Orthoreoviruses
Terence S. Dermody
John S. L. Parker
Barbara Sherry
Introduction and History
Basic Features of the Reoviridae
Members of the Reoviridae form nonenveloped virions composed of 1 to 3 concentric protein shells. The genomes of these viruses consist of 9 to 12 discrete segments of double-stranded RNA (dsRNA). Much of what is known about the Reoviridae comes from studies of orbiviruses, orthoreoviruses, and rotaviruses. Although these and other Reoviridae members display important structural and biological differences, there are several properties held in common across the family.
The dsRNA gene segments of the Reoviridae viruses are not thought to leave the inner capsid particle. Enzymes that catalyze RNA transcription, capping, and replication are contained within the inner capsid, which serves as the site for the RNA synthetic activities. The messenger RNA (mRNA) transcripts are capped but not polyadenylated. The infectious cycle is entirely cytoplasmic and occurs for the most part in neo-organelles that are termed viral factories, viral inclusion bodies, or viroplasms. These viruses are sensed by the innate immune system and activate signaling cascades that induce an antiviral state or evoke apoptosis.
Discovery of Mammalian Reoviruses
Reoviruses were first isolated from stool specimens of children during the 1950s by Albert Sabin, Leon Rosen, and their colleagues.412,423 Reovirus is an acronym for respiratory enteric orphan virus; infections of humans by reovirus usually involve the respiratory and intestinal tracts with minimal or no associated disease symptoms (therefore the virus is an “orphan”). In the early 1960s, Peter Gomatos and Igor Tamm noticed that the inclusion bodies in reovirus-infected cells fluoresce greenish-yellow when stained with acridine orange, whereas single-stranded RNA (ssRNA) fluoresces orange.198 Using chemical
and physical studies, the viral genome was demonstrated to consist of dsRNA.40,197,347 Reoviruses were the first dsRNA viruses to be described.
and physical studies, the viral genome was demonstrated to consist of dsRNA.40,197,347 Reoviruses were the first dsRNA viruses to be described.
Table 44.1 Reoviridae Viruses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
The Reoviridae now includes 15 genera of dsRNA viruses that infect a wide variety of plants, animals, fungi, and protozoa (Table 44.1). The mammalian orthoreoviruses are the type species of the Orthoreovirus genus, which also contains viruses that infect birds and reptiles. The term “reovirus” refers to the mammalian orthoreoviruses and will be used as such in this chapter.
Pathogenic Reoviridae Viruses
Some members of the Reoviridae are pathogenic. Rotaviruses are responsible for gastroenteritis in animals and humans; they cause a large proportion of childhood gastroenteritis and are a major cause of infant illness and death in the developing world (see Chapter 45). Orbiviruses are transmitted by arthropod vectors and replicate in both mammalian and arthropod hosts (see Chapter 46). The best-studied orbivirus is bluetongue virus, an economically significant pathogen of sheep and cows named for a symptom affecting sick animals. Coltiviruses are also transmitted by arthropod vectors, and the prototype member, Colorado tick fever virus, can cause neurologic disease in humans. Aquareoviruses infect fish and mollusks. Fusogenic orthoreoviruses appear to cause pulmonary disease in humans.
Reovirus as a Model for Studies of dsRNA Virus Replication and Pathogenesis
Reoviruses are useful experimental models for studies of viral replication and pathogenesis. Studies of reovirus RNA synthesis were the first to describe the methylated cap structure that modifies most eukaryotic mRNA molecules and is required for normal translation;178,179,180 this work was done using RNAs synthesized in vitro from reovirus cores. Genetic approaches used to study the pathogenesis of reovirus in mouse models of infection and disease led to the discovery that phenotypic differences in specific steps in the virus–host encounter segregate with specific gene segments.254,498,515 Reovirus was the first virus shown to traverse the intestinal mucosa by transcytosis across microfold (M) cells534 and undergo proteolytic disassembly catalyzed by endosomal cathepsins.156 More recent developments have led to the deployment of reovirus as an oncolytic adjunct to cancer therapy475,493 and the potential use of reovirus as a vaccine vector through the application of plasmid-based reverse genetics.261
Taxonomy
dsRNA Viruses
There are nine taxonomic families of dsRNA viruses: the Birnaviridae, Chrysoviridae, Cystoviridae, Endornaviridae, Hypoviridae, Partitiviridae, Picobirnaviridae, Reoviridae, and Totiviridae. The Birnaviridae, Picobirnaviridae, and Reoviridae infect vertebrates; the Picobirnaviridae and Reoviridae infect mammals, including humans. The nine families are distinguished by genome organization, coding strategy, particle structure, and host range, among other properties. Differences in RNA-dependent RNA polymerase (RdRp) sequences suggest independent evolutionary lineages of dsRNA viruses from positive-sense RNA virus ancestors.267 Similarities beyond the existence of dsRNA genomes also suggest common ancestry, most notably in the organization of the capsid components. Interestingly, not all dsRNA viruses encode capsids. Members of the Hypoviridae transfer their genomes vertically during propagation of their fungal hosts.126
Family Reoviridae
The Reoviridae is the largest and most diverse family of dsRNA viruses. The viral particles display icosahedral symmetry with a diameter of 65 to 80 nm. The protein capsid is organized as one, two, or three concentric capsid layers, which surround the dsRNA segments of the viral genome. Members of some genera have filamentous attachment proteins, but most do not. The viral RNA species are usually monocistronic, although some segments have two or three in-frame initiation codons that lead to expression of additional open-reading frames (ORFs). Alternative out of frame ORFs or nonoverlapping ORFs are occasionally present. Proteins are encoded by only one strand of each duplex (the mRNA species).
The 15 genera of the Reoviridae are divided into two groups based on particle morphology (Table 44.1). The Sedoreovirinae subfamily includes genera containing “smooth” viruses that are almost spherical in appearance, including Cardoreovirus, Mimoreovirus, Orbivirus, Phytoreovirus, Rotavirus, and Seadornavirus. The Spinareovirinae subfamily includes genera containing viruses with large “spikes” or “turrets” at the 12 icosahedral vertices of the virus or core particle, including Aquareovirus, Coltivirus, Cypovirus, Dinovernavirus, Fijivirus, Idnoreovirus, Mycoreovirus, Orthoreovirus, and Oryzavirus. The overall fold of several of the major inner and outer capsid proteins of Reoviridae members is similar, despite the absence of primary sequence identity.
The inner protein layer of the Sedoreovirinae (smooth) viruses has T = 2 symmetry and is relatively fragile. This structure is surrounded by an additional protein layer, which has T = 13 symmetry, to form the transcriptionally active core particle. These double-layered particles are surrounded by an
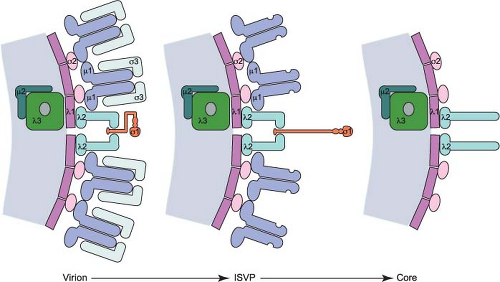
outer-capsid shell, giving rise to triple-layered particles. In contrast, the transcriptionally active core particle of the Spinareovirinae (turreted) viruses appears to contain only a single complete capsid layer, which has been interpreted as having T=1 or T=2 symmetry, to which the spikes are attached. Despite these different interpretations, the structure of the innermost complete capsid layer is almost identical for members of both the Sedoreovirinae and Spinareovirinae. In most cases, the core is surrounded by an additional protein layer with T=13 symmetry that forms the outer capsid, which is penetrated by the spikes that arise from the core surface. These virus particles are therefore usually regarded as double shelled (Fig. 44.1).
outer-capsid shell, giving rise to triple-layered particles. In contrast, the transcriptionally active core particle of the Spinareovirinae (turreted) viruses appears to contain only a single complete capsid layer, which has been interpreted as having T=1 or T=2 symmetry, to which the spikes are attached. Despite these different interpretations, the structure of the innermost complete capsid layer is almost identical for members of both the Sedoreovirinae and Spinareovirinae. In most cases, the core is surrounded by an additional protein layer with T=13 symmetry that forms the outer capsid, which is penetrated by the spikes that arise from the core surface. These virus particles are therefore usually regarded as double shelled (Fig. 44.1).
The innermost protein layer of Reoviridae viruses has an internal diameter of approximately 50 to 60 nm surrounding the 9 to 12 linear dsRNA gene segments. In the Sedoreovirinae viruses, the enzymatically active proteins of the virion, RdRp, NTPase, guanylyltransferase, and transmethylase, are also situated within this central structure attached to the inner surface at the fivefold symmetry axes. In the Spinareovirinae viruses, some of these enzymes form the turrets on the surface of the core. These hollow projections also act as conduits for the exit of nascent mRNAs synthesized by the core-associated enzymes.
Genus Orthoreovirus
The genus Orthoreovirus contains the nonfusogenic mammalian orthoreoviruses and the fusogenic orthoreoviruses, which infect a variety of vertebrate hosts. Common features of the members of the genus Orthoreovirus include: (1) two protein shells with core spikes at the icosahedral vertices; (2) 10 dsRNA segments including three large (L), three medium (M), and four small (S) size-class RNA gene segments; (3) three λ, three μ, and four σ primary translation products; (4) additional small gene products encoded by a polycistronic S1 gene segment; and (5) common serologic characteristics. Related members of the genus Orthoreovirus can exchange gene segments following co-infection of cells, producing viral progeny termed “reassortant viruses” containing mixtures of gene segments derived from both parents. The capacity of two strains to produce viable reassortant progeny can serve as the basis for designating species of virus within the genus.153,322 Nonfusogenic isolates can exchange gene segments with each other, as can fusogenic isolates, but reassortment between nonfusogenic and fusogenic viruses has not been reported.
Mammalian orthoreovirus was the first species of the family Reoviridae to be isolated and identified.423 The prefix “ortho-” was added to the name of the initial isolates (orthoreoviruses) and the corresponding genus (Orthoreovirus) to distinguish them from other members of the family, which also could be called “reoviruses.” However, in common usage the term “reovirus” refers to the mammalian orthoreoviruses.
Nonfusogenic Orthoreoviruses
Reoviruses have a wide geographic distribution, and virtually all mammals, including humans, serve as hosts for infection. Based on sequence analysis of reovirus strains isolated from a variety of mammalian hosts, there is no evidence that any of the gene segments confer host range restriction.88,510 Because of their near ubiquity, reoviruses are used as a marker for mammalian fecal contamination of water sources.314 Human infections
with reovirus are common, with most children demonstrating serologic evidence of infection by the age of 5 years.482 However, reovirus is rarely associated with disease, except in the very young. There is evidence suggesting an association between reovirus and infantile biliary atresia,393,499 but a causal link has not been demonstrated. Newborn mice are exquisitely sensitive to reovirus infection and have been used as the preferred experimental model for studies of reovirus pathogenesis.510
with reovirus are common, with most children demonstrating serologic evidence of infection by the age of 5 years.482 However, reovirus is rarely associated with disease, except in the very young. There is evidence suggesting an association between reovirus and infantile biliary atresia,393,499 but a causal link has not been demonstrated. Newborn mice are exquisitely sensitive to reovirus infection and have been used as the preferred experimental model for studies of reovirus pathogenesis.510
Three mammalian reovirus serotypes have been identified by classical neutralization and hemagglutination-inhibition (HAI) tests. Four virus strains isolated from children in the 1950s—type 1 Lang (T1L), type 2 Jones (T2J), type 3 Abney (T3A), and type 3 Dearing (T3D)—are used most frequently as prototype strains.423 A distinct reovirus strain was isolated from a mouse in the Cameroon18 and may represent a fourth mammalian reovirus serotype. Nucleotide sequences indicate that nonfusogenic mammalian isolates are a distinct phylogenetic group within the genus Orthoreovirus.153 General features of the nonfusogenic mammalian orthoreoviruses are provided in Table 44.2.
Table 44.2 General Features of Nonfusogenic Mammalian Orthoreoviruses | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Fusogenic Orthoreoviruses
The fusogenic orthoreoviruses infect birds, mammals, and reptiles. In contrast to the nonfusogenic orthoreoviruses, fusogenic reoviruses induce the formation of large, multinucleated syncytia.127 Fusion activity is mediated by virus-encoded, fusion-associated small transmembrane (FAST) proteins, which are not capsid components but instead expressed on the surface of infected cells.453 FAST proteins are small (<20 kDa), basic, acylated proteins that induce fusion in transfected cells in the absence of other viral proteins.62 Fusogenic members of the related genus, Aquareovirus, also encode FAST proteins.386 Extensive syncytium formation results in apoptosis and enhanced release of infectious virions.426 Expression of the p14 FAST protein by a recombinant vesicular stomatitis virus increases neuropathology.69 The FAST proteins therefore likely contribute to the natural pathogenicity of the fusogenic orthoreoviruses. Other than this fusion activity, the replication strategies of fusogenic and nonfusogenic orthoreoviruses are similar.42
The fusogenic orthoreoviruses are divided into four species: avian reovirus (ARV), Nelson Bay reovirus (NBV), reptilian reovirus (RRV), and baboon reovirus (BRV).88 ARVs infect a variety of avian hosts and are important pathogens in the poultry industry, causing gastroenteritis, hepatitis, malabsorption, myocarditis, and pneumonia.249 Birds that survive acute systemic infection can develop joint and tendon inflammation that resembles the pathology of human rheumatoid arthritis. RRVs have been isolated from numerous reptile species, and infected animals often develop pneumonia and neurologic symptoms.519 BRV was isolated from baboons with meningoencephalomyelitis in a colony in Texas.280 Therefore, it is an example of a fusogenic mammalian orthoreovirus. BRV virions, like those of the aquareoviruses, lack the classical sigma-class fiber protein used for cell adhesion by other reoviruses.537 NBV is the prototype orthoreovirus species isolated from bats; it is more closely related to ARV than to BRV, the other fusogenic mammalian orthoreovirus species.127
Additional fusogenic bat reoviruses have been isolated,381 one of which represents a new species that also lacks a sigma-class fiber attachment protein.487 Three fusogenic reoviruses isolated from humans with acute respiratory infections also may be of bat origin.96,97,98 These viruses appear capable of human-to-human transmission, raising concern that fusogenic bat reoviruses may be a potential source of emerging infections in humans.
Particles
Particle Function
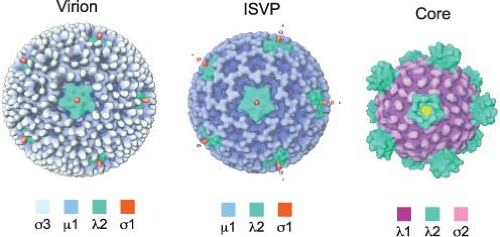
Reovirus virions are spherical, nonenveloped particles approximately 85 nm in diameter as assessed by cryoelectron microscopy (cryo-EM) image reconstructions151 (Fig. 44.2). Virions are formed by two concentric capsid protein shells that surround and protect the dsRNA genome. The inner-capsid shell or core particle encloses the genome, which is present in a
condensed liquid crystalline form.394 The outer-capsid shell surrounds the core particle, which protrudes radially into the outer-capsid layer at each of the fivefold axes of symmetry.151
condensed liquid crystalline form.394 The outer-capsid shell surrounds the core particle, which protrudes radially into the outer-capsid layer at each of the fivefold axes of symmetry.151
Several different morphotypes of reovirus particles are found within infected cells or can be generated in vitro. These include genome-containing virions, infectious or intermediate subvirion particles (ISVPs), and core particles (Figs. 44.1 and 44.2) as well as their genome-lacking counterparts. In addition, less well-defined particles have been purified from infected cells and are characterized by the capacity to transcribe the genome in vitro (“transcriptase” particles) or synthesize a negative-stranded RNA copy of particle-associated viral mRNA (“replicase” particles).338,557 Physical properties of virions, ISVPs, and cores are described in Table 44.3.
Table 44.3 Characteristics of Common Reovirus Particle Forms | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Virions
Reovirus virions are relatively stable in the environment198,208 and maintain infectivity for years when stored below 4°C.44 Large quantities of infectious virions can be purified from infected mouse L929 cells by Freon extraction and centrifugation in CsCl gradients.44
Virions are composed of a genome-containing inner T = 1 icosahedral core surrounded by a T = 13 laevo outer-capsid layer (Fig. 44.2). The outer capsid consists of 200 heterohexameric complexes of the M2-encoded μ1 (76 kDa, 708 aa) and S4-encoded σ3 (41 kDa, 365 aa) proteins. These complexes form a fenestrated shell that overlies the core. The outer-capsid shell is perforated at the icosahedral fivefold symmetry axes by turret-like structures formed by pentamers of the L2-encoded
λ2 protein (144 kDa, 1288–1289 aa). The S1-encoded σ1 attachment protein (49–51 kDa, 455–470 aa) forms filamentous trimers that protrude from each of the λ2 turrets. In addition to encapsidating the 10 dsRNA gene segments, virions also contain short ssRNA oligonucleotides that can constitute up to 25% of the RNA in purified virions.443 Most of these oligonucleotides (70%) are 2- to 9-residue products of abortive transcription and terminate with 5′-GC(U)(A).536 The remainder (approximately 30%) are oligoadenylates that range in length from 2 to 20 residues.455 Complete or partial atomic resolution structures are available for σ1 trimers,92,395 σ3 dimers,368 and μ1:σ3 heterohexamers.289 Cryo-EM image reconstructions of reovirus virions, ISVPs, and cores151 (Fig. 44.2), and an atomic resolution structure of the core394 have been determined.
λ2 protein (144 kDa, 1288–1289 aa). The S1-encoded σ1 attachment protein (49–51 kDa, 455–470 aa) forms filamentous trimers that protrude from each of the λ2 turrets. In addition to encapsidating the 10 dsRNA gene segments, virions also contain short ssRNA oligonucleotides that can constitute up to 25% of the RNA in purified virions.443 Most of these oligonucleotides (70%) are 2- to 9-residue products of abortive transcription and terminate with 5′-GC(U)(A).536 The remainder (approximately 30%) are oligoadenylates that range in length from 2 to 20 residues.455 Complete or partial atomic resolution structures are available for σ1 trimers,92,395 σ3 dimers,368 and μ1:σ3 heterohexamers.289 Cryo-EM image reconstructions of reovirus virions, ISVPs, and cores151 (Fig. 44.2), and an atomic resolution structure of the core394 have been determined.
Infectious Subvirion Particles (ISVPs)
Treatment of virions with chymotrypsin or trypsin using controlled conditions in vitro58 or virion disassembly during infection of cells480 or in the murine intestine29,54 generates ISVPs (Figs. 44.1 and 44.2). These particles lack σ3 but retain σ1 and a proteolytically cleaved version of μ1 (see Replication—Disassembly). The loss of σ3 and cleavage of μ1 imparts a twofold increase in infectivity of ISVPs relative to virions for reovirus strain T1L. In contrast, in vitro generation of ISVPs from strain T3D virions leads to a 10-fold decrease in infectivity as a consequence of additional proteolytic cleavage of σ1.349 This cleavage is controlled by a sequence polymorphism in σ1 at amino acid 24987 and likely explains the decreased growth in the intestine and avirulence of T3D after peroral inoculation of newborn mice.53 An additional intermediate particle, the ISVP*, is generated from ISVPs during infection of cells.81 ISVP*s lack the σ1 attachment protein, have an altered conformer of μ1, and tend to aggregate. In contrast to virions and ISVPs, ISVP*s are transcriptionally active.79 ISVP*s are formed following generation of ISVPs and thought to interact with cell membranes leading to membrane penetration and release of the core particle into the cytosol (see Replication—Membrane Penetration).
Core Particles
Like ISVPs, cores can be generated in vitro by treating virions with chymotrypsin or trypsin using specific conditions.60 Such treatment removes all of the outer-capsid proteins (μ1, σ3, and σ1) and exposes prominent turrets formed by pentamers of λ2. These turrets protrude from the capsid shell at each of the fivefold symmetry axes (Figs. 44.1 and 44.2) and form hollow cylinders that open to the interior of the particle. In cryo-EM image reconstructions of virions and ISVPs, the cavity within each turret is partially filled by a bulb-like structure that corresponds to a portion of the σ1 protein151 (Fig. 44.2). The σ1 bulb is held in place by flap domains present on each of the five λ2 proteins. In cores, the flap domains of λ2 open outwards. The interior surface of the pore formed by the λ2 turret possesses guanylyltransferase and methyltransferase activities, which allow the turrets to function as mRNA capping complexes394 (see Replication—Transcription). Cores are approximately 1,000,000-fold less infectious than virions84 as a consequence of the loss of the outer-capsid proteins, which are required for virus attachment, entry, and membrane penetration.
The T = 1 core shell is formed by 120 copies of the L3-encoded λ1 protein (142 kDa, 1275 aa) arranged in parallel asymmetric dimers (sometimes referred to as T = 2). Five copies of λ1 (conformer λ1A) radiate from the fivefold axes of symmetry. These molecules interdigitate with a second set of five λ1 proteins (conformer λ1B) to form decamers; 12 decamers form the core shell.394 The λ1 shell is stabilized by 150 copies of the S2-encoded σ2 protein (47 kDa, 418 aa), which clamps onto λ1 at three distinct sites within each icosahedral asymmetric unit of the shell.394 Channels permeate the shell, with those at the fivefold symmetry axes allowing entry of NTP substrates and exit of newly synthesized viral mRNAs.
The reovirus core is described as a “molecular machine” owing to its capacity to synthesize capped viral mRNA transcripts in vitro when incubated with suitable substrates. During infection, particles thought to be similar to in vitro–generated cores initiate viral transcription after being deposited in the cytoplasm. Core particles contain 12 copies of the L1-encoded λ3 polymerase (142 kDa, 1267 aa). Each λ3 protein is anchored to three λ1 proteins on the inner surface of the core and overlaps with, but is eccentric from, each of the icosahedral symmetry axes.550 In addition, cores contain approximately 24 copies of the M1-encoded μ2 protein (83 kDa, 736 aa),114 a polymerase co-factor. The precise location of μ2 within the core particle is not known, but one or two copies of μ2 are thought to associate with each copy of λ3.
Empty Particles (“Top Component” Particles)
Purification of reovirus particles from infected cells in vitro yields a substantial proportion of particles that lack genomic dsRNA.463 These empty particles have a lower buoyant density in CsCl gradients and band closer to the top of the gradient in comparison to genome-containing particles, hence the designation “top component”.44 Protease treatment of such particles can generate “empty” ISVPs and cores.
Recombinant Particles
Recombinant core-shell particles can be formed by co-expression of the λ1 and σ2 proteins or the λ1, λ2, and σ2 proteins.535 Purified viral core particles prepared by in vitro digestion of virions with chymotrypsin or trypsin can be recoated with outer-capsid proteins μ1, σ3, and σ1 prepared using baculovirus expression systems.84 Such recoated particles display infectivity similar to native virions and are useful tools for defining the consequences of lethal mutations in outer-capsid proteins on early steps in viral infection (see Genome and Coding Strategies—Genetic Complementation).
Genome and Coding Strategies
Physical Characteristics
The reovirus genome consists of 10 discontiguous segments of dsRNA. The [+] and [−] strands of genomic dsRNA are collinear and complementary.40,197 The genomic dsRNA has an A-form structure with a right-handed double helix (10 base pairs per turn, 30 Å pitch, nucleotides oriented at 75° to 80° to the long axis).17
Gene Segments and Nomenclature
The 10 reovirus dsRNA gene segments are present in equimolar amounts in viral particles.444 The segments are grouped by size into large (L), medium (M), and small (S) classes according
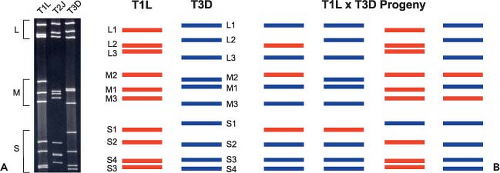
to their migration in polyacrylamide gels.195,330,444,513 There are three large (L1, L2, L3), three medium (M1, M2, M3), and four small (S1, S2, S3, S4) segments. Homologous gene segments from different reovirus isolates often migrate with different mobilities in polyacrylamide gels despite being of similar or identical length388 (Fig. 44.3). This property allows gene segments from different isolates to be unambiguously distinguished, which is essential for the assignment of gene segments in reassortant viruses to specific parental strains (see Genome and Coding Strategies—Reassortment). The gene segments of prototype strain T3D are numbered according to their relative mobilities in Tris/acetate-buffered gels.441 Each gene segment encodes at least one protein product (Table 44.4).
to their migration in polyacrylamide gels.195,330,444,513 There are three large (L1, L2, L3), three medium (M1, M2, M3), and four small (S1, S2, S3, S4) segments. Homologous gene segments from different reovirus isolates often migrate with different mobilities in polyacrylamide gels despite being of similar or identical length388 (Fig. 44.3). This property allows gene segments from different isolates to be unambiguously distinguished, which is essential for the assignment of gene segments in reassortant viruses to specific parental strains (see Genome and Coding Strategies—Reassortment). The gene segments of prototype strain T3D are numbered according to their relative mobilities in Tris/acetate-buffered gels.441 Each gene segment encodes at least one protein product (Table 44.4).
Modifications to the Genomic RNA
The [+] strand RNA of each gene segment has a dimethylated cap-1 structure on its 5′ terminus,95,179,332 which is added by viral guanylyltransferase and methyltransferase activities present in the λ2 protein.106,295 The viral mRNA cap is recognized by translation initiation factor eIF4E61,346 and protects the free 5′ ends of the viral mRNA from 5′ exonucleases.177 The 5′ terminus of the [−] strand RNA of each gene segment has an unblocked diphosphate.24,95 The terminal γ-phosphate of the [−] strand is removed by a phosphohydrolase activity (possibly mediated by the μ2 or λ1 protein) within reovirus particles.24,51,95,256 The 3′ end of each gene segment has an unblocked hydroxyl group.330 The viral [+] strand RNA does not contain a polyadenylate tract.
Sequence Features
Full-length sequences of the entire genomes of prototype mammalian orthoreovirus strains T1L, T2J, and T3D as well as those of several fusogenic orthoreovirus isolates have been determined (National Center for Biotechnology Information). The total length of the genome sequence is approximately 23,500 base pairs. The length of individual gene segments varies from 1,196 (T3D S4 gene segment) to 3,916 (T3D L2 gene segment) nucleotides. Most homologous gene segments of different isolates display little (±4 base pairs) or no variation in segment length. However, the S1 gene segment varies in length to a greater extent (1,463, 1,440, and 1,416 base pairs for the T1L, T2J, and T3D S1 gene segments, respectively).154,350
Protein-Coding Strategies and Nomenclature
Twelve proteins are translated from the 10 T3D reovirus gene segments (Table 44.4). Eight of these segments encode a single protein, and two encode two proteins each. Eight of the 12 proteins are structural components of virions (λ1, λ2, λ3, μ1, μ2, σ1, σ2, and σ3), and four are nonstructural (μNS, μNSC, σ1s, and σNS). The nonstructural protein μNS was originally designated μ0.558 The assignment of individual proteins to specific gene segments was determined by analysis of viral reassortants344,345,390,518 and in vitro translation of viral mRNAs.315
The names of the proteins encoded by each of the gene segments are designated by a Greek symbol that corresponds to large (λ), medium (μ), and small (σ) sizes, which are encoded by gene segments of the L, M, and S classes, respectively. For historical reasons, the numbering of gene segments does not always correspond to the numbering of the proteins (Table 44.4).
During reovirus infection, the [−] strand of each gene segment is transcribed to yield a full-length [+] strand RNA copy that is capped as it is released from the transcribing core particle. The viral mRNAs are not polyadenylated, but they are efficiently translated. ORFs of each gene segment are flanked by short nontranslated regions (NTRs). The AUG start codon for each gene segment is usually the first to be encountered by a scanning ribosome and, in most cases, it is found in an excellent context for translation initiation.269 The S1 gene segment encodes a second protein, σ1s, in an ORF overlapping that of the larger σ1 protein.160,241,429 The σ1s protein is translated from the first out-of-frame start codon within the σ1 ORF.343 Like the S1 gene segment, the M3 gene segment encodes a second protein, which is called μNSC. Translation of μNSC initiates from the second or third in-frame start codon of the
m3 RNA.316 There is no evidence that any reovirus protein is translated from a genomic [−] strand.
m3 RNA.316 There is no evidence that any reovirus protein is translated from a genomic [−] strand.
Table 44.4 Functions of Reovirus Proteins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Terminal Nontranslated Regions
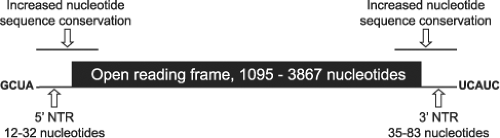
The reovirus gene segments contain short terminal NTRs (Fig. 44.4). The 5′ NTRs range in length from 12 to 32 nucleotides, and their relative sizes may affect their translation efficiency.406 The 3′ NTRs are a little longer and vary in length from 35 to 83 nucleotides. In general, NTR length of homologous gene segments is conserved between different virus isolates.316
The [+] strand of all reovirus gene segments contains the 5′ sequence GpCpUpA and the 3′ sequence UpCpApUpC.14 Longer regions of sequence near the RNA termini and extending into the ORFs display less variability between isolates than sequences farther from the ends90,202,253 (Fig. 44.4). RNA-folding predictions suggest that sequences near the ends of the reovirus [+] strands can form panhandle structures, which may be important for certain RNA functions.90,286,553 For example, the 5′ and 3′ NTRs include sequences and structural elements important for RNA packaging.405,408,409
Genetics
Reassortment
Gene segment exchange (or reassortment) among reovirus strains occurs during mixed infections of cultured cells or animal hosts. The progeny of such mixed infections are commonly referred to as reassortant viruses. Many reovirus strains are distinguishable by signature electrophoretic profiles of their dsRNA gene segments in acrylamide gels (Fig. 44.3A). Co-infection of cells with such strains produces a collection of reassortant viruses in which the parental origin of each gene segment can be determined following electrophoresis of viral genomic dsRNA (Fig. 44.3B). Studies of reassortant viruses have been used to assign biological polymorphisms displayed
by different reovirus strains to specific viral gene segments. Thus, a phenotypic difference between two parental strains can be genetically mapped by screening the reassortant viruses in appropriate assays and correlating expression of the phenotype with a specific parental gene segment.
by different reovirus strains to specific viral gene segments. Thus, a phenotypic difference between two parental strains can be genetically mapped by screening the reassortant viruses in appropriate assays and correlating expression of the phenotype with a specific parental gene segment.
Reassortment of reovirus gene segments also occurs in vivo. Infection of mice perorally with strains T1L and T3D yields reassortant progeny in the stool and at sites of secondary replication.521 In at least one case, a reassortant virus arising during such a mixed infection displays virulence properties that differ from both parental strains,451 suggesting the selection of new mutations in vivo. Beyond experimental infection of animals, additional support for the hypothesis that reovirus gene segments reassort in nature is provided by analysis of nucleotide sequences of the L1,277 M1,541 S1,137 S2,90 S3,202 and S4253 gene segments derived from a panel of field-isolate strains isolated in the 1950s and 1960s by Leon Rosen.413,414,417 The topologies of phylogenetic trees constructed from protein-coding sequences of these gene segments are distinct, providing convincing evidence that these segments reassort in nature.
Reassortment is not entirely random. In a study of 83 reassortant viruses derived from strains T1L and T3D, statistically significant nonrandom associations of parental alleles were observed in the L1–L2, L1–M1, L1–S1, and L3–S1 gene segment pairs.352 It is possible that these gene segments or their protein products cannot accommodate exchange with gene segments from another strain without the selection of fitness-compromising mutations. Additionally, the majority of progeny viruses resulting from mixed infections retains the genome constellation of the parental strains,169 suggesting that viral replication is compartmentalized in some way.
Selection of Mutants: “Forward Genetics”
Temperature-Sensitive Mutants
Reovirus mutants with reduced capacity to replicate at elevated temperature, usually defined as 39°C or 40°C115,169,214 as opposed to 31°C, have been selected using a variety of conditions. Temperature-sensitive mutants mapping to seven of the 10 genetic groups (gene segments) of strain T3D were isolated after chemical mutagenesis with nitrous acid (groups B [L2], D [L1], and E [S3]), nitroso-guanidine (groups C [S2], F [M3], and G [S4]), or proflavin (group A [M2]).169,233 Mutants from the other three groups were isolated from T3D stocks passaged serially at high multiplicity of infection (MOI) (group H [M1]) or from persistently infected cultures (groups I [L3] and J [S1]).5,6,8 The genetic groups were assigned to discrete gene segments by reassortant analyses or the capacity to complement temperature-sensitive mutants from other groups.344,387,390 Although the prototype isolates of groups F, H, and J have not been confirmed to be temperature-sensitive,113 a new strain for temperature-sensitive group H (M1 gene segment) has been isolated.113 Sequence analysis reveals coding changes in mutants from groups A,214 C,527 E,526 G,120 and H.113 Reversion of reovirus temperature-sensitive mutants has been documented to occur through a change at the site of the original mutation (true reversion)115 or as a consequence of a mutation in a second gene that suppresses the defect produced by the original mutation (extragenic suppression).389,391
Deletion Mutants
Some reovirus isolates lose portions of their genomes during serial high-MOI passage.7,70,358,360,434 Deletions in the L1 and L3 gene segments occur most commonly, but L2 and M1 deletions also occur. Analysis of a series of internally deleted M1 gene segments indicates that the minimum sizes for retained 5′- and 3′-terminal regions of message-sense RNA are 132–135 and 182–185 bases, respectively,553 suggesting that these regions contain signals required for RNA synthesis or gene segment packaging. Differences in the capacity of strains T1L and T3D to accumulate deletions segregates with the L2 gene segment, while the M3 gene segment is a determinant of which viral gene segments incur deletions.70 Functions of the L2 and M3 gene products λ2 and μNS, respectively, in synthesis or packaging of reovirus RNA may influence the generation or amplification of deletion mutations.
Cell-Adapted Mutants
Although usually cytolytic, reoviruses are capable of establishing persistent infections in many types of cells in culture.136 These cultures are maintained by horizontal transmission of virus from cell to cell and can be cured of persistent infection by passage in the presence of reovirus-specific antibodies. During maintenance of L-cell cultures persistently infected with
reovirus, mutations are selected in both cells and viruses.4,139 Mutant cells selected during persistent infection have alterations in the expression of cathepsin L and do not support disassembly of wild-type virus.22 These cells in turn select mutant viruses (termed PI viruses) that have gained the capacity to bypass cellular blocks to infection by virtue of mutations in outer-capsid proteins σ1532 and σ3,524 which promote more efficient disassembly. Studies of the virus-cell co-evolution that occurs during persistent reovirus infections have thus enhanced an understanding of reovirus entry (see Replication—Disassembly).
reovirus, mutations are selected in both cells and viruses.4,139 Mutant cells selected during persistent infection have alterations in the expression of cathepsin L and do not support disassembly of wild-type virus.22 These cells in turn select mutant viruses (termed PI viruses) that have gained the capacity to bypass cellular blocks to infection by virtue of mutations in outer-capsid proteins σ1532 and σ3,524 which promote more efficient disassembly. Studies of the virus-cell co-evolution that occurs during persistent reovirus infections have thus enhanced an understanding of reovirus entry (see Replication—Disassembly).
Nonsialic acid–binding reovirus strains have been adapted to growth in murine erythroleukemia (MEL) cells by serial passage.91 These variants have single amino-acid substitutions in the sialic acid–binding domain of σ1 protein and have gained the capacity to bind sialylated glycans.91 Serial passage also has been used to select reovirus mutants for resistance to ammonium chloride,99 which inhibits endosomal acidification, and E64,157 which inhibits cysteine proteases. These mutants have alterations in σ3 protein that enhance kinetics of viral disassembly. Other reovirus mutants have been selected during chronic infections of immunocompromised mice; these viruses are adapted to replicate more efficiently in the organs from which they were isolated.206
Mutants Resistant to Denaturants
Treatment of reovirus with harsh denaturants (e.g., heat, extremes of pH, ethanol, phenol, or SDS) results in substantial losses in infectivity.148 Strain-specific differences in susceptibility to inactivation by heat and ethanol have been mapped using reassortant viruses to the σ3-encoding S4 gene and μ1-encoding M2 gene,149 respectively. Concordantly, reovirus mutants selected for resistance to ethanol have mutations in the M2 gene.226,522 These mutants display increased thermostability and a decreased capacity to penetrate host-cell membranes.226
Directed Mutagenesis of the Genome: “Reverse Genetics”
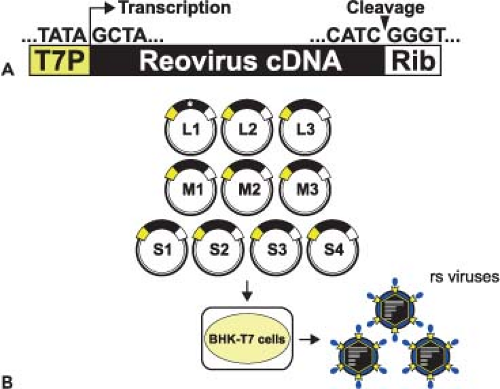
The ability to engineer viruses that contain specific sequence modifications was first accomplished for mammalian reoviruses using a strategy involving message-sense RNA transcribed in vitro and a helper virus.410 This system has been used to recover infectious particles containing gene segments derived from cloned DNA copies of single reovirus gene segments.405,407 Development of a plasmid-based reverse genetics system for members of the Birnaviridae family,539 which contain two genomic dsRNA segments, suggested that delivery of viral positive-strand RNA alone could launch successful production of viral progeny for a dsRNA virus. This approach proved possible with the development of an entirely plasmid-based reverse genetics system for reovirus in which viable viruses are generated from a set of 10 cloned cDNAs representing the complete viral genome261 (Fig. 44.5). Gene segment cDNAs corresponding to strains T1L264 and T3D261 were introduced into plasmids at sites flanked by the promoter sequence for T7 RNA polymerase and the hepatitis delta virus (HDV) ribozyme (Fig. 44.5A). Neither helper virus nor co-expression of viral replication proteins is required for recovery of infectious virus.
Reovirus can be recovered from cells in which T7 RNA polymerase is delivered transiently by infection with a recombinant vaccinia virus236 or from cells that constitutively express the enzyme73 (Fig. 44.5B). Since T7 RNA polymerase initiates transcription at a defined guanosine residue329 and all reovirus RNAs contain the nucleotide sequence, GCUA, at the 5′ terminus of the message-sense strand, T7 RNA polymerase produces transcripts with native reovirus 5′ termini. Self-cleavage of the HDV ribozyme generates RNAs with native 3′ termini.407 In cells transfected with plasmids encoding all 10 reovirus gene segments, viral gene products join with reovirus RNAs to form viral replication complexes, which mediate negative-strand synthesis and ultimately generate infectious particles. Inclusion of expression plasmids encoding reovirus proteins does not enhance recovery of recombinant reoviruses.57 However, the efficiency of virus recovery is increased by using baby hamster kidney cells constitutively expressing T7 RNA polymerase73 and reducing the plasmid number from 10 to four by combining reovirus gene segment cDNAs into multicistronic vectors.264
Plasmid-derived wild-type virus recapitulates properties of native virus in all cell-culture and in vivo models of reovirus infection studied to date.261,264 This reverse genetics approach has been used to introduce changes into the virion capsid and replication proteins to define the roles of individual amino acids, functional domains, and structural motifs in receptor utilization,259,395 virion disassembly,147,261 membrane penetration,121,123,430 replication biochemistry,369 inclusion
formation,239,263 interferon (IFN) expression,235,556 apoptosis induction,121,123 viral growth and spread in vivo,55,56,261 neurovirulence,121,123 and myocardial injury.235 The same strategy also can be exploited to engineer recombinant reoviruses for vaccine and oncolytic applications.
formation,239,263 interferon (IFN) expression,235,556 apoptosis induction,121,123 viral growth and spread in vivo,55,56,261 neurovirulence,121,123 and myocardial injury.235 The same strategy also can be exploited to engineer recombinant reoviruses for vaccine and oncolytic applications.
Genetic Complementation
A variety of complementation strategies have been developed to enable studies of viral mutations that would otherwise compromise viral viability. Stable expression in cultured cells of wild-type forms of temperature-sensitive mutant proteins allows replication of the corresponding temperature-sensitive mutant virus at nonpermissive temperature. This approach has been used to complement the replication of tsE (which segregates with the σNS-encoding S3 gene segment) and tsH (which segregates with the μ2-encoding M1 gene segment) by stable expression of the σNS33 and μ2554 proteins, respectively. These complementing cell lines and temperature-sensitive mutant viruses could be used to define sequences in σNS and μ2 required for viral replication.
Recombinant reovirus outer-capsid proteins μ1, σ1, and σ3 can bind and “recoat” purified subvirion particles (ISVPs or cores) in vitro to generate infectious virion-like particles.83,84,244 These recoated particles have been used to identify sequences in μ1, σ1, and σ3 proteins that function during cell entry and disassembly.81,84,243,531
Reovirus replication is blocked in cell lines stably expressing small interfering RNAs (siRNAs) specific for reovirus transcripts.262 RNAi-mediated inhibition of viral gene expression can be complemented by vectored expression of viral genes in which the siRNA target sequence has been altered to prevent siRNA-mediated degradation. This approach has been used to identify sequences in μ2, μNS, and σNS proteins that function in viral inclusion formation and RNA synthesis.16,77,262,263 Limitations of this strategy include the efficiency of viral RNA knockdown and transfection efficiency of the complementation plasmids.
Replication
Attachment
Reovirus Attachment Protein σ1
The S1-encoded σ1 protein (49–51 kDa, 455–470 aa) mediates viral binding to cellular receptors27,89,279,514 and influences target-cell selection in the infected host.28,515,517 It is a homotrimer present in 36 copies per virion.114,478 Structural information is available for the σ1 protein of prototype type 3 strain, T3D. The 455 amino acids of T3D σ1 fold into a filamentous trimer approximately 480 Å long and 90 Å wide at its broadest point, with a globular C-terminal head, a central body, and a slender N-terminal tail92,174,395 (Fig. 44.6A). Residues 310 to 455 comprise the head, which is constructed from two Greek-key motifs that assemble into an eight-stranded β-barrel.92,431 With the exception of the loop connecting β-strands D and E (D-E loop), which contains a 310 helix, loops connecting individual strands of the β-barrel are very short. Residues 170 to 309 form the body domain, which is constructed primarily from repeating units of two antiparallel β-strands connected by short loops. Three such units assemble into a triple β-spiral, which is a motif observed to date only in viral fibers, including the adenovirus fiber,505 bacteriophage PRD1 P5,321 avian reovirus attachment protein σC,203 and mammalian reovirus T3D σ1.92,395 The body domain features four β-spiral repeats at the N-terminus (β1–β4, residues 170–235), a short α-helical coiled-coil (residues 236–251), and three additional β-spiral repeats (β5–β7, residues 252–309) at the C-terminus (Fig. 44.6A). The short α-helical coiled-coil corresponds to a narrowing in the body domain in a composite image of negative-stained electron micrographs of purified σ1.174 Residues 1 to 160 form the tail domain, the structure of which is unknown. However, a repeating heptad sequence motif suggests that this region forms an amphipathic α-helix, which likely assembles into an α-helical coiled-coil in the trimer.92,350 The extreme N-terminus of σ1 does not contain any obvious sequence motifs. It is hydrophobic and anchors the protein into the pentameric turret formed by λ2 in the reovirus virion.151,176,283 This symmetry mismatch (a trimer of σ1 engaged to a pentamer of λ2) suggests an interaction of limited strength, which may aid in σ1 release during viral disassembly.472
The σ1 molecule possesses discrete regions of flexibility92,174,395 (Fig. 44.6A, arrows). One site of substantial flexibility in T3D σ1 is contributed by a four-residue insertion between the two most C-terminal β-spiral repeats (β6 and β7).78,92 Sequence alignments suggest that σ1 of reovirus prototype strains T1L and T2J each contain a six-residue insertion at the same position.92 This insertion appears to correspond to a region of flexibility observed just below the σ1 head in electron micrographic images.174 A second region of flexibility is observed at the midpoint of σ1 and corresponds to the transition between the predicted α-helical coiled-coil region of the tail and the β-spiral–containing body. A final region of flexibility close to the N-terminus likely represents the virion insertion domain.92,174,350
Glycan Binding by σ1
Reoviruses display the capacity to agglutinate erythrocytes of several mammalian species.284 For type 3 reoviruses, hemagglutination is mediated by interactions of the σ1 protein with terminal α-linked sialic acid residues on several glycosylated erythrocyte proteins such as glycophorin A.183,376 Strain T3D exhibits a reduced capacity to agglutinate erythrocytes following treatment with neuraminidase, which removes terminal sialic acid moieties.183 Preincubation of L cells with neuraminidase or virus with sialosides also significantly diminishes T3D binding.184,375 Sialic acid residues linked in α2,3 or α2,6 configurations effectively block type 3 reovirus binding to L cells.375 Reovirus T3D binds to sialoglycophorin, but not to asialoglycophorin, with an avidity of approximately 5 × 10−9 M,26 which is a property mediated by the σ1 protein.89
Binding to sialic acid is essential for reovirus infection of some types of cultured cells, such as MEL cells.91,422 Sequence polymorphisms within the σ1 body domain at residues 198, 202, and 204 determine the capacity of field-isolate reovirus strains to bind sialic acid and infect MEL cells.138,421 Furthermore, nonsialic acid–binding type 3 variants can be adapted to growth in MEL cells during serial passage. These variants have gained the capacity to bind sialic acid and contain sequence changes within the polymorphic region of the σ1 body. In addition, sialic acid binding also serves an important role in reovirus tropism and pathogenesis in vivo. A sialic acid–binding strain of reovirus, but not a nonsialic acid–binding strain, causes bile
duct injury in newborn mice and exhibits 1,000-fold greater binding capacity for human cholangiocarcinoma cells28 (see Pathogenesis and Immunity—Hepatobiliary System).
duct injury in newborn mice and exhibits 1,000-fold greater binding capacity for human cholangiocarcinoma cells28 (see Pathogenesis and Immunity—Hepatobiliary System).
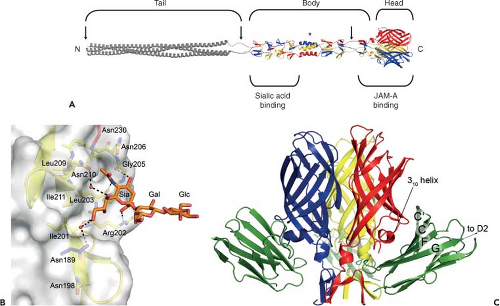 Figure 44.6. Structure of reovirus σ1 in complex with its receptors sialic acid and junctional adhesion molecule-A. A: Full-length model of σ1. A full-length depiction of σ1 was generated by modeling a predicted trimeric α-helical coiled coil and linking it to the N-terminus of the largest crystallized fragment of σ1 (residues 170–455 of strain type 3 Dearing [T3D] σ1).395 The three monomers of the crystallized fragment are shown in blue, red, and yellow; the model is shown in grey. Tail, body, and head regions are indicated. Regions of flexibility are marked by arrows. A sequence polymorphism that confers susceptibility to cleavage by intestinal proteases is indicated with an asterisk. The approximate locations of binding sites for sialic acid and junctional adhesion molecule-A (JAM-A) are shown. Amino (N)- and carboxy (C)-termini are indicated. B: The T3D σ1 protein in complex with α-2,3-sialyllactose.395 Residues in the binding region are drawn in stick representation, while the remainder of the protein is shown as a ribbon drawing. The side chain of Asn189 (blue) is contributed by a neighboring σ1 monomer. The α-2,3-sialyllactose is shown in stick representation, with carbons in orange, oxygens in red, and nitrogens in blue. The sugar moieties are labeled Sia (sialic acid), Gal (galactose), and Glc (glucose). Bridging waters are shown as orange spheres. Hydrogen bonds and salt bridges are represented with broken lines. C: The T3D σ1 protein in complex with JAM-A. Ribbon drawing of a complex formed between the trimeric σ1 head domain and monomeric JAM-A D1, viewed perpendicular to the threefold symmetry axis.259 Beta-strands C, C′, F, and G of JAM-A D1 are indicated on the right. Many of the residues in σ1 that engage JAM-A reside in the loop that connects β-strands D and E and its 310 helix. |
The structure of a T3D σ1 construct encompassing the body and head domains in complex with α-2,3-sialyllactose has been determined.395 The oligosaccharide binds in a shallow groove next to the loop connecting the second and third β-spiral repeats (β2 and β3). The σ1 protein contains three identical binding sites, one on each chain, and all three are occupied by α-2,3-sialyllactose molecules in the crystal structure, with the sialic acid making identical and extensive contacts in each chain (Fig. 44.6B). Remarkably, σ1 also can form complexes with α-2,6-sialyllactose and α-2,8-di-siallylactose.395 In each case, the terminal sialic acid forms almost all of the contacts with σ1 in an identical manner, while the remaining components of the oligosaccharides make little or no contacts. Studies using point-mutant viruses indicate that Asn198, Arg202, Leu203, Pro204, and Gly205 are required for hemagglutination and infection of MEL cells,395 suggesting that these residues serve a functional role in T3D σ1–sialic acid interactions.
Type 1 reoviruses also appear to bind sialic acid in some contexts. T1L, but not T3D, binds the apical surface of microfold (M) cells, but not enterocytes, in tissue sections of rabbit Peyer patches.217 Binding is inhibited by preincubation of the tissue sections with neuraminidase or with lectins that specifically recognize α-2-3–linked sialic acid. The capacity of T1L to bind the apical surface of M cells segregates with the S1 gene segment using reassortant genetics and with reovirus particles recoated with recombinant σ1 protein. The interaction between T1L σ1 and sialic acid is especially intriguing as type 1 reoviruses are
incapable of infecting MEL cells, a property dependent on sialic acid binding that segregates with the S1 gene segment,422 and are insensitive to the growth-inhibitory effects of neuraminidase treatment of L cells.349 These findings suggest that the functional glycans engaged by types 1 and 3 reoviruses differ, even though such glycans appear to terminate in α-2,3–linked sialic acid. Concordantly, studies using expressed protein suggest that T1L engages carbohydrates using a domain that differs from that used by T3D to bind carbohydrates.89
incapable of infecting MEL cells, a property dependent on sialic acid binding that segregates with the S1 gene segment,422 and are insensitive to the growth-inhibitory effects of neuraminidase treatment of L cells.349 These findings suggest that the functional glycans engaged by types 1 and 3 reoviruses differ, even though such glycans appear to terminate in α-2,3–linked sialic acid. Concordantly, studies using expressed protein suggest that T1L engages carbohydrates using a domain that differs from that used by T3D to bind carbohydrates.89
Type 2 reoviruses also are capable of producing hemagglutination,284 but little is known about the basis for this property.
JAM-A Binding by σ1
Reovirus strains of all three serotypes use junctional adhesion molecule-A (JAM-A, also known as F11R/JAM/JAM1), a member of the immunoglobulin superfamily,27,309,529 as a receptor.27,76,382 JAM-A was identified as a reovirus receptor using a genetic screen and subsequently shown to bind directly to the σ1 head domain with nanomolar affinity.27,431 Human and murine homologs of JAM-A, but not JAM family members JAM-B or JAM-C, serve as receptors for all reovirus serotypes and strains tested to date.27,76,382 The role of JAM-A as a reovirus receptor in vivo has been examined using JAM-A–null mice13 (see Pathogenesis and Immunity – Spread of Virus). Following peroral inoculation, JAM-A is dispensable for reovirus growth in the intestine. However, it is required for infection of vascular endothelial cells and promotes efficient hematogenous dissemination of reovirus to sites of secondary infection. Thus, JAM-A serves as a high-affinity reovirus receptor in cultured cells and in vivo. Receptors for reovirus at other sites within the host, including the central nervous system (CNS), have not been conclusively identified.
Structural and biochemical studies highlight the regions and specific interactions that mediate reovirus engagement of JAM-A76,92,173,204,259,382 (Fig. 44.6C). The largest area of conserved residues in σ1 forms the D-E and F-G loops in the head domain.76,92 The crystal structure of the σ1 head domain in complex with the JAM-A D1 domain reveals that residues in this region, centered at the D-E loop and its 310 helix, form the largest area of JAM-A contact.259 Interactions in this area are polar and involve residues Thr380, Gly381, and Asp382. A second area of JAM-A contact includes residues within the σ1 body, just N-terminal to the head domain. Interactions in this region are largely hydrophobic and involve Arg316 in β-strand A of the head, β-spiral residues Arg297 and Tyr298, the α-helical turn that connects the β-spiral with the β-barrel, and Pro377.
The D1 domain of JAM-A is required for high-affinity binding to σ1.173,204,382 Mutation of individual JAM-A D1 domain residues Arg59, Glu61, Lys63, Leu72, Tyr75, and Asn76 that lie in or adjacent to the dimer interface diminishes or abolishes σ1 binding and reovirus infectivity.204 Concordantly, the structure of the σ1–JAM-A complex shows that each σ1 trimer binds three independent JAM-A monomers. Contacts primarily involve the JAM-A dimer interface and a conserved region at the base of the σ1 head92,259 (Fig. 44.6C). In addition, the structure of the σ1-JAM-A complex also identifies residues bound by σ1 that are found just outside the dimer interface of JAM-A.259 These residues may serve as initial contact points for σ1 and facilitate disruption of the JAM-A homodimer to allow interaction of σ1 with the JAM-A dimer interface. It is also possible that a cavity in the JAM-A dimer interface renders the homodimer intrinsically unstable, thereby promoting its disruption by σ1. Regardless of the mechanism, the σ1-JAM-A interaction is thermodynamically favored, as the KD is ∼ 1,000-fold lower than the KD of the JAM-A homodimer interaction.204,259,506
The presence of discrete receptor-binding domains in σ1 suggests that reoviruses employ a multiple-step binding process similar to that used by some herpesviruses109,465 and retroviruses.46,503 Binding studies using isogenic point-mutant viruses T3SA+ and T3SA-, which vary only in the capacity to engage sialic acid,26 support this hypothesis. Kinetic analyses using inhibitors of sialic acid and JAM-A binding demonstrate that sialic acid is engaged first in the adsorption process, as the inhibitory effect of sialic acid analogs on infection by T3SA+ occurs at early but not late time points.26 However, a σ1-specific monoclonal antibody that blocks virus binding to JAM-A inhibits viral infectivity at both early and late times during adsorption.26 Thus, reovirus binding to sialic acid enhances virus attachment through rapid adhesion of virus to the cell surface where access to JAM-A is thermodynamically favored. It is not known whether binding to sialic acid induces structural changes in σ1 that affect its capacity to interact with JAM-A. However, it is clear that sialic acid binding is not a necessary prerequisite for JAM-A binding, as reoviruses incapable of binding sialic acid are capable of binding JAM-A.27 It is possible that engagement of JAM-A by the σ1 head leads to conformational changes in other regions of σ1 or in other capsid components.168
Cleavage of σ1 by Intestinal Proteases
Treatment of virions of some reovirus strains with intestinal proteases in vitro to generate ISVPs leads to cleavage of the σ1 protein.87,349 Strain-specific differences in cleavage susceptibility segregate with a single amino acid polymorphism in the short α-helical coiled-coil in the body domain of σ1 (Fig. 44.6A, asterisk). Strains like T3D with a threonine at position 249 in σ1 are susceptible to cleavage by trypsin after Arg245, whereas those with an isoleucine at position 249 are resistant to cleavage.87 It is possible that Thr249 disrupts the α-helical coiled-coil and allows access to sites that otherwise would be shielded from protease.87,349 Conversion of T3D virions to ISVPs by intestinal proteases leads to loss of the σ1 head domain and a resultant 90% reduction in infectivity.349 Since cleavage of σ1 occurs C-terminal to the sialic acid–binding pocket, residual infectivity of T3D ISVPs is dependent on this carbohydrate.349 Susceptibility of the T3D σ1 protein to cleavage likely accounts for the attenuated virulence of this strain after oral inoculation.53,254,419
Determinant of Neutralizing Antibody Responses
In addition to its function in receptor engagement, the σ1 protein is the target of serotype-specific neutralizing antibodies.75,469,516 For example, serotype-specific monoclonal antibodies 5C6508 and 9BG575 neutralize infectivity of type 1 and type 3 reovirus strains, respectively, and bind the σ1 head domain in a serotype-specific fashion.89 Variants of T1L selected for resistance to neutralizing monoclonal antibodies have mutations at
Ala415, Gln417, Asn445, and Gly447,216 which are located in the σ1 head. Similarly, variants of T3D selected for resistance to neutralizing monoclonal antibodies have mutations at Asp340 and Glu419,30 which are also located in the σ1 head. Interestingly, for both T1L and T3D, the sites in σ1 altered in the neutralization-resistant variants are in close proximity on adjacent monomers,92,216 suggesting an epitope that spans two monomers in the σ1 trimer. Neutralization-resistant variants of T3D display diminished neurovirulence and altered CNS tropism in mice,467,468 perhaps as a consequence of altered σ1 receptor recognition in the murine CNS.
Ala415, Gln417, Asn445, and Gly447,216 which are located in the σ1 head. Similarly, variants of T3D selected for resistance to neutralizing monoclonal antibodies have mutations at Asp340 and Glu419,30 which are also located in the σ1 head. Interestingly, for both T1L and T3D, the sites in σ1 altered in the neutralization-resistant variants are in close proximity on adjacent monomers,92,216 suggesting an epitope that spans two monomers in the σ1 trimer. Neutralization-resistant variants of T3D display diminished neurovirulence and altered CNS tropism in mice,467,468 perhaps as a consequence of altered σ1 receptor recognition in the murine CNS.
Entry
Internalization by Clathrin-Dependent Endocytosis
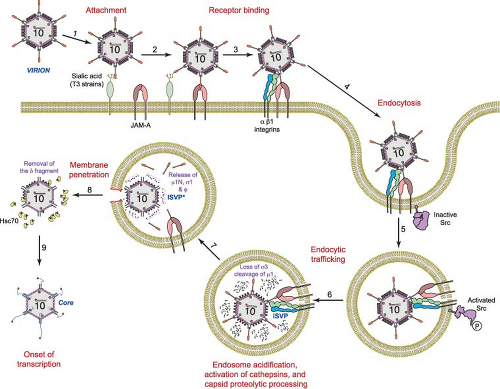
Following attachment to cell-surface glycans and JAM-A, reovirus is internalized by receptor-mediated endocytosis (Fig. 44.7). Thin-section electron micrographs show virions in structures that appear to be clathrin-coated pits on the cell surface and in clathrin-coated vesicles in the cytoplasm,59,60,298,421,480 suggesting clathrin-dependent uptake. This finding was confirmed using video fluorescence microscopy in which reovirus virions and clathrin are observed to colocalize during internalization.158 Treatment of cells with chlorpromazine, which impairs clathrin-mediated endocytosis, inhibits reovirus internalization and infection,298 suggesting a functional role for clathrin in reovirus entry. However, there is mounting evidence that both clathrin- and caveolin-dependent mechanisms can be employed by some viruses to enter host cells.275,385 Although there are no published reports of clathrin-independent uptake strategies for reovirus, a role for caveolae or other entry mechanisms in reovirus cell entry has not been conclusively excluded.
Regardless of the internalization mechanism, reovirus particles are sorted into vacuoles resembling endosomes and lysosomes during the entry process.59,60,158,298,421,480
Regardless of the internalization mechanism, reovirus particles are sorted into vacuoles resembling endosomes and lysosomes during the entry process.59,60,158,298,421,480
β1 Integrins Promote Entry and Sorting in the Endocytic Compartment
Expression of a JAM-A truncation mutant lacking a cytoplasmic tail allows reovirus to infect nonpermissive cells,297 suggesting that molecules other than JAM-A mobilize the internalization apparatus that promotes reovirus cell entry. The L2-encoded λ2 protein (144 kDa, 1288–1289 aa) contains conserved integrin-binding motifs, RGD and KGE,64,438 suggesting that reovirus employs integrin-dependent internalization mechanisms to enter cells. The RGD and KGE sequences are displayed on surface-exposed loops of λ2,394 where they could interact with integrins to promote internalization. However, direct interactions between λ2 and integrins have not been reported. Interestingly, the L2 gene segment is genetically linked to viral shedding in infected mice and spread to littermates,254 suggesting a role for λ2 in reovirus-induced disease.
Treatment of cells with antibodies specific for β1 integrin reduces reovirus infection, while antibodies specific for other integrin subunits expressed on permissive cells, including those specific for α integrin subunits, have no effect.297 However, antibodies specific for β1 integrin do not alter infection by in vitro–generated ISVPs,297 which directly penetrate the plasma membrane and do not require endocytosis.226,294 Concordantly, β1 integrin–specific antibodies do not alter the binding of reovirus to JAM-A.297 These findings suggest that β1 integrin blockade inhibits endocytic uptake of virions following JAM-A engagement. In comparison to β1 integrin-expressing cells, β1-null cells are substantially less susceptible to infection by reovirus virions, while infection by ISVPs is equivalent in both cell types.297 Diminished reovirus replication in β1-null cells correlates with diminished viral uptake, indicating that β1 integrin is required for efficient reovirus cell entry.
NPXY motifs in the β1 integrin cytoplasmic tail play a key role in sorting reovirus within the endocytic compartment. NPXY motifs are found in the cytoplasmic domains of many receptors94,125,367 and recruit adaptor protein 2 or disabled protein 2341,367 to initiate clathrin assembly at the plasma membrane. Substitution of a tyrosine with a phenylalanine residue in either or both β1 integrin NPXY motifs (NPXF) results in inefficient internalization of reovirus virions and diminished infectivity.298 Infection of cells expressing NPXF β1 integrin results in distribution of virions to lysosomes where they are degraded, suggesting that the β1 integrin NPXY motifs target reovirus to the precise endocytic organelle that permits functional disassembly.
Src Kinase Regulates Endosomal Sorting of Reovirus
The Src family of kinases contains eight members, Blk, Fgr, Fyn, Hck, Lck, Lyn, Src, and Yes, three of which—Fyn, Src, and Yes—are expressed in most cell types.491 Src-family kinases regulate numerous cellular processes including proliferation, differentiation, migration, adhesion, and cytoskeletal rearrangements491 and transduce signals from a variety of receptors.145,182,308,427 In a small-molecule inhibitor screen, genistein, a broad-spectrum tyrosine-kinase inhibitor, and PP2, a specific Src-family-kinase inhibitor, were found to diminish reovirus infectivity by inhibiting an early step in the viral life cycle.299 Although neither inhibitor impedes internalization of reovirus virions, both inhibitors target virions to lysosomes. At early times following infection, Src co-localizes with reovirus virions and is phosphorylated at the activation residue, Tyr416, suggesting that reovirus induces activation of Src during entry and traffics with Src during sorting in the endocytic compartment. Reduction of Src expression by RNAi decreases reovirus infectivity.299 These findings suggest that Src is part of a signaling network that targets reovirus to endocytic organelles for viral disassembly and thus promotes functional entry into host cells. The precise mechanism by which Src mediates these effects is unknown.
Disassembly
Cleavage of σ3: The First Step in the Reovirus Disassembly Cascade
In cellular endosomes, reovirus virions undergo stepwise disassembly to form discrete intermediates, the first of which is the ISVP21,60,85,456,480 (Fig. 44.7). ISVPs are characterized by the loss of σ3, a conformational change in σ1, and cleavage of μ1 to form δ and φ. The rate-limiting step in reovirus disassembly is the proteolytic removal of the S4-encoded σ3 protein (41 kDa, 365 aa).21,480 In some cell types, proteolysis of σ3 is dependent on acidic pH139,480 and endocytic cysteine proteases.21 Murine L929 fibroblast cells treated with inhibitors of endosomal acidification, such as ammonium chloride480 or bafilomycin A1,310 or inhibitors of cysteine proteases, such as E64,21 do not support reovirus replication when infection is initiated by virions but do so when infection is initiated by ISVPs. Thus, the block to reovirus replication imposed by these inhibitors in murine L929 cells occurs following internalization but prior to disassembly, which is coincident with the proteolysis of σ3.
Cathepsins B and L, which reside in late endosomes and lysosomes,86 catalyze reovirus disassembly in fibroblasts.156 Both enzymes are optimally active at acidic pH and serve functions in extracellular matrix formation, antigen presentation, and apoptosis.86 These enzymes also mediate cell entry of several other viruses, including Ebola virus82 and SARS coronavirus.229 Cathepsin S, an acid-independent cysteine protease required for processing internalized antigens,401 and the acid-independent serine protease neutrophil elastase can mediate uncoating of some reovirus strains in monocyte and macrophage cell lines.191,193 It is possible that the broad tissue tropism displayed by reovirus is determined in part by the multiple host proteases capable of mediating its disassembly.247
Proteolytic enzymes also are required for reovirus infection following peroral inoculation of mice29,54 (see Pathogenesis and Immunity—Entry into the Host). Reovirus virions are converted to ISVPs in the intestinal lumen by the resident serine proteases chymotrypsin and trypsin. ISVPs produced in this fashion enter intestinal M cells to allow systemic dissemination of reovirus in the host.12 ISVPs generated by chymotrypsin or trypsin in vitro or in the gut lumen29,54 display a protein profile (loss of σ3 and cleavage of μ1) indistinguishable from that of ISVPs generated in the endocytic compartment of cells.22,156
Sequences in σ3 that influence its susceptibility to proteolysis have been identified through studies of viruses selected
during persistent infection (PI viruses) or mutant viruses selected for resistance to either cysteine protease inhibitor E64 (D-EA viruses)157 or ammonium chloride (ACA-D viruses).99 These viruses exhibit accelerated kinetics of disassembly and harbor a tyrosine-to-histidine mutation at amino acid 354 (Y354H) near the C-terminus of the protein99,157,524 (Fig. 44.8). Cryo-EM image analysis of a PI virus with an isolated Y354H mutation reveals a structural alteration in σ3 at a hinge region located between its two major domains.531 These findings suggest that the C-terminus of σ3 influences susceptibility of the protein to cleavage. Interestingly, the σ3 amino acid sequence of strain T3A differs from that of T3D at eight positions, including Y354H. However, a glycine-to-glutamate substitution at position 198 in T3A σ3 suppresses the disassembly-enhancing effects of His354,147 suggesting the existence of an allosteric network regulating σ3 cleavage.
during persistent infection (PI viruses) or mutant viruses selected for resistance to either cysteine protease inhibitor E64 (D-EA viruses)157 or ammonium chloride (ACA-D viruses).99 These viruses exhibit accelerated kinetics of disassembly and harbor a tyrosine-to-histidine mutation at amino acid 354 (Y354H) near the C-terminus of the protein99,157,524 (Fig. 44.8). Cryo-EM image analysis of a PI virus with an isolated Y354H mutation reveals a structural alteration in σ3 at a hinge region located between its two major domains.531 These findings suggest that the C-terminus of σ3 influences susceptibility of the protein to cleavage. Interestingly, the σ3 amino acid sequence of strain T3A differs from that of T3D at eight positions, including Y354H. However, a glycine-to-glutamate substitution at position 198 in T3A σ3 suppresses the disassembly-enhancing effects of His354,147 suggesting the existence of an allosteric network regulating σ3 cleavage.
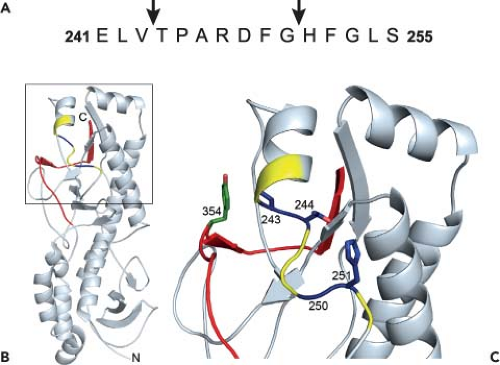 Figure 44.8. Structure of reovirus σ3 with cleavage sites. A: The amino acid sequence of σ3 from amino acids 241 to 255 is shown. Arrows highlight cathepsin L cleavage sites identified by N-terminal sequencing of σ3 cleavage products following treatment of reovirus strain type 1 Lang (T1L) with cathepsin L in vitro. B: Cathepsin L cleavage sites are highlighted in the crystal structure of σ3. A ribbon diagram of the crystal structure of type 3 Dearing σ3368 is displayed on the left. The cathepsin L cleavage sites in T1L are depicted in blue between amino acids 243 and 244 and between 250 and 251. Surrounding residues, from amino acids 241 to 253, are shown in yellow. The C-terminal residues of σ3, from amino acids 340 to 365, are in red. Amino acid 354, which is altered in a variety of mutant viruses selected for accelerated disassembly kinetics, is in green. The virion–distal end of σ3 is at the top of the figure, and the virion-proximal end and N-terminus are at the bottom. C: An enlarged view of the boxed region of σ3 indicated in B is shown using the same color scheme. Side chains of amino acids 243, 244, 250, 251, and 354 are depicted in stick representation. |
The σ3 C-terminus also dictates strain-specific differences in the susceptibility of σ3 to proteolytic attack.243,244 The σ3 protein of strain T1L is cleaved more rapidly than that of T3D. Analysis of ISVPs recoated with chimeric σ3 proteins generated from T1L and T3D revealed that the C-terminus is primarily responsible for the rate of σ3 proteolysis. Moreover, sequence polymorphisms at residues 344, 347, and 353 in σ3 contribute to this effect.243
Treatment of reovirus virions in vitro with either cathepsin B or cathepsin L leads to an initial cleavage of σ3 at a terminus.156 Since sequence polymorphisms in the σ3 C-terminus determine susceptibility to proteolysis, the initial cleavage of σ3 probably occurs in this region. During proteolysis by cathepsin L, subsequent cleavages occur between residues 243 to 244 and 250 to 251156 (Fig. 44.8A). These cleavage sites are physically located near the C-terminus in the σ3 crystal structure368 (Fig. 44.8B and C). Because of this proximity, the small end fragment released following initial cathepsin L cleavage likely exposes the cleavage sites between residues 243 to 244 and 250 to 251, rendering them sensitive to proteolysis. The C-terminus therefore appears to control access to internal, proteolytically sensitive sites in σ3. Because reovirus disassembly in some cell types is acid-dependent,139,480 the C-terminus might be primed for movement at acidic pH. Mutations near the C-terminus, like Y354H, may alter the conformation of the protein to allow improved access to these cleavage sites and thus accelerate outer capsid disassembly.531
Conformational Changes in σ1
The disassembly of reovirus virions to ISVPs is accompanied by a dramatic conformational change in σ1. Electron micrographs of negatively stained reovirus virions and ISVPs reveal filamentous projections extending up to 400 Å from the surface of ISVPs but not virions.176 These images suggest that σ1 adopts a compact form in the virion and a more extended one in the ISVP. Cryo-EM image reconstructions of virions and cores lack a discernable density corresponding to σ1 at the icosahedral vertices151,323 (Fig. 44.2). However, cryo-EM image analysis of
ISVPs demonstrates discontinuous density for σ1 extending ∼ 100 Å from each vertex (Fig. 44.2). Presumably, the full length of σ1 is not visible in reovirus particles because the molecule is flexible and icosahedral averaging was employed for the cryo-EM image reconstructions. As further evidence of the conformational alterations in σ1 during viral disassembly, proteolytic cleavage of T3D σ1 during virion-to-ISVP conversion increases viral hemagglutination capacity,349 suggesting enhanced access to the sialic acid–binding region as a consequence of extension of the σ1 tail and body domains. Interdomain regions of flexibility in σ1174 (Fig. 44.6) may be required to allow such conformational rearrangements.
ISVPs demonstrates discontinuous density for σ1 extending ∼ 100 Å from each vertex (Fig. 44.2). Presumably, the full length of σ1 is not visible in reovirus particles because the molecule is flexible and icosahedral averaging was employed for the cryo-EM image reconstructions. As further evidence of the conformational alterations in σ1 during viral disassembly, proteolytic cleavage of T3D σ1 during virion-to-ISVP conversion increases viral hemagglutination capacity,349 suggesting enhanced access to the sialic acid–binding region as a consequence of extension of the σ1 tail and body domains. Interdomain regions of flexibility in σ1174 (Fig. 44.6) may be required to allow such conformational rearrangements.
Electron micrographs of purified σ1 show molecules with either single- or multilobed head domains,174 suggesting another type of structural alteration in σ1 in which the head can exist in both “open” and “closed” conformations. Although neither the mechanism nor the functional significance of this σ1 conformational change is understood, structural studies of σ1 provide clues about how this change might occur. A cluster of six conserved aspartic acid residues on a rigid β-hairpin at the base of the σ1 head, sandwiched between hydrophobic residues that block access to solvent, forms the main contact area between monomers in the trimer.92,431 Of the two aspartic acid residues contributed by each monomer, one (Asp346) is neutralized by a salt-bridge interaction with a nearby residue, while the other (Asp345) is not.92,431 The three Asp345 side chains closely appose each other at the center of the trimer in an otherwise hydrophobic environment. Since accumulation of negative charge in this region is predicted to destabilize the trimer,78 and an aspartate-to-asparagine mutation results in σ1 trimers with a structure indistinguishable from wild-type,431 it is likely that Asp345 is protonated in the σ1 crystal structure,92 thus representing the “closed” conformation of σ1. This conformation might form during crystallization at near-neutral pH and physiologically in conditions of low pH, similar to those encountered in the endocytic compartment during reovirus entry.431 Thus, the aspartic acid sandwich motif may contribute to σ1 conformational rearrangements by acting as a molecular switch that mediates the oligomeric state of the σ1 head, depending on environmental pH.431
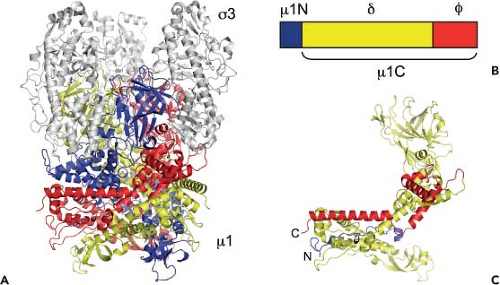 Figure 44.9. Structure of reovirus μ1 with cleavage sites. A: Ribbon diagram of the crystal structure of the μ1-σ3 heterohexamer from strain type 1 Lang (T1L).289 The μ1 monomers are in blue, red, and yellow; the σ3 monomers are in grey. B: Cleavage products of the μ1 protein. (The C-terminal cleavage is not depicted.) C: Ribbon diagram of the crystal structure of the T1L μ1 monomer showing regions corresponding to the cleavage products. The domain color code is as depicted in B. |
Membrane Penetration
Critical Function of the μ1 Protein
Studies to assess the capacity of reovirus entry intermediates to penetrate artificial lipid bilayers, model membranes of erythrocytes, or membranes of cells that support reovirus infection indicate that ISVPs but not virions or cores mediate membrane penetration.59,80,83,84,226,294,492 Such studies led to the idea that ISVPs or related subviral particles are the membrane-active intermediates in the reovirus entry pathway. Since ISVPs differ from cores by the presence of outer-capsid proteins σ1 and μ1,114,151 and because cores recoated with μ1 and σ3 in vitro and then treated with chymotrypsin to remove σ3 are capable of membrane penetration,83 these findings point to a role for the M2-encoded μ1 protein (76 kDa, 708 aa) in membrane penetration. This biochemical evidence also is supported by several genetic studies. Differences in membrane-penetration efficiency displayed by reovirus strains T1L and T3D segregate with the M2 gene segment.79,294,430 Additionally, viruses selected for resistance to denaturants such as ethanol contain mutations within the M2 gene segment and display alterations in membrane penetration capacity.79,123,226,522 Together, these data demonstrate a function for the μ1 protein in membrane penetration.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree