitor, may have been indigenous in wild birds, and its appearance or evolution as a “new” disease entity may have been associated with the increasing scale of poultry farming at that time. Nine distinct serotypes of avian PIVs (now usually called avian paramyxoviruses, or APMVs) are now recognized, of which NDV constitutes serotype 1.5 Another PIV was recovered in 1952 in Japan from mice inoculated with an autopsy specimen from an infant with respiratory disease.216,264 The natural history of this virus, Sendai virus (SeV), is not well understood, but it appears to be a murine virus that is closely related to HPIV1 but is not a human pathogen. Its antigenic relatedness to HPIV1 led to some confusion when it was used in serologic studies of patients with acute respiratory disease. Bovine parainfluenza type 3 (BPIV3), a close bovine relative of HPIV3, was isolated in 1959 from cattle with respiratory tract disease called shipping fever.1,2 PIV5, previously known as simian virus 5 (SV5), was first isolated in 1954 as a common contaminant of primary monkey kidney tissue cultures (hence its name).163 This was at a time when these cultures were being used to prepare poliovirus vaccine material, and a number of new viruses were recovered and identified from the primary tissue. PIV5 was shown to be related to HPIV2 and was identified as a cause of croup (“kennel cough”) in dogs.21,22,44 Simian virus 41 (SV41) was isolated in 1961, also as a contaminant of primary monkey kidney cell culture.251 SV41 was found to be even more closely related to HPIV2 than PIV5.377 Therefore, there are four known human PIVs (HPIV1–4) and 13 known animal PIVs (SeV, BPIV3, PIV5, SV41, NDV/APMV1, and APMV2–9). This number may increase: virus that appears to represent a 10th APMV serotype was recently isolated from penguins from the Falkland Islands.250
The name parainfluenza originally was coined because some of the disease signs are influenza-like and because, like influenza, the particle is medium-sized, has a lipid envelope, and has hemagglutination and neuraminidase activities. This name was first used in 1959 for the four viruses now known as HPIV1, HPIV2, HPIV3, and SeV.10 Therefore, the term parainfluenza refers to the four serotypes of HPIV and their
close animal relatives, and also includes the APMVs, even though these lack close human relatives. Another common human virus, mumps virus (MuV, Chapter 35), is related to the PIVs (most closely to HPIV2) and shares their physical and morphologic properties, but its hallmarks of parotitis and orchitis render it distinct.
close animal relatives, and also includes the APMVs, even though these lack close human relatives. Another common human virus, mumps virus (MuV, Chapter 35), is related to the PIVs (most closely to HPIV2) and shares their physical and morphologic properties, but its hallmarks of parotitis and orchitis render it distinct.
Because they can readily be grown to high titer, the animal PIVs SeV, PIV5, and NDV have been used extensively in studies spanning several decades that have defined many of the basic molecular and biological properties of Family Paramyxoviridae: this information is described in detail in Chapter 33. The present chapter focuses on PIV biology and in particular the HPIVs.
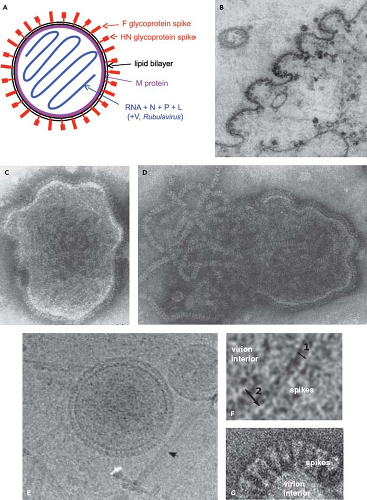 Figure 34.1. Schematic diagram (A) and electron photomicrographs (B–G) of parainfluenza virus (PIV) virions. A: Idealized diagram of a PIV virion, not to scale and not intended to imply relative molar amounts or exact spatial relationships. The V protein (not shown) is found as a structural protein only in Rubulavirus.284 The C protein (not shown) of the Respirovirus Sendai virus (SeV) also has been reported to be present associated with the virion nucleocapsid (not shown).403 B: PIV5 virions budding from the surface of a cultured cell.58 Intact (C) and disrupted (D) HPIV2 virions that were fixed and negatively stained; envelope spikes can be seen in both C and D, and the helical nucleocapsid is evident in D.156 E and F: Cryomicrographs of ice-embedded PIV5.369 E: Intact PIV5 virions and free nucleocapsids (arrow), and (F) higher-magnification images showing the thickness of the lipid bilayer (double arrow 1) and areas of the lipid bilayer with underlying matrix M protein (double arrow 2).369 G: Negatively stained cryomicrographs of a portion of a PIV5 virion showing distinct envelope spikes.237 |
Infectious Agent
Classification, Relationships, and Diversity
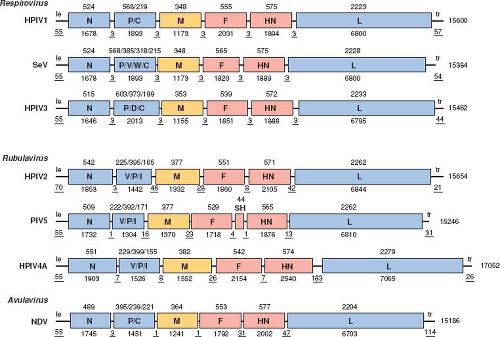
The PIVs are enveloped, cytoplasmic viruses (Fig. 34.1) with single-stranded, nonsegmented, negative-sense RNA genomes of 14.9 to 17.3 kb (Fig. 34.2). They are distributed among three genera (namely Respirovirus, Rubulavirus, and Avulavirus)
of subfamily Paramyxovirinae, family Paramyxoviridae, order Mononegavirales.218 (The other subfamily is Pneumovirinae, which contains HRSV, human metapneumovirus [HMPV], and their relatives). HPIV1 and HPIV3, and their respective murine and bovine relatives SeV and BPIV3, constitute the genus Respirovirus (Fig. 34.3). HPIV2, its relatives PIV5 and SV41, and HPIV4 are part of the genus Rubulavirus. Rubulavirus is a diverse genus that also contains related viruses that are not considered PIVs, such as MuV, Mapuera virus, and porcine rubulavirus. The various APMV serotypes constitute the genus Avulavirus.
of subfamily Paramyxovirinae, family Paramyxoviridae, order Mononegavirales.218 (The other subfamily is Pneumovirinae, which contains HRSV, human metapneumovirus [HMPV], and their relatives). HPIV1 and HPIV3, and their respective murine and bovine relatives SeV and BPIV3, constitute the genus Respirovirus (Fig. 34.3). HPIV2, its relatives PIV5 and SV41, and HPIV4 are part of the genus Rubulavirus. Rubulavirus is a diverse genus that also contains related viruses that are not considered PIVs, such as MuV, Mapuera virus, and porcine rubulavirus. The various APMV serotypes constitute the genus Avulavirus.
Table 34.1 Percent Amino Acid Sequence Identity Between the F Proteins of the Indicated PIVsa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The relationships between the PIVs are illustrated in Figure 34.3 by alignment of the amino acid sequences of the L proteins. Representative viruses from Morbillivirus and Henipavirus, two other genera of the subfamily Paramyxovirinae, are included for comparison. This shows that the PIVs as a whole are broadly divergent and are not clearly demarcated by sequence relatedness as a group distinct from the non-PIV members of Paramyxovirinae. This comparison also shows the close relatedness between some of the human and animal PIVs, such as between HPIV1 and SeV and between HPIV3 and BPIV3. It is likely that these closely related viruses arose from transmission across host species.
The relationships between the PIVs also are illustrated in Tables 34.1 to 34.3 by the percent amino acid sequence identity for the two major surface antigens, namely the fusion F glycoprotein (Table 34.1) and the hemagglutinin-neuraminidase HN glycoprotein (Table 34.2), as well as for the large polymerase L protein (Table 34.3). This illustrates, for example, that the HPIV serotype distinctions are associated with
amino acid sequence identities of less than 50% for F and HN. The APMV serotypes are not shown in these tables, but they also usually (but not always) have less than 50% amino acid sequence identity between F and HN of the different serotypes.43,172,213,214,265,318,353,401,402 For example, the percent amino acid sequence identity between APMV5 versus serotypes 1, 2, 3, 4, 6, 7, 8, and 9 is, respectively: 41, 47, 31, 33, 55, 37, 46, and 37 for the F protein; and 35, 42, 33, 30, 56, 43, 41, and 31 for the HN protein.319
amino acid sequence identities of less than 50% for F and HN. The APMV serotypes are not shown in these tables, but they also usually (but not always) have less than 50% amino acid sequence identity between F and HN of the different serotypes.43,172,213,214,265,318,353,401,402 For example, the percent amino acid sequence identity between APMV5 versus serotypes 1, 2, 3, 4, 6, 7, 8, and 9 is, respectively: 41, 47, 31, 33, 55, 37, 46, and 37 for the F protein; and 35, 42, 33, 30, 56, 43, 41, and 31 for the HN protein.319
Antigenic reactivity based on binding assays can be detected between PIVs within a genus with polyclonal sera and, less frequently, with monoclonal antibodies (MAbs).167,203,271 A lower level of reactivity between genera sometimes is detected with polyclonal sera, although there is no group antigen encompassing the three PIV genera.
HPIV4 has been segregated into two variants, A and B, based on antigenic differences detected by hemadsorption-inhibition (HI) and MAb reactivity.204 Sequence analysis shows that these two subgroups are very closely related: the percent identity between the F, HN, and L proteins is 95, 87, and 97, respectively (Tables 34.1 to 34.3), and they likely would not be distinguishable in neutralization assays with postinfection sera. Variation within the other HPIV serotypes appears to be somewhat less. For HPIV2, the percent amino acid sequence
identity between the V94 strain versus the V98 and Greer strains was, respectively, 98 and 99 for F, 95 and 96 for HN, and 99 and nearly 100 for L.337 For HPIV3, comparison of the F protein sequence of prototype strain Washington/47885/57 with seven clinical strains revealed 98% or more identity,62 and comparison of HN with six clinical strains revealed 97% or more identity.385 For HPIV1, comparison of 40 strains showed that the percent amino acid sequence identity for the HN protein was 95% or greater.146 For NDV, comparison of 50 strains from various times and places of isolation showed that the percent identity between F and HN was 91% and 90% or greater, respectively.317 Some of the other animal PIVs, such as APMV2,352 APMV3,214 APMV6,402 and BPIV3,152 have been found to have somewhat greater diversity (i.e., intraserotype amino acid sequence identity for F and HN of 75% to 79% for APMV2, 70% to 73% for APMV3, 81% to 86% for APMV6, and 86% to 89% for BPIV3), resulting in distinct genotypes or subgroups within a serotype. In the case of APMV2, APMV3, and APMV6, this has been shown to be associated with a modest degree of antigenic difference detectable with postinfection sera.
identity between the V94 strain versus the V98 and Greer strains was, respectively, 98 and 99 for F, 95 and 96 for HN, and 99 and nearly 100 for L.337 For HPIV3, comparison of the F protein sequence of prototype strain Washington/47885/57 with seven clinical strains revealed 98% or more identity,62 and comparison of HN with six clinical strains revealed 97% or more identity.385 For HPIV1, comparison of 40 strains showed that the percent amino acid sequence identity for the HN protein was 95% or greater.146 For NDV, comparison of 50 strains from various times and places of isolation showed that the percent identity between F and HN was 91% and 90% or greater, respectively.317 Some of the other animal PIVs, such as APMV2,352 APMV3,214 APMV6,402 and BPIV3,152 have been found to have somewhat greater diversity (i.e., intraserotype amino acid sequence identity for F and HN of 75% to 79% for APMV2, 70% to 73% for APMV3, 81% to 86% for APMV6, and 86% to 89% for BPIV3), resulting in distinct genotypes or subgroups within a serotype. In the case of APMV2, APMV3, and APMV6, this has been shown to be associated with a modest degree of antigenic difference detectable with postinfection sera.
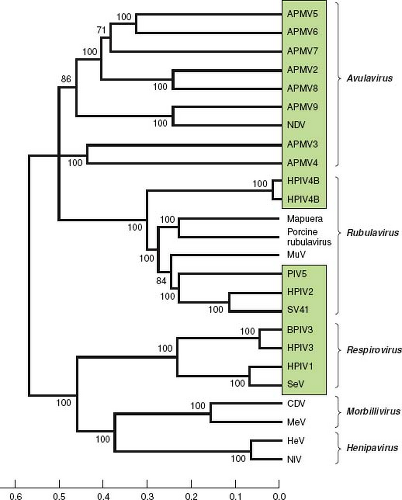 Figure 34.3. Phylogenetic analysis of the amino acid sequences of the L proteins of the parainfluenza viruses (PIVs) and other selected members of Paramyxovirinae (genera are indicated on the right). PIVs are boxed. The scale at the bottom indicates evolutionary distance as the number of substitutions per site. The analysis is based on the neighbor-joining method313 and was performed with Molecular Evolutionary Genetics Analysis (MEGA)4.361 The numbers at branch points indicate the percentage in which the associated taxa clustered together in the bootstrap test (500 replicates). The L protein sequence was chosen for analysis because it is one of the more conserved proteins, accounts for a substantial part of the viral coding sequence, and is similar in size for each virus. The sequences were as in Figure 34.2 or were from the following: avian paramyxovirus 2 (APMV2), EU338413; APMV3, EU403085; APMV4, EU877976; APMV5, GU206351; APMV6, EU622637; APMV7, FJ231524; APMV8, FJ215863; APMV9, EU910942; Mapuera virus, NC_009489; porcine rubulavirus, NC_009640; MuV, NC_002200; Simian virus 41 (SV41), NC_006428; BPIV3, NC_002161; SeV, NC_001552; CDV, canine distemper virus, NC_002728; MeV, measles virus, AF266288; HeV, hendra virus, NC_001906; NiV, Nipah virus, NC_001906. This analysis was kindly provided by Drs. Sachin Kumar and Siba Samal, University of Maryland at College Park. |
Table 34.2 Percent Amino Acid Sequence Identity Between the HN Proteins of the Indicated PIVsa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 34.3 Percent Amino Acid Sequence Identity Between the L Proteins of the Indicated PIVsa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NDV is notable because the many highly related naturally occurring isolates and strains that have been recovered for this single serotype exhibit a broad spectrum of virulence, ranging from nonvirulent or mildly virulent (lentogenic), to moderately virulent (mesogenic), to highly virulent (velogenic). Lentogenic strains are associated with subclinical infection or can cause mild respiratory tract disease, and the more attenuated natural isolates are used as live vaccines. At the other extreme, velogenic strains can be highly virulent and, depending on the strain, can cause hemorrhagic lesions in the intestines (viscerotropic) or neurologic disease (neurotropic).317 In contrast, there is no evidence of differences in virulence among the various isolates of each of the four serotypes of HPIV, although this has not been studied extensively. Little is known about the possible diversity of disease within the other animal PIVs.
Virion Morphology and Activities
The virions of PIVs are medium-sized particles of 150 to 200 nm. Fixed, negatively stained virions typically appear in electron micrographs as pleomorphic (irregularly shaped) round particles (Fig. 34.1).58,156,253,324,369 Filamentous virions have been described in some cases, such as for HPIV2.405 Cryoelectron microscopy of ice-embedded SeV and PIV5 provided images of virions as predominantly perfect spheres of varied diameters.153,233,369 This suggests that the irregular shapes of particles observed with conventional electron microscopy are artifacts of sample fixation and dehydration, whereas the variations in size were observed using both methods and thus may be authentic. Seventy-one percent of the ice-embedded PIV5 virions consisted of spheres of 129 to 360 nm (average 217 nm), and the remainder were elongated particles of up to 445 nm.369
PIVs replicate in the cytoplasm and bud through the plasma membrane (Fig. 34.1B). The virion consists of a nucleocapsid that is packaged in a lipid envelope derived from the host cell plasma membrane during budding (Fig. 34.1). In the nucleocapsid, the viral genome is tightly bound along its entire length with the nucleoprotein N at a ratio of one protein molecule per six nucleotides (2,484 to 2,877 protein molecules, depending on the length of the viral genome). Associated with the nucleocapsid in the virus particle are approximately 300 copies of the phosphoprotein P and approximately 40 copies of the major polymerase protein L, based on studies with SeV.200,219 Rubulaviruse virions contain an additional nucleocapsid-associated protein called V,284 and the virion of the Respirovirus SeV has been reported to contain 40 copies of a small protein called C, also associated with the nucleocapsid.403 Electron micrographs of nucleocapsids released from PIV virions indicate a length of approximately 1.0 to 1.1 μm.65,233 The nucleocapsid with its associated proteins has RNA-dependent RNA polymerase activity.134 Purified virions can be activated for cell-free transcription by disruption of the envelope with detergent and can transcribe the viral genome in its entirety into messenger RNAs (mRNAs).64
The envelope bears spike-like surface projections composed of homotrimers and tetramers of the F and HN glycoproteins, respectively. Based on cryoelectron microscopy, PIV5 virions were estimated to contain approximately 2,000 glycoprotein spikes per 200 nm particle, with an average spike length of 14.2 nm.369 PIV5 and AMPV6 have a third, small transmembrane protein, SH (“small hydrophobic”). The nonglycosylated matrix M protein is associated with the inner surface of the envelope. The hemagglutination activity of the HN protein mediates adsorption of virus to the host cell to initiate infection. The cellular receptor for the PIVs is N-acetylneuraminic
acid (sialic acid) in a terminal linkage to cellular glycoproteins and glycolipids.356,411 In the case of HPIV3, cell surface nucleolin also has been reported to serve as a receptor co-factor.29 Viral attachment can be measured experimentally by the agglutination of erythrocytes by virus in suspension (hemagglutination) or by the adsorption of erythrocytes to infected cell monolayers expressing HN (hemadsorption) as was used in the original detection of the HPIVs. Late in infection, the neuraminidase activity of HN cleaves sialic acid to facilitate release of progeny virions. Neuraminidase activity can be quantified using sialic acid derivatives as substrates in a colorimetric or fluorometric assay. The F protein mediates fusion between the viral envelope and the host cell plasma membrane, an activity that can be measured in vitro by lysis of erythrocytes (hemolysis).
acid (sialic acid) in a terminal linkage to cellular glycoproteins and glycolipids.356,411 In the case of HPIV3, cell surface nucleolin also has been reported to serve as a receptor co-factor.29 Viral attachment can be measured experimentally by the agglutination of erythrocytes by virus in suspension (hemagglutination) or by the adsorption of erythrocytes to infected cell monolayers expressing HN (hemadsorption) as was used in the original detection of the HPIVs. Late in infection, the neuraminidase activity of HN cleaves sialic acid to facilitate release of progeny virions. Neuraminidase activity can be quantified using sialic acid derivatives as substrates in a colorimetric or fluorometric assay. The F protein mediates fusion between the viral envelope and the host cell plasma membrane, an activity that can be measured in vitro by lysis of erythrocytes (hemolysis).
RNA
The PIV genome is a single strand of negative-sense RNA that ranges in length from 14,904 (APMV2) to 17,262 (APMV5) nucleotides (nt). The differences in genome length between the different PIVs are mostly due to differences in the lengths of noncoding sequences rather than substantial differences in the lengths of open reading frames (ORFs). The PIV genome is not capped or polyadenylated. It contains, in 3′ to 5′ order: a short 3′ extragenic leader region of 55 nt (except in the case of HPIV2, for which the leader region is 70 nt), followed by six genes encoding the N, P, M, F, HN, and L proteins, followed by an extragenic trailer region of 21 to 291 nt (Fig. 34.2; note that the longest, 291-nt trailer region is that of APMV-3 and is not shown in this figure). Sequences of the leader regions of selected PIVs are shown in e-Fig. 34.1A. As noted below, the P gene also encodes one or more accessory proteins—namely C, V, W, I, and D—depending on the virus (Fig. 34.2). PIV5 and APMV-6 each contain a seventh small gene that is located between F and HN and encodes the SH protein. In the case of Respirovirus, the PIV genes are separated by intergenic (IG) regions that are conserved trinucleotides (usually 3′-GAA in genome-sense); in the case of Rubulavirus and Avulavirus the IG regions have nonconserved sequences of variable length (0 to 183 nt) (Fig. 34.2; also, see e-Fig. 34.1B for gene junction sequences of selected PIVs).
Transcription and RNA replication occur in the cytoplasm and follow the Mononegavirales model. Briefly, the genes are transcribed sequentially in their 3′ to 5′ order to yield separate nonoverlapping mRNAs that are polyadenylated, capped, and methylated. RNA synthesis also yields short nonpolyadenylated and noncapped transcripts of the leader and trailer regions. Transcription is guided by short conserved gene-start (GS) and gene-end (GE) transcription signals that flank each gene (see e-Fig. 34.1B for gene junction sequences of selected PIVs). For RNA replication, the polymerase ignores the GS and GE signals and produces a complete positive-sense copy of the genome that is called the antigenome. Like the genome, the antigenome is not capped or polyadenylated. Both the genome and antigenome are completely bound with N protein.199 Encapsidation of nascent genomes and antigenomes is thought to drive chain elongation during RNA replication. The tightly encapsidated nature of the nucleocapsid likely shields the uncapped and nonpolyadenylated genome/antigenome from degradation. It also likely shields the genome/antigenome from recognition by the cytoplasmic helicases retinoic acid-inducible gene 1 (RIG-I) and Melanoma Differentiation-Associated protein 5 (MDA5), which detect triphosphorylated RNA and double-stranded RNA (dsRNA) and initiate signaling to activate the cellular transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor kappa B (NF-κB) to induce type I interferon (IFN) and proinflammatory cytokines. This also reduces activation of protein kinase R (PKR), which is triggered through dsRNA to activate NF-κB as well as to phosphorylate eukaryotic translation initiation factor eIF-2a and thereby inhibit translational initiation as part of host defense. As another example of how viral RNA can affect host cell responses, one of the products of SeV RNA replication is a 55-nt aborted RNA representing the 5′ trailer region that contains a U-rich sequence that inhibits apoptosis by binding to the proapoptotic factor T-cell intracellular antigen 1 related (TIAR).166
The nucleotide lengths of the genomes and antigenomes of the PIVs (and of all of subfamily Paramyxovirinae) are even multiples of six. This property is essential for efficient RNA replication and is called the “rule of six”.199–200,201,337 This is thought to reflect an obligatory nucleocapsid organization in which each N protein monomer associates with exactly six nucleotides. In experiments to recover recombinant HPIV2 and HPIV3 viruses whose nucleotide lengths were designed to not be even multiples of six, the recovered viruses contained genomes that had mutated to conform to the rule.336,337
Each PIV gene encodes—via transcribed mRNA—a single major protein, with the exception of the P gene that can encode additional proteins in two ways that are described briefly here and in greater detail for the HPIVs in e-Fig. 34.2. First, all of the PIVs in the genus Respirovirus contain a C ORF that initiates near the 5′ end of the P mRNA, closely overlapping the start of the P ORF. Depending on the virus, the C ORF has from one to four different translational start sites that are utilized to give rise to up to four carboxy–co-terminal C proteins. Rubulavirus and Avulavirus do not have a C ORF. Second, the P genes of most PIVs encode additional proteins by “RNA editing”.199,201,388 This involves the co-transcriptional insertion of 1 or more G residues into the nascent mRNA by polymerase stuttering at an editing motif midway along the P gene. An array of mRNAs is produced: they include the unedited form as well as subpopulations that contain 1, 2, or more G residues inserted at the editing site. The insertion of 1 G residue (or 3+1, and so on) or 2 residues (or 3+2, and so on) creates frameshifts that access ORFs in the two other reading frames. For the PIVs of Respirovirus and Avulavirus, the unedited mRNA encodes the P protein,286,348,388 and the addition of a single G by RNA editing fuses the upstream half of the P ORF to an internal ORF encoding a domain with a conserved cysteine-rich domain: the resulting protein is called V. The addition of 2 G residues fuses the upstream half of the P ORF to an ORF in the third reading frame: this downstream ORF encodes only a few added amino acids and results in a protein called W, except in the case of HPIV3 and BPIV3 in which the number of added amino acids is substantially more and the resulting protein is called D (e-Fig. 34.2). HPIV1 is an exception because it does not appear to engage in RNA editing.245,307 In addition, although HPIV3 does engage in RNA editing, the V ORF is separated from the editing site by two or more (depending on the strain) stop codons in the same reading frame that may preclude expression of V (see e-Fig. 34.2).108 For the PIVs of Rubulavirus, the exact-copy mRNA encodes the V protein, whereas an edited version containing
two inserted G residues encodes P.192,208,276,342,370 An edited version containing one inserted residue encodes the I protein, which is the Rubulavirus equivalent of W. [See the ebook for more information on coding assignments and RNA editing.)
two inserted G residues encodes P.192,208,276,342,370 An edited version containing one inserted residue encodes the I protein, which is the Rubulavirus equivalent of W. [See the ebook for more information on coding assignments and RNA editing.)
Several factors control the relative efficiency of transcription of the various PIV genes. As is typical for Mononegavirales, there is a gradient of transcription in which promoter-proximal genes are expressed somewhat more efficiently than promoter-distal genes.64,138 This is thought to be due to polymerase fall-off at the gene junctions.168 However, with the exception of L, the gradient of expression is not continuous or steep; in the case of SeV, for example, the P, M, F and HN mRNAs accumulate at 0.30, 1.15, 0.61, and 0.38 times the level of N.148 Accumulation of the L mRNA is much lower (0.02 that of N). Differences in transcription signals also influence transcription. In PIV5, the efficiency of transcription across the different gene junctions, measured by the relative level of expression of the downstream versus upstream gene, was found to vary over a fourfold range, indicative of regulation at the level of the termination/re-initiation at the gene junctions.138 However, the HN-L junction was not associated with a particularly high level of fall-off.138 This suggests that the low level of expression of L relative to the other genes is due to some other factor such as polymerase fall-off during L gene transcription or instability of the L mRNA.
A number of PIVs have evolved mechanisms for downregulating expression of the F gene. The M GE signal of HPIV3 contains an apparent eight-nucleotide insertion that causes increased M-F readthrough (see e-Fig. 34.1B).344 The M GE signal of PIV5 contains a single nucleotide substitution that has the same effect.300 In SV41, the M GE signal is lacking altogether and M is expressed solely as an M-F readthrough mRNA.376 Interestingly, the F gene of SV41 also is expressed as a monocistronic mRNA by initiation at its GS signal, but this occurs at a reduced level because the majority of the polymerase molecules are already engaged in reading across the M-F junction. In HPIV1, the same effect of increased production of M-F mRNA at the expense of monocistronic F mRNA was observed, and studies with recombinant viruses mapped the effect to a combination of features, namely the intergenic sequence, the F GS signal, and the long upstream nontranslated region of the F gene.31 Therefore, various features in these different viruses result in the synthesis of an M-F readthrough mRNA at the expense of a monocistronic F mRNA. In these M-F readthrough mRNAs, the F ORF would not be efficiently accessed by ribosomes due to its internal position.211 Finally, SeV downregulates expression of its F gene by yet another mechanism, namely through a suboptimal GS signal.188 Therefore, each of these strategies results in reduced expression of this fusogenic factor. In the case of SeV, this was shown to reduce the virulence of the virus.188 It might be that, by reducing morbidity and mortality in the host, the virus increases its opportunities for shedding and spread.
Proteins
All PIVs encode six common proteins: N, P, M, F, HN, and L, all of which are essential for virus replication. All members encode at least one additional protein from the P gene (C, V, D, W, and I, depending on the virus). PIV5 and APMV-6 also encode a small hydrophobic transmembrane SH protein.
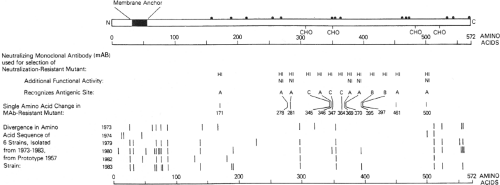 Figure 34.4. Linear diagram and antigenic organization of the human parainfluenza virus 3 (HPIV3) HN glycoprotein (strain 47885/5761). • Denotes cysteine; CHO denotes a potential site for N-glycan; HI (hemagglutinin-inhibiting) and NI (neuraminidase-inhibiting) denote positions of amino acid substitutions identified in neutralization-resistant mutants selected with HI and NI monoclonal antibodies (MAbs), and the amino acid positions and antigenic sites (A-C) are indicated; bars indicate positions of amino acid variability among natural isolates. |
HN Glycoprotein
The PIV HN glycoprotein (Fig. 34.4) mediates attachment by binding to host cell sialic acid. This activity is responsible for the ability of the virus to agglutinate erythrocytes. HN also functions late in infection to cleave sialic acid residues on the virus and nearby cell surface proteins to facilitate release of progeny virions. The dual hemagglutinin/neuraminidase functions of HN appear to be modulated by halide ion concentration and pH.248 Hemagglutination activity appears to be favored by the halide ion concentration and pH of the extracellular environment, consistent with the role of HN in binding to extracellular receptors, whereas neuraminidase activity is optimal at lower pH and halide ion concentration, consistent with the role of HN in stripping sialic acid from newly formed
viral and host cell glycoproteins in intracellular vesicles during transport to the cell surface. The HN proteins of PIVs and indeed most of Paramyxovirinae also play an essential role by interacting with the F protein to promote fusion.66,256,290,292,311
viral and host cell glycoproteins in intracellular vesicles during transport to the cell surface. The HN proteins of PIVs and indeed most of Paramyxovirinae also play an essential role by interacting with the F protein to promote fusion.66,256,290,292,311
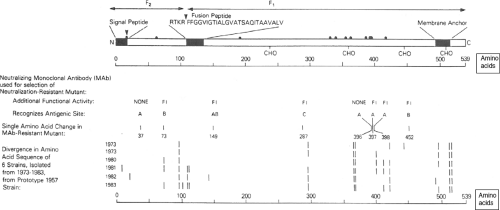 Figure 34.5. Linear diagram and antigenic organization of the human parainfluenza virus 3 (HPIV3) F protein (strain 47885/5761). • Denotes cysteine; CHO denotes a potential site for N-glycan; FI (fusion-inhibiting) denotes amino acid substitutions identified in neutralization-resistant mutants selected with FI MAbs, and the amino acid positions and antigenic sites are indicated; bars indicate positions of amino acid variability among natural isolates. |
HN is a type II glycoprotein that contains an uncleaved signal/anchor sequence located near the N-terminus (Fig. 34.4). HN assembles into homotetramers that contain a stalk that is sensitive to trypsin cleavage and a globular head that represents most of the extracellular domain. The globular head retains the HA and NA biologic activities and the major antigenic sites. On a gross level, the globular head has a box-shaped structure in which the four identical subunits exhibit fourfold symmetry. Crystal structures have been determined for the HN proteins of HPIV3, NDV, and PIV5, both free and complexed with its receptor or inhibitors32,69,222,312,409: these are described in Chapter 33.
The HN protein of some avirulent strains of NDV is synthesized as a longer precursor, HN0, in which the hemagglutinin and neuraminidase are inactive.263,316 Activation requires an endoproteolytic cleavage that results in the loss of a small, 9-kD glycopeptide from the carboxy terminus and a change in conformation.198,263 Like the F0 precursor protein of the avirulent strains (see below), HN0 is resistant to intracellular cleavage in most cell types and presumably is cleaved by extracellular secretory proteases, but unlike F0 it does not have a marked trypsin-like specificity and can be activated in vitro by a variety of proteases.263 It might be that the shorter HN proteins of virulent NDV strains that lack this extension and do not require cleavage arose evolutionarily from longer cleaved ancestral ORFs by the introduction of translational stop codons. This is suggested by the finding that the ORFs of certain virulent NDV strains retain the apparent relic of an in-frame C-terminal extension beyond the nonsense codon terminating the current ORF.249 A counterpart to HN0 has not been described for any other PIV, although the sequence of the HN gene of HPIV4 has been interpreted as containing a relic of such an extension.13
F Glycoprotein
The fusion (F) glycoprotein (Fig. 34.5) mediates penetration of the host cell by fusion of the viral envelope to the plasma membrane. Late in infection, when newly synthesized F glycoprotein has accumulated on the surface of the infected cell, it also can mediate fusion with contiguous uninfected cells. This results in the formation of syncytia, a prominent cytopathic effect in monolayer cultures in vitro. At least in the case of SeV, the F protein also can act as an auxiliary attachment protein that binds to cells via the hepatocyte-specific asialoglycoprotein receptor,24 although the significance of this in vivo is not known.
F is a typical type I glycoprotein (Fig. 34.5), with a cleaved N-terminal hydrophobic signal peptide and a C-proximal membrane anchor. The F protein is synthesized as an inactive precursor, F0, which is converted into the fusogenic form by cleavage by a host endoprotease to yield two subunits: F2, which contains the N-terminal 20% of the molecule, and F1, which contains the remainder of the molecule and is anchored in the membrane. F1 and F2 remain linked by a disulfide bond.170,198 The F1 amino terminus created by cleavage is a hydrophobic region called the fusion peptide that is thought to insert into the target membrane to initiate fusion. Crystal structures have been determined for the F proteins of HPIV3, NDV, and PIV5.52,357,406,407 The structure and function of F is described in detail in Chapter 33.
Cleavage of F0 is a prerequisite for PIV infectivity and can be an important determinant of tissue tropism and pathogenesis for NDV and possibly other PIVs (see below and Pathogenesis and Pathology).262,374 Most velogenic (highly virulent) and mesogenic (moderately virulent) strains of NDV have a cleavage site with the sequence R/K–R-Q-R/K–R↓F (Table 34.4). This multibasic
(basic residues are underlined) cleavage site conforms to the favored cleavage site R-X-R/K–R↓ for the ubiquitous intracellular protease furin,308 providing for efficient intracellular cleavage. Cleavage by furin or a furin-like protease allows the virus to replicate in cell culture without the need to supply exogenous protease in the culture medium. In vivo, it provides the potential for systemic spread and replication in a wide range of tissues, resulting in increased virulence. In comparison, the cleavage site sequence found in most avirulent NDV strains, G/E-K/R-Q-G/E-R↓L, has fewer basic residues and does not conform to the furin cleavage site. These strains are not cleaved by furin and require added protease (typically trypsin or allantoic fluid added to the culture medium) for replication in vitro, and are restricted in vivo to mucosal tissue of the lungs or intestines where secreted protease capable of cleaving the F0 precursor is found. Furin reportedly also may cleave at a “minimal” motif R-X-X-R↓,308 but apparently does not do so for the avirulent NDV strains.
(basic residues are underlined) cleavage site conforms to the favored cleavage site R-X-R/K–R↓ for the ubiquitous intracellular protease furin,308 providing for efficient intracellular cleavage. Cleavage by furin or a furin-like protease allows the virus to replicate in cell culture without the need to supply exogenous protease in the culture medium. In vivo, it provides the potential for systemic spread and replication in a wide range of tissues, resulting in increased virulence. In comparison, the cleavage site sequence found in most avirulent NDV strains, G/E-K/R-Q-G/E-R↓L, has fewer basic residues and does not conform to the furin cleavage site. These strains are not cleaved by furin and require added protease (typically trypsin or allantoic fluid added to the culture medium) for replication in vitro, and are restricted in vivo to mucosal tissue of the lungs or intestines where secreted protease capable of cleaving the F0 precursor is found. Furin reportedly also may cleave at a “minimal” motif R-X-X-R↓,308 but apparently does not do so for the avirulent NDV strains.
Table 34.4 Cleavage Sites of the F0 Proteins of Selected PIVs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Prototype strains of most of the PIVs have F0 cleavage sites that contain the furin cleavage motif, including HPIV3, BPIV3, HPIV2, SV41, and PIV5 (examples are shown in Table 34.4), and these viruses do not require added protease for replication in vitro. On the other hand, the F0 proteins of HPIV1, SeV, and HPIV4 lack the furin motif, and these viruses do require added protease in vitro. However, for viruses other than NDV, the lack of intracellular cleavage by furin does not necessarily indicate reduced virulence; for example HPIV1 and SeV can be highly virulent in vivo despite the lack of furin cleavage. Analysis of clinical isolates of HPIV3 showed that, although five of seven isolates contained the consensus furin motif observed in the prototype strains, two other isolates had the sequence D-P-R-T-E-R↓,62 which has the same arrangement of basic residues as avirulent NDV strains. However, these strains were fully competent for replication and the production of infectious virus in vitro without added protease, and did not exhibit any restriction for replication in the respiratory tract of rhesus monkeys. Similarly, several clinical isolates of HPIV2 were found to have the cleavage site sequence T/A-T/P-R-Q-E-R↓,12 which does not match the preferred furin motif. In this case, restricted growth by these clinical strains in vitro was observed in simian Vero cells but not in primary cultures of primate cells. Therefore, the presence of the preferred furin cleavage motif is not essential for intracellular cleavage or virulence in HPIVs.
Nucleocapsid-Associated N, P, and L Proteins
The N, P, and L proteins, together with the RNA genome, are the viral components that are necessary and sufficient to assemble the nucleocapsid and to direct transcription and RNA replication.94,134,365
The N protein is one of the more conserved PIV proteins. It associates with genomic and antigenomic RNAs to form highly stable, RNase-resistant helical nucleocapsids. Monomeric N is maintained in a soluble complex with the P protein prior to assembly into nucleocapsids. The N-terminal 75% of N is the more highly conserved part and is involved in forming the soluble complex with P as well as in subsequently associating with other N monomers and with RNA to form the nucleocapsid. The more variable C-terminal 25% of the molecule is not required to form the nucleocapsid but is essential for it to function as a template.34,199
The P protein is not highly conserved within a genus and has little or no significant sequence identity between genera. The P protein consists of N- and C-terminal functional modules separated by a divergent spacer that spans the RNA editing site.199 P is found as a homotetramer.366 P is the most heavily phosphorylated viral protein, although the bulk of constitutive phosphorylation can be ablated by mutation in recombinant SeV without effect.159 The N-terminal module of P is responsible for binding to free N protein and maintaining it as a soluble monomer necessary for nucleocapsid formation during RNA replication.74,149 The C-terminal module contains the homo-oligomerization domain and the polymerase co-factor domain, and is the only region of P necessary for transcription. This C-terminal module
mediates binding of P to the nucleocapsid. It also binds L protein and mediates its association with the nucleocapsid.149,199
mediates binding of P to the nucleocapsid. It also binds L protein and mediates its association with the nucleocapsid.149,199
The L protein is a large multifunctional protein responsible for nucleotide polymerization and mRNA capping and methylation.275 The N-terminal half of L contains blocks of highly conserved amino acids that are thought to be polymerase domains.287,288 The L protein forms a complex with the P protein that appears to serve as the RNA polymerase.149,199
The matrix M protein is a conserved, nonglycosylated species that is the most abundant virion protein and is located on the inner face of the virion envelope. In the infected cell, M associates with the inner face of the plasma membrane and plays key roles in virion assembly, budding, and release.326,360 Depending on the virus, expression of M alone (e.g., HPIV1) or together with N and HN or F (e.g., PIV5) triggers the formation and release of virus-like particles.67,354 The M protein of PIV5 was recently shown to contain a domain that mediates interaction with the host ubiquitin-proteasome pathway during the late stage of budding.326 M may also play a role in directing the transport of viral components to the plasma membrane.277,360
Accessory C, V, D, W, and I Proteins
These are products of the P gene, and the various PIVs differ as to which of these proteins are expressed, with the general pattern being genus specific. These proteins are not essential for virus replication (and thus are termed accessory), although the C and V proteins in particular can substantially increase the efficiency of growth in vitro and in vivo. As noted, C is encoded by a separate ORF in the P gene of Respirovirus, and is not found in Rubulavirus or Avulavirus. The V, D, W, and I proteins are produced by various PIVs (see Fig. 34.2) by frame shifts introduced by RNA editing (except in the case of Rubulavirus, where the V protein is produced from unedited mRNA while P depends on editing). These proteins are summarized below, and additional information on the expression and functions of these proteins in HPIVs is in e-Fig. 34.2. (The complexity of the proteins encoded by the P gene is even greater for SeV, for which the last ∼95 codons of the P ORF also are translated independently to yield a small nonstructural protein called X73,76; this protein is almost equimolar to P in infected cells, but its function is unknown and it will not be considered further.)
The C protein is an abundant small basic protein whose sequence is not well conserved between viruses. C is expressed into one or more carboxy-co-terminal forms, depending on the virus, by utilization of one or more translational start sites in the ORF: for example, SeV and HPIV1 produce four C proteins (C′, C, Y1, and Y2, in order of decreasing size), whereas HPIV3 produces one C protein (see e-Fig. 34.2). The different forms of the C proteins of SeV have been reported to have functional differences.75,111,112,220 C has historically been considered to be nonstructural, but the C protein of SeV was reported to co-localize with nucleocapsids in the infected cell and to be tightly associated with the virion-bound nucleocapsid, at 40 molecules per nucleocapsid.403 The functions of the C proteins have been investigated in detail for SeV. Deletion or mutation of the SeV C proteins results in strong induction of type I IFN and the establishment of an IFN-mediated antiviral state that restricts viral replication in IFN-competent cell culture and in vivo.110,215 The SeV C proteins were reported to inhibit activation of the transcription factors IRF-3 and NF-κB that leads to induction of IFN-β.206 The C proteins also inhibit signaling from the type I IFN receptor by binding to the signal transducer and activator of transcription protein 1 STAT1 and inhibiting phosphorylation of both STAT1 and STAT2.112,122,207,359 Another function of the SeV C proteins is to downregulate production of viral RNA at the level of transcription75 and RNA replication.37,150,364 By preventing overly robust RNA synthesis, this regulatory activity appears to prevent the formation of dsRNA and unencapsidated triphosphorylated replicative RNAs during SeV infection, thus reducing activation of MDA-5/RIG-I and PKR involved in innate immunity.358 This regulatory activity also prevents the overproduction of antigenomes, which otherwise can result in the packaging of antigenomes into progeny virions that would be noninfectious.165 The SeV C proteins inhibit apoptosis209 and have been reported to play a role in budding.136,315,354 Expression and functions of the C proteins of the HPIVs are described in e-Figure 34.2.
The V protein consists of the N-terminal half of P fused to a C-terminal V-specific domain that contains a sequence motif that is highly conserved in Paramyxovirinae and includes seven invariant cysteine residues (e-Fig. 34.2).140,272,299,322 The cysteine-rich domain has been shown to coordinate with two zinc atoms per protein molecule.227,284,347 V is a structural component of the nucleocapsid in the case of Rubulavirus, whereas V does not appear to be a structural component in Respirovirus virions and may be present in small amounts in Avulavirus virions.72,284,348 The clearest characterization of the functions of the V protein has come from studies with PIV5 and HPIV2, in which the absence of C protein facilitates evaluation. V has been shown to bind to MDA-5 and inhibit induction of IFN-β, whereas it did not appear to inhibit RIG-I.8,54,289,322 In addition, the V protein inhibits IFN-mediated signaling by mediating degradation of STAT1 or STAT2, depending on the virus and the host cell.9,161,272,320 PIV5-mediated degradation of STAT1 has been studied in detail and involves the V-protein binding to ubiquitin ligase and hijacking this cellular complex to target STAT1 for ubiquitination and proteosome-dependent degradation.85,227 The cysteine-rich domain must be present in order for V to inhibit IFN induction and signaling.140 The V protein also delays apoptosis during viral infection,355 and downregulates viral transcription and RNA replication.230 The mechanism for the effect on RNA synthesis was studied with minireplicons of SeV and HPIV2 and was found to be different for the two viruses.151,270 With SeV, the presence of the N-terminal domain of P allows the V protein to bind to soluble N protein and thus interfere with nucleocapsid assembly,151 whereas with HPIV2, the inhibitory activity of the V protein was associated with binding to the L protein, and involved the unique C-terminal domain of V.270 The V protein of PIV5 also has been shown to slow progression of the cell cycle.228
Therefore, the PIV C and V proteins have a number of similarities in their general effects, even though they are completely distinct proteins that appear to operate by distinct mechanisms. Two major common functions involve interference with host innate immunity—especially the type I IFN response—and downregulation of viral RNA synthesis. These functions may be related: as noted above for the C protein, reducing viral RNA synthesis can reduce activation of MDA-5/RIG-I, PKR, and other sensors that trigger innate immunity. The V protein is particularly important for members of Rubulavirus and Avulavirus given their lack of C proteins. For Respiroviruses, which encode the potent C proteins, some of the
host-antagonist functions of the V protein may be redundant or less robust. For example, although the V protein of SeV has been shown to bind MDA-5 and inhibit induction of IFN-β,53,54,206,289 the magnitude of this effect may be minor.123,314,350 Nonetheless, loss of expression of the SeV V protein significantly reduces the efficiency of viral replication in vivo, indicating a contribution that is additional to that of the C proteins.189,190 Exactly what this contribution is remains unclear.314 For the human Respiroviruses, V is more dispensable: as noted, HPIV1 does not encode a V protein due to a lack of RNA editing and the presence of translational stop codons within the V ORF, and HPIV3 likely expresses, at most, only low levels of V due to the presence of stop codons upstream of the V domain (see e-Fig. 34.2 for details on the expression and functions of the V proteins of the human PIVs). The presence of relict V ORFs interrupted by stop codons suggests that predecessors of the present HPIV1 and HPIV3 expressed V proteins, but that this ability became compromised by mutations that introduced these stop codons. Their animal relatives, SeV and BPIV3, respectively, retain the ability to efficiently express V.
host-antagonist functions of the V protein may be redundant or less robust. For example, although the V protein of SeV has been shown to bind MDA-5 and inhibit induction of IFN-β,53,54,206,289 the magnitude of this effect may be minor.123,314,350 Nonetheless, loss of expression of the SeV V protein significantly reduces the efficiency of viral replication in vivo, indicating a contribution that is additional to that of the C proteins.189,190 Exactly what this contribution is remains unclear.314 For the human Respiroviruses, V is more dispensable: as noted, HPIV1 does not encode a V protein due to a lack of RNA editing and the presence of translational stop codons within the V ORF, and HPIV3 likely expresses, at most, only low levels of V due to the presence of stop codons upstream of the V domain (see e-Fig. 34.2 for details on the expression and functions of the V proteins of the human PIVs). The presence of relict V ORFs interrupted by stop codons suggests that predecessors of the present HPIV1 and HPIV3 expressed V proteins, but that this ability became compromised by mutations that introduced these stop codons. Their animal relatives, SeV and BPIV3, respectively, retain the ability to efficiently express V.
The W (present in SeV and Avulavirus), I (Rubulavirus), and D (HPIV3 and BPIV3) proteins are created when RNA editing fuses the upstream end of the P ORF to a short internal ORF in the remaining reading frame. In the case of W and I, this internal ORF adds only a few amino acids; in the case of the D proteins of HPIV3 and BPIV3 the extension is longer (see e-Fig. 34.2). In general, the functions of the W, I, and D proteins are poorly understood. In the case of SeV, the W protein (like V, as noted above) was reported to downregulate viral genome replication in a reconstituted minireplicon system, an effect that was mediated by its P-related domain.71,151 The HPIV3 D protein was shown to accumulate in the nucleus of HPIV3-infected cells, but the significance of this is unclear.398
SH Protein
Among the PIVs, only PIV5 and AMPV6 encode SH proteins, which are 44 and 142 amino acids in length, respectively. MuV (Chapter 36) and all members of subfamily Pneumovirinae (Chapter 38) also encode SH proteins. In each case, SH is a transmembrane virion envelope protein with an externally oriented C-terminus. SH can be deleted without much effect on the magnitude of virus replication in vitro. However, deletion of SH from recombinant PIV5 resulted in increased cytopathology in cell culture due to increased apoptosis, although overall replication was not reduced, and the virus was attenuated in vivo.139,229 Further results indicated that infection with the ΔSH virus was associated with increased production of, and signaling by, tumor necrosis factor α, leading to the observed increase in apoptosis.229
Antigenic Composition and Determinants
Postinfection sera from animals and humans contain antibodies against most or all of the major PIV proteins. However, the HN and F proteins are the only antigens that have been shown to induce antibodies that neutralize infectivity, and they have been shown to be major independent protective antigens. In vivo, the parenteral administration of polyclonal or monoclonal antibodies specific to SeV HN or F mediated resistance to challenge with SeV.302 Sera obtained from children following HPIV3 infection that contain antibodies specific to the HN and F proteins have been shown to have virus-neutralizing activity.185 Infection of rodents with vaccinia virus recombinants expressing the HPIV3 HN or F glycoprotein, or immunization with purified HN and F glycoprotein, showed that either protein induced a high level of resistance to HPIV3 challenge, with HN being more protective than F.7,33,303,345
The “internal” PIV proteins also induce a protective response. This was demonstrated in experiments in hamsters using a recombinant version of HPIV3 in which the HN and F surface antigen genes were replaced by those of HPIV1. This made it possible to compare the relative contributions of the “internal” proteins and the surface glycoproteins to protection.363 The HN and F proteins induced a high level of protection (in this case specific to HPIV1) that was long-lived. In contrast, the HPIV3-specific protection attributed to the internal proteins—which presumably was mediated by major histocompatibility class I-restricted, CD8+ cytotoxic T lymphocytes (CTLs)—was weaker and waned over a period of several months.363 This suggests that cellular immunity can contribute significantly to protection for a short period following infection, but is not effective in providing long-term protection.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree