Orthomyxoviridae
Megan L. Shaw
Peter Palese
Influenza viruses were probably responsible for the disease described by Hippocrates in 412 BC,275 and thus they have been with us for a long, long time. Influenza remains a major cause of morbidity and mortality worldwide, and large segments of the human population are affected every year. In addition, many animal species can be infected by influenza viruses, and some of these viruses may give rise to pandemic strains in humans, as in the case of the 2009 H1N1 pandemic. Most threatening is the possibility of another pandemic similar to that experienced in 1918, which is estimated to have caused on the order of 50 million deaths worldwide.319
Classification
The family of Orthomyxoviridae is defined by viruses that have a negative-sense, single-stranded, and segmented RNA genome. The definition of negative-sense RNA viruses came from work by David Baltimore, who showed that the packaged genome of this class of viruses is complementary to the messenger RNA (mRNA), which is defined as positive.21 There are six different genera in the family of Orthomyxoviridae: the Influenzaviruses A, B, and C; Thogotovirus; Isavirus; and a new genus, Quaranfilvirus526 (Fig. 40.1). Members belonging to any of the three different genera of influenza viruses can undergo genetic reassortment (see below), and thus readily exchange genetic information. However, reassortment between members of different genera (types) has never been reported. This absence of genetic exchange between viruses of different genera (types) is one manifestation of speciation as a result of evolutionary divergence.
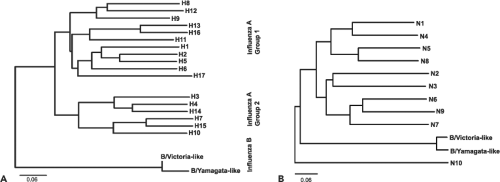
Different influenza virus strains are named according to their genus (type), the species from which the virus was isolated (omitted if human), location of the isolate, the number of the isolate, the year of isolation, and, in the case of the influenza A viruses, the hemagglutinin (H) and neuraminidase (N) subtypes. For example, the 220th isolate of an H5N1 subtype virus isolated from chickens in Hong Kong in 1997 is designated influenza A/chicken/Hong Kong/220/97(H5N1) virus. There are now 17 different hemagglutinin (H1 to H17) subtypes and 9 different neuraminidase (N1 to N9) subtypes for influenza A viruses as well as a new N10 neuraminidase,186,668,721 while the influenza B virus hemagglutinins and neuraminidases are each classified into two lineages (Fig. 40.2).
Virion Structure
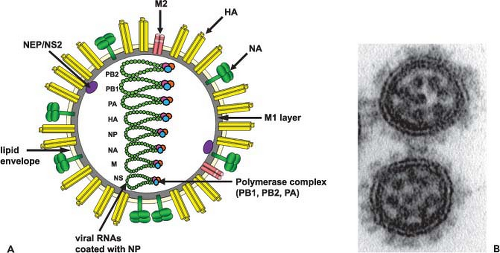
Influenza A viruses have a complex structure and possess a lipid membrane derived from the host cell (Fig. 40.3A). This envelope harbors the hemagglutinin (HA), the neuraminidase (NA), and the M2 proteins that project from the surface of the virus. The matrix protein (M1) lies just beneath the envelope, and the core of the virus particle is made up of the RNP (ribonucleoprotein) complex, consisting of the viral RNA segments, the polymerase proteins (PB1 [polymerase basic 1], PB2 [polymerase basic 2], and PA [polymerase acid]), and the nucleoprotein (NP).601 The NEP/NS2 (nuclear export protein/nonstructural protein 2) protein is also present in purified viral preparations.556 The overall composition of virus particles is about 1% RNA, 5% to 8% carbohydrate, 20% lipid, and approximately 70% protein.2,116,188 However, these results will have to be revisited using more quantitative approaches. Specifically, it will be important to get more quantitative data on the presence of individual viral components as well as cellular components that are packaged into the virus.602 Excellent analyses have already been performed using electron
microscopy (EM) of negatively stained or frozen-hydrated (cryoelectron microscopy) particles and tomographic reconstructions.80,192,256,464,489 The morphology of influenza A virus particles is characterized by distinctive spikes that are readily observable in electron micrographs of negatively stained virus particles (Fig. 40.3B). These spikes, made up of HA and NA, have lengths of ∼10 to 14 nm, with an approximate ratio of four HA to one NA. Influenza viruses are pleomorphic. The spherical particles have a diameter of about 100 nm, but filamentous particles with elongated viral structures (more than 300 nm) have frequently been observed, particularly in fresh clinical isolates102 and in preparations of viruses with specific M1 or M2 proteins (e-Fig. 40.1).55,77,159,571
microscopy (EM) of negatively stained or frozen-hydrated (cryoelectron microscopy) particles and tomographic reconstructions.80,192,256,464,489 The morphology of influenza A virus particles is characterized by distinctive spikes that are readily observable in electron micrographs of negatively stained virus particles (Fig. 40.3B). These spikes, made up of HA and NA, have lengths of ∼10 to 14 nm, with an approximate ratio of four HA to one NA. Influenza viruses are pleomorphic. The spherical particles have a diameter of about 100 nm, but filamentous particles with elongated viral structures (more than 300 nm) have frequently been observed, particularly in fresh clinical isolates102 and in preparations of viruses with specific M1 or M2 proteins (e-Fig. 40.1).55,77,159,571
Less is known about the internal structures of influenza viruses. However, the underlying M1 layer can be visualized and reveals a helical superstructure.80,575,577 The RNP complexes were first separated by Duesberg147 on sucrose gradients and were visualized by electron microscopy using positive staining with uranyl acetate.115 These RNP structures appear to consist of a strand that is folded back on itself to form a double-helical arrangement.115 Most recently, attempts have been made to visualize RNPs or individual RNA segments by electron microscopy of thin sectioned virus particles489 (for review see (488)).
Influenza B viruses are mostly indistinguishable from the A viruses by electron microscopy. They have four proteins inserted in their lipid envelopes: the HA, NA, NB, and BM2.38,62,491,757 The M1 and the RNP complexes make up the interior of the particle. It has also been shown that the influenza B virus NEP/NS2 is associated with purified virus preparations.304
Influenza C viruses have been found to possess hexagonal reticular (net-like) structures on the surface10 and to form unusually long (500 μm) cord-like structures on the surface of infected cells.461,484 Influenza C viruses also contain a core of three polymerase proteins and the NP, which are associated with seven RNA segments. The influenza C virus M1 appears to have a similar role to those of influenza A and B viruses. The major glycoprotein, HEF (hemagglutinin-esterase-fusion), combines the functions of the HA and NA (and thus influenza C viruses contain one less RNA segment than do the A and B viruses). The HEF is inserted into the lipid membrane, as is the glycosylated CM2, which is structurally analogous to the M2 of influenza A viruses and the NB of influenza B viruses.466,467,524
Genome Structure and Organization
Influenza Viruses
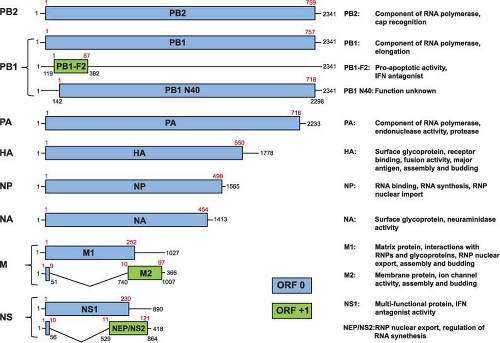
All A- and B-type influenza viruses possess eight RNA segments, whereas influenza C viruses only have seven RNAs (Fig. 40.4 and e-Figs. 40.2 to 40.4). This was first shown by using polyacrylamide gel electrophoresis of isolated RNAs from two parent influenza A virus strains and their reassortants. Identifying the derivation of an RNA segment (by gel electrophoresis) in a reassortant and simultaneous protein analysis (by serologic or gel analysis) allowed the assignment of individual RNAs to specific viral proteins508,513,514 (for review, see (509)) (Fig. 40.5 and e-Fig. 40.4). Interestingly, influenza A viruses increase the coding capacity of their genomes via both splicing and use of alternative open reading frames. The M and NS genes each give rise to a spliced mRNA encoding the M2 and the NEP/NS2 proteins, respectively.354,355 The PB1-F2 and PB1 N40 proteins are expressed from alternative open reading frames
within the PB1 gene93,726 (Fig. 40.4), although not all influenza A virus strains encode these proteins, making them true accessory proteins. Each viral segment contains noncoding regions at both the 5′ and 3′ ends. The extreme ends are conserved among all segments of influenza A viruses and this is followed by a segment-specific noncoding region.
within the PB1 gene93,726 (Fig. 40.4), although not all influenza A virus strains encode these proteins, making them true accessory proteins. Each viral segment contains noncoding regions at both the 5′ and 3′ ends. The extreme ends are conserved among all segments of influenza A viruses and this is followed by a segment-specific noncoding region.
The influenza B virus genome is similar to that of influenza A virus. Again, eight RNA segments code for one or more viral proteins (e-Figs. 40.2 and 40.4) with the three largest RNAs coding for the polymerase proteins, the fourth RNA for the HA, and the fifth and sixth RNAs for the NP and NA, respectively.550 The NA gene codes for the NB protein as well as for the NA. The NB protein is encoded by a −1 open reading frame seven nucleotides upstream (…AUGAACAAUG…) of the NA coding frame.603 The seventh RNA codes for the M1 protein (248 amino acids in length). Its termination codon (…UAAUG) overlaps with the initiation codon (UAAUG…) for the BM2 (109 amino acids in length), which allows for a “stop–start” translation mechanism.288 The eighth RNA codes for the NS1 as well as for the NEP/NS2 protein, the latter via a spliced mRNA. Cognate PB1-F2 and PB1 N40 proteins have not yet been identified in influenza B viruses. The noncoding regions of the influenza B virus genome are longer than those in influenza A virus.
The genome of influenza C viruses has only seven RNA segments, with the three largest RNAs each coding for one of the polymerase proteins (e-Figs. 40.3 and 40.4). The PB1 and PB2 proteins are homologous to the corresponding influenza A and B virus proteins. The third influenza C virus polymerase protein is named P3 because it does not display acid charge features at neutral pH, as do the corresponding PA proteins of influenza A and B viruses.740 The fourth RNA codes for the HEF protein,271 combining the hemagglutinin, receptor-destroying, and fusion activities. The NP is encoded by the fifth RNA. The sixth RNA codes for the M protein, which is expressed from a spliced mRNA, and from the unspliced mRNA a long precursor is translated (p42), which is then processed by signal peptide cleavage into CM2. This 115-amino acid (aa)-long protein consists of an amino-terminal extracellular domain (with a carbohydrate chain), a hydrophobic transmembrane domain, and an
intracellular cytoplasmic tail.523,524,741 Finally, RNA 7 codes for the NS1 protein (246 amino acids),467 and via a spliced mRNA, for the NEP/NS2 protein (182 amino acids).5,284
intracellular cytoplasmic tail.523,524,741 Finally, RNA 7 codes for the NS1 protein (246 amino acids),467 and via a spliced mRNA, for the NEP/NS2 protein (182 amino acids).5,284
Evolutionarily, influenza A, B, and C viruses have a common precursor, and it is also likely that influenza A and B viruses diverged from each other more recently than influenza C viruses. Based on comparative sequencing studies using the hemagglutinin molecules, it has been postulated that the influenza A viruses diverged about 2,000 years ago, and the influenza B and C viruses about 4,000 and 8,000 years ago, respectively.653 Clearly, these numbers are based on a series of unproveable assumptions, including a steady rate of evolution over time, and therefore must be taken cum grano salis.
Thogoto Virus
The genomes of Thogoto viruses possess only six single-stranded RNA segments of negative polarity, with a total coding capacity for seven proteins. As with the influenza viruses, three proteins make up the RNA-dependent RNA polymerase complex. The NP, the glycoprotein (G), the matrix protein (M), and one nonessential accessory protein (ML) are coded for by the remaining three RNAs. The M and ML proteins are both encoded by the shortest RNA, with the M protein being derived from a spliced mRNA.242,339 The 304-aa-long ML protein has been shown to possess interferon antagonist activity69,242,316 and is virion associated.241 It appears that Thogoto viruses do not possess a nonstructural protein.
Infectious Salmon Anemia Virus
The genome of infectious salmon anemia virus consists of eight negative-sense, single- stranded RNA segments.122,447 Segment 1 most likely encodes a protein analog of the influenza virus PB2.626 The second segment appears to code for a PB1 analog because it carries the PB1-specific polymerase motifs.626 Segments 3 and 4 code for the NP and PA proteins, respectively.15,169 Segment 5 encodes the 50-kD F (fusion) protein, and segment 6 encodes the HA (hemagglutinin-acetylesterase) protein.16,169,526 The 42-kD HA has been demonstrated to bind to 4-O-acetylated sialic acid (i.e., to use it as a receptor) and also to hydrolyze the acetyl group.267 This activity is similar to the binding and receptor-destroying enzyme (RDE) activities previously observed for the HEF protein of influenza C viruses and the HE glycoprotein of coronaviruses.267,271,685,686,687 Segment 7 encodes two proteins via an alternative splicing mechanism.40 The larger protein is expressed from the unspliced transcript and has interferon antagonist activity.211,430 Segment 8 encodes two proteins of 27.6 kD and 22 kD, the larger of which has interferon antagonist activity.122,211 The smaller protein is a structural protein, presumed to be the equivalent of the influenza virus matrix protein.169
Quaranfil Virus
The genome of Quaranfil virus, which is the type species of the new Quaranfilvirus genus, consists of six negative-sense, single-stranded RNA segments.526 The ends of each segment are conserved and partially complementary. Segments 1, 2, and 3 encode the PB2, PA, and PB1 polymerase subunits, respectively, while segment 5 codes for a protein that is distantly related to the Thogoto virus glycoprotein, and thus is likely the attachment protein.544 The predicted protein products of segments 4 and 6 do not show any homology with known proteins.
Stages of Viral Replication
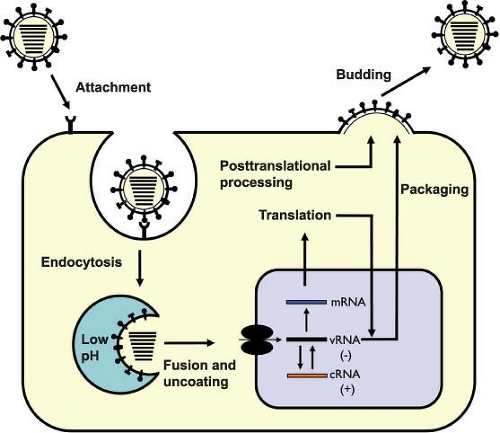
An overview of the influenza virus life cycle is illustrated in Figure 40.6 and in the following pages we will discuss each stage of this life cycle in order.
Mechanism of Attachment
Influenza viruses bind to neuraminic acids (sialic acids) on the surface of cells to initiate infection and replication. The interaction of influenza viruses with a ubiquitous molecule such as sialic acid is constrained by the fact that the HAs of viruses that replicate in different species show specificity toward sialic acids with different linkages. Human viruses preferentially bind to N-acetylneuraminic acid attached to the penultimate galactose sugar by an α2,6 linkage (SAα2,6Gal), whereas avian viruses mostly bind to sialic acid with an α2,3 linkage120 (e-Fig. 40.5). In agreement with this finding is the fact that human tracheal epithelial cells contain mostly SAα2,6Gal, while the gut epithelium from ducks possesses mostly SAα2,3Gal sugar moieties.123,307 It should be noted, however, that this viral specificity is not absolute and that avian and human cells can contain both neuraminic acid linkages (2,3 as well as 2,6). Studies on ciliated cells in the human airway epithelium have shown that sialylated proteins with α2,3 linkages are present and that these cells can be infected with avian influenza viruses.425 Furthermore, glycan structure is far more complex than just the terminal sialic acid
linkage, and evidence suggests that factors such as the type of backbone, chain length, and branching pattern as well as sulfation and fucosylation may also influence the interactions with HA.99,639 Glycan microarrays that contain a wide spectrum of glycan structures are now being used as tools to better understand the specificity of receptor binding.638 Also, when viruses are passaged in a particular host, they can adapt to that host by mutating the receptor-binding site in the viral HA.202,448 In a study of the A/New York/1/1918 virus HA, it was shown that the binding specificity can be changed by a single amino acid mutation (D190E) to a preference for α2,3-linked sialic acids. It is thus hypothesized that the HA gene of the 1918 influenza virus has its origin in avian species and that a single amino acid change (E190D) allowed the hemagglutinin to recognize the α2,6-linked sialic acids prevalent in human cells.222,637
linkage, and evidence suggests that factors such as the type of backbone, chain length, and branching pattern as well as sulfation and fucosylation may also influence the interactions with HA.99,639 Glycan microarrays that contain a wide spectrum of glycan structures are now being used as tools to better understand the specificity of receptor binding.638 Also, when viruses are passaged in a particular host, they can adapt to that host by mutating the receptor-binding site in the viral HA.202,448 In a study of the A/New York/1/1918 virus HA, it was shown that the binding specificity can be changed by a single amino acid mutation (D190E) to a preference for α2,3-linked sialic acids. It is thus hypothesized that the HA gene of the 1918 influenza virus has its origin in avian species and that a single amino acid change (E190D) allowed the hemagglutinin to recognize the α2,6-linked sialic acids prevalent in human cells.222,637
Mechanism of Entry
While some viruses (e.g., paramyxoviruses and herpes viruses) can enter cells directly through the plasma membrane by a pH-independent fusion process, influenza viruses require a low pH to initiate fusion and are therefore internalized by endocytic compartments. There are four internalization mechanisms: (a) via clathrin-coated pits; (b) via caveolae; (c) through nonclathrin, noncaveolae pathways; and (d) through macropinocytosis (for review, see (119,353,443,613)). Clathrin-mediated endocytosis has traditionally been the model for influenza virus entry.424 However, a non-clathrin, non-caveolae-mediated internalization mechanism has also been described for influenza viruses.612 The latter pathway is dependent on low pH and trafficking to late endosomes, as it requires protein kinase C, Rab5, and Rab7 functions.611 More recently, through the use of specific inhibitors and RNA interference (RNAi), it has been shown that in addition to entering via a dynamin-dependent, clathrin-driven pathway, influenza viruses can also enter via a dynamin-independent pathway that is characteristic of macropinocytosis.133 Potential differences in the entry pathways defined in polarized versus nonpolarized cells should also be appreciated, as, for example, the actin cytoskeleton appears to be critical for uptake of influenza viruses into polarized cells but not nonpolarized cells.651 The requirement of specific host proteins during influenza virus entry will help to further define the cellular pathways utilized by the virus. Genome-wide RNAi screens have identified multiple factors that are required for efficient entry mediated by the influenza virus glycoproteins341; however, the ability of the virus to enter via different routes means that this approach is unlikely to capture factors that are specific to one endocytic route. A more focused approach, such as that which shows the requirement of epsin 1 for clathrin-mediated uptake of influenza virus, is needed.90 The epidermal growth factor receptor (EGFR) has also been demonstrated to play a role during influenza virus entry,154 and it is thought that virus attachment to the cell stimulates EGFR, which leads to activation of signaling cascades such as phosphatidylinositol 3 kinase (PI3K) signaling, which is known to promote influenza virus entry.152 There are also questions over whether sialic acid is the only attachment molecule. Lec-1 cells, which are deficient in N-linked glycosylation (but still contain sialylated proteins), are unable to internalize influenza viruses.103 Similarly, cells deficient in sialic acid can be made to support influenza virus entry if they express one of the C-type lectins, DC-SIGN or L-SIGN.398
Mechanism of Fusion and Uncoating
Influenza viruses and other enveloped viruses (including rhabdo-, flavi-, bunya-, and filoviruses) require low pH to fuse
with endosomal membranes. After binding to the target cell surface and endocytosis, the low pH of the endosome activates fusion of the viral membrane with that of the endosome. This fusion activity is induced by a structural change in the HA of influenza viruses, but in order for this to occur, the HA0 precursor must first be cleaved into two subunits, HA1 and HA2. Once in the acid environment of the endosome, the cleaved HA molecule undergoes a conformational change and this exposes the fusion peptide at the N-terminus of the HA2 subunit, enabling it to interact with the membrane of the endosome (for review, see (128,258,632) and for details see the following section on Hemagglutinin). The transmembrane domain of the HA2 (inserted into the viral membrane) and the fusion peptide (inserted into the endosomal membrane) are in juxtaposition in the low pH–induced HA structure. The concerted structural change of several hemagglutinin molecules then opens up a pore, which releases the contents of the virion (i.e., viral RNPs) into the cytoplasm of the cell. The precise timing and the location of uncoating (maturity of the endosome) depends on the pH-mediated transition of the specific HA molecule involved.
with endosomal membranes. After binding to the target cell surface and endocytosis, the low pH of the endosome activates fusion of the viral membrane with that of the endosome. This fusion activity is induced by a structural change in the HA of influenza viruses, but in order for this to occur, the HA0 precursor must first be cleaved into two subunits, HA1 and HA2. Once in the acid environment of the endosome, the cleaved HA molecule undergoes a conformational change and this exposes the fusion peptide at the N-terminus of the HA2 subunit, enabling it to interact with the membrane of the endosome (for review, see (128,258,632) and for details see the following section on Hemagglutinin). The transmembrane domain of the HA2 (inserted into the viral membrane) and the fusion peptide (inserted into the endosomal membrane) are in juxtaposition in the low pH–induced HA structure. The concerted structural change of several hemagglutinin molecules then opens up a pore, which releases the contents of the virion (i.e., viral RNPs) into the cytoplasm of the cell. The precise timing and the location of uncoating (maturity of the endosome) depends on the pH-mediated transition of the specific HA molecule involved.
The uncoating of influenza viruses in endosomes is blocked by changes in pH caused by weak bases (e.g., ammonium chloride or chloroquine) or ionophores (e.g., monensin) (for review, see (418)). Effective uncoating is also dependent on the presence of the M2 protein, which has ion channel activity.532,534 Early on it was recognized that amantadine and rimantadine inhibit replication immediately following virus infection.617 Later it was found that the virus-associated M2 protein allows the influx of H+ ions from the endosome into the virus particle, which disrupts protein–protein interactions and results in the release of RNP free of the M1 protein424,767 (for review, see (534)). Amantadine and rimantadine have been shown to block the ion channel activity of the M2 protein and thus uncoating.100,281,532,648,699 The HA-mediated fusion of the viral membrane with the endosomal membrane and the M2-mediated release of the RNP result in the appearance of free RNP complexes in the cytoplasm. This completes the uncoating process.421 The time frame for the uncoating process was examined by inhibiting virus penetration with ammonium chloride. The majority of virus particles showed a half time for penetration of about 25 minutes (after adsorption). Barely 10 minutes later (half time of 34 minutes after adsorption) RNP complexes are found in the nucleus.421 The process for uptake of RNP molecules through nuclear pores is an active one, involving the nucleocytoplasmic trafficking machinery of the host cell (for details, see Nuclear Import of RNPs).
Much less is known about the uptake and uncoating of influenza B and C viruses. Influenza B viruses are more like influenza A viruses as both recognize N-acetylneuraminic acid as receptors, while influenza C viruses bind to 9-O-acetylated neuraminic acid derivatives.272 Like influenza A viruses, the B- and C-type viruses go through an endosome-mediated uncoating process, which requires proteolytic activation (cleavage) of the HA or HEF proteins and subsequent fusion of the viral and endosomal membranes. Although the viral glycoproteins of both viruses are dependent on a low pH–triggered fusion process, the influenza C virus HEF-mediated fusion/uncoating may occur at a higher pH in early endosomes.184,767 At this point, the role of CM2 in uncoating of influenza C viruses is less well established than that of the BM2 protein of influenza B viruses, which is the homolog of the influenza A virus M2 protein.193,253,262,283,455,701
The Hemagglutinin
Structural Features
Much is known about the HA molecule and excellent reviews are available.150,204,258,613,618,633,658 In fact, the influenza virus HA has become a model for studies of protein folding and trafficking, protein quality control, membrane fusion, protein– receptor interactions, and antigen–antibody complexes and, last but not least, for investigating how the immune system reacts to a foreign protein. The major functions of the HA are the receptor-binding and fusion activities, but there may also be a structural role for the HA in budding and particle formation. The HA is a trimeric rod-shaped molecule with the carboxy terminus inserted into the viral membrane and the hydrophilic end projecting as a spike away from the viral surface (i.e., a type I integral membrane protein). Early on it was shown that (a) posttranslational modifications of the precursor molecule (i.e., glycosylation and palmitoylation), (b) cleavage of the signal peptide in the endoplasmic reticulum (ER), and (c) cleavage of the HA0 precursor into HA1 and HA2 subunits are required for the full activity of the molecule.335,363,542
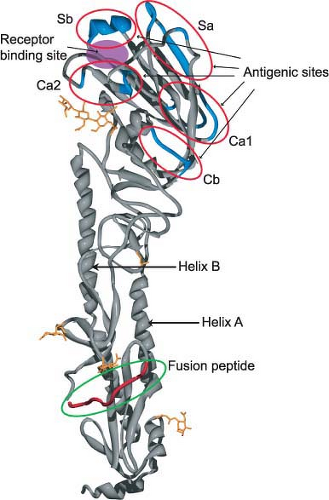
The first x-ray crystallographic structure of an HA (the ectodomain released from the virus by bromelain treatment) was resolved in 1981 by Wilson et al.725 At that time the HA (from A/Aichi/68 [H3N2] virus) was the largest biological molecule for which a structure had been resolved, and it started an unprecedented drive to study the structure/function relationships of biologically important molecules, which continues unabated to this day. The structures of numerous HAs have now been resolved, including the subtype 1 HA molecules of the 1918 and 2009 pandemic influenza viruses203,640,735 and that of an H2 subtype HA736 (Fig. 40.7). Remarkably, even though the overall amino acid sequence identity can be less than 50%, the structure and functions of these HAs are highly conserved (e-Fig. 40.6). This represents a case of evolution and sequence variation proceeding to an extreme level while structure and function have remained conserved. Even more surprising is that the structure of the influenza B virus HA is similar to that of influenza A virus HA despite sharing only 25% sequence identity.703,704
The crystallographic structure of the uncleaved influenza A virus HA is superimposable onto that of the cleaved HA1 and HA2, with the exception of the amino acids adjacent to the cleavage site. The major features of the structure are (a) a long fibrous stem, which is made up of a triple-stranded coiled coil of α-helices derived from the three HA2 parts of the molecule (helix A and helix B; Fig. 40.7 and e-Fig. 40.6), and (b) the globular head, which is also made up of three identical domains whose sequences are derived from the HA1 portions of the three monomers. The first major function of HA is binding to receptors and the receptor-binding site lies within the globular head of the molecule. This site has been defined through crystallization and structure analysis of HA–receptor complexes, as well as by mutational analysis. In the H3-subtype viruses, it appears that within the receptor-binding pocket of an avian HA, a glutamine in position 226 preferentially accommodates the 2,3-linked sialic acid, whereas a leucine in that position in human H3 HAs preferentially accommodates the 2,6-linked sialic acid239 (e-Fig. 40.7). (See the previous section on Attachment for more details).
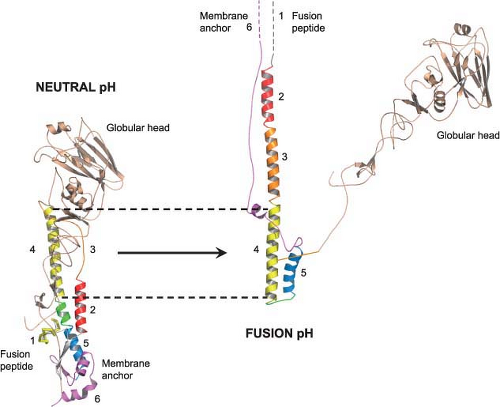
The second major function of the HA is acid pH–triggered fusion, which is required for the uncoating process. Low pH treatment changes the structure of the HA dramatically. The molecule becomes susceptible to protease digestion, and the disulfide bond linking the HA1 and HA2 subunits becomes susceptible to mercaptoethanol.232,615 However, the important feature of the acid pH–mediated change is that the fusion peptide becomes aligned antiparallel to the membrane anchor of the HA2 (Fig. 40.8). The end result is that the fusion peptide brings the endosomal membrane into juxtaposition with the viral membrane, leading to fusion. The presence of more than one hemagglutinin then leads to the formation of a fusion pore through which the RNP can enter the cytoplasm (Fig. 40.9). Structures of the postfusion HA as well as an early fusion intermediate have helped to reveal the molecular details of the changes that occur during the transition from pre- to postfusion state.75,737 Structures of the fusion peptide in lipid environments have been resolved by nuclear magnetic resonance (NMR) and show that the peptide forms a tight helical hairpin structure that angles back on itself252,351,399 (e-Fig. 40.8). This hook structure may help to pull the endosomal membrane close to the viral membrane, resulting in the initiation of the actual fusion process. These studies represent the first foray into the characterization of the transmembrane region, which so far has proved difficult to analyze.
The third major structural element of the HA, the cytoplasmic tail, is highly conserved among all subtypes. There are three cysteines that are palmitoylated (with one of them located in the transmembrane domain). The role of this cytoplasmic tail (and the palmitates attached to the cysteines) is not entirely clear due to subtype- and cell/host-specific differences.89,318,439,696,774
Antigenic Determinants
In addition to having an important role in receptor binding, fusion, and assembly, the influenza virus HA is also the major determinant recognized by the adaptive immune system of the host. Following infection and replication, a vigorous immune response is induced, which usually results in the formation of neutralizing antibodies. These antibodies then lead to the selection of “antibody escape” variants. The amino acids undergoing change are almost exclusively on the HA1 (and on the outside of the molecule). Many of these changes get fixed (accumulate over time), defining the antigenic drift of influenza viruses (e-Fig. 40.9). Fab antibody fragments have been shown to bind to different regions of the HA1 (e-Fig. 40.10) and, interestingly, not in all cases do three Fabs bind to one trimeric spike (3:1 ratio). Examples have been found where only one Fab molecule binds to one HA spike (1:1 ratio) or where just two Fab molecules bind to a trimeric spike (2:1 ratio) (e-Fig. 40.10).336 In the latter case, the two Fab fragments cross-link the two monomers so that the HA molecule cannot undergo an acid pH–induced conformational change.336 Attempts have also been made to measure antigenic evolution of influenza virus strains by pairwise comparison of hemagglutination inhibition assays.187,622
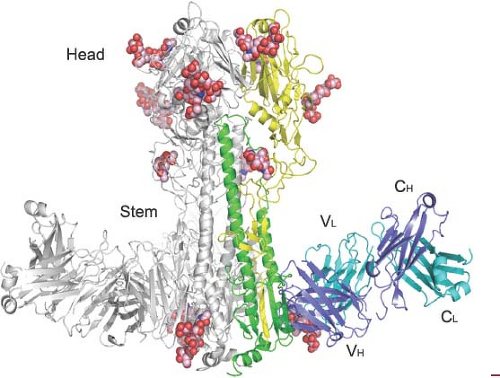
Unexpectedly, broadly cross-reactive monoclonal antibodies have been identified, which do not bind to the tip of the HA molecules but also have neutralizing activity.121,157,327,495,650,663,730 These antibodies are directed against the conserved stem region of the HA spike and recognize the membrane-proximal part of HA1 in combination with HA2 or the HA2 alone (Fig. 40.10).707 In general, these antibodies recognize the HAs within either group 1 (for review see (706)) or group 2,158,707 but recently a human monoclonal antibody was described that binds to the stem of both group 1 and group 2 HAs.121 The most likely mechanism by which these cross-reactive antistem antibodies neutralize influenza viruses is by blocking conformational rearrangements associated with membrane fusion (antifusion vs. hemagglutination inhibition activity). A broadly neutralizing human monoclonal antibody that recognizes the highly conserved sialic acid–binding site of H1 HAs has also been described.343 It is hoped that the conserved epitopes identified by these cross-protective antibodies could be utilized as immunogens with the possibility of developing novel vaccine constructs that will result in effective and safe universal influenza virus vaccines.47,631,708
The HEF of Influenza C Viruses
In contrast to the HA of influenza A and B viruses, the major glycoprotein of the C viruses has a receptor-destroying activity.
In contrast to the neuraminidase activity of the NA proteins of influenza A and B viruses (see Neuraminidase section), HEF has esterase activity, which cleaves off an acetyl group at position 9 of the neuraminic (sialic) acid receptor, eliminating the ligand (for review, see (644)). This activity is important for entry of the virus, implying a role in releasing the incoming virus from the receptor so that the uncoating process can begin.688 Thus, in addition to receptor-binding (hemagglutination) and fusion activities, the molecule also has esterase activity, hence the name HEF. Although there is only about a 12% sequence identity between HAs and HEF, the overall structure of the molecule is similar, as was shown by x-ray crystallographic analysis.569 Even more surprising are structural and sequence similarities between the esterases of influenza C viruses and some coronaviruses.758
In contrast to the neuraminidase activity of the NA proteins of influenza A and B viruses (see Neuraminidase section), HEF has esterase activity, which cleaves off an acetyl group at position 9 of the neuraminic (sialic) acid receptor, eliminating the ligand (for review, see (644)). This activity is important for entry of the virus, implying a role in releasing the incoming virus from the receptor so that the uncoating process can begin.688 Thus, in addition to receptor-binding (hemagglutination) and fusion activities, the molecule also has esterase activity, hence the name HEF. Although there is only about a 12% sequence identity between HAs and HEF, the overall structure of the molecule is similar, as was shown by x-ray crystallographic analysis.569 Even more surprising are structural and sequence similarities between the esterases of influenza C viruses and some coronaviruses.758
The M2 Protein
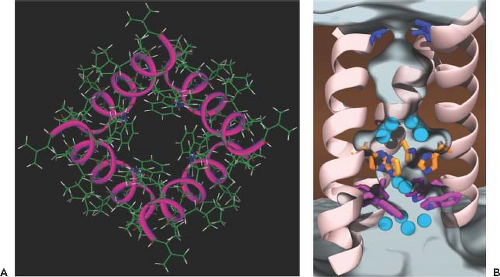
The M2 protein of influenza A viruses is a tetrameric type III (lacking a signal peptide sequence) integral membrane protein. It has a short ecto-domain, a transmembrane domain, and a cytoplasmic domain with palmitate and phosphate modifications (for review, see (531,534)). M2 has been shown to possess ion channel activity, and its major role is thought to be that of conducting protons from the acidified endosomes into the interior of the virus to dissociate the RNP complex from the rest of the viral components, thus facilitating the uncoating process (see earlier section on Fusion and Uncoating). The structural and genetic analysis of the M2 protein has revealed that the ion channel is acid gated (but not voltage gated) and highly selective for H+ ions.86,100,453,454,485 The structures of the transmembrane regions of M2 and of those that include the cytoplasmic sequences reveal a good understanding of the mechanism of proton conductance, which is controlled by the histidine-37 and tryptophan-41 cluster.1,79,291,482,589,599,643 The transmembrane region, when viewed from the top, shows four helices that sit at an angle in the lipid bilayer, forming a pore (Fig. 40.11). Structural studies on M2 in complex with adamantine drugs indicate two potential sites of interaction. In the x-ray structure, a single drug molecule binds to the core of the pore.643 In contrast, the NMR structure shows four drug molecules binding to the lipid-exposed surface of the channel close to the cytoplasmic ends of the helices.589 More recent data confirm the existence of these two interaction sites and propose that both drug-binding mechanisms may have physiologic
significance.79,342 The structure and the precise function of the extracellular portion of the M2 protein remain to be resolved. This external portion of M2 has been considered as the basis of a universal influenza virus vaccine approach because the M2 protein maintains a highly conserved sequence over long periods of time.590
significance.79,342 The structure and the precise function of the extracellular portion of the M2 protein remain to be resolved. This external portion of M2 has been considered as the basis of a universal influenza virus vaccine approach because the M2 protein maintains a highly conserved sequence over long periods of time.590
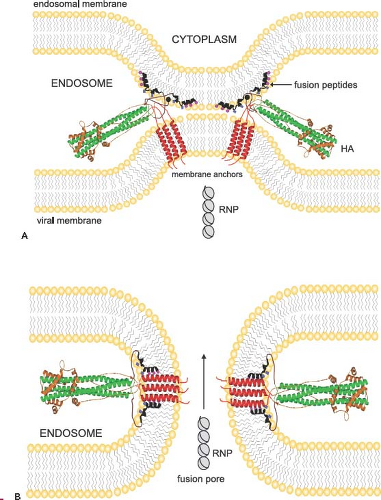 Figure 40.9. Model for juxtaposition of viral and endosomal membranes resulting in formation of a fusion pore and release of ribonucleoproteins (RNPs). Structures of influenza virus hemagglutinins in their postfusion state modeled into a possible fusion intermediate (A) and into a fusion pore (B). The fusion peptides are shown inserted into the endosomal membrane, while the transmembrane domains remain anchored in the viral membrane. Note the large conformational changes of the ecto- and fusion domains when compared to their prefusion structures (see Figs. 40.7 and 40.8). The small spheres (pink) on the fusion peptide denote glycines that may mediate helix interactions and the small squares (blue) denote glutamates that may be responsible for the pH dependence of the fusion peptide penetration into lipid bilayers. Following the formation of a pore, the RNPs are released from the interior of the virus particle into the cytoplasm, completing the uncoating process. (Courtesy of Lukas Tamm.) |
The ion channel activity of M2 has also been implicated in stabilizing HAs from premature low pH transitions in the trans-Golgi network, but this second function may only come into play for viruses carrying highly acid-sensitive HAs.104 This is the case for H5 and H7 HAs, which have a multibasic cleavage site that can be cleaved by ubiquitous proteases and are therefore more susceptible to a premature low pH–induced conformational change. Further functions attributed to the M2 protein include roles in particle morphology,231,310,564,571 genome packaging,231,310,432 membrane scission572 (see Assembly and Release), and inhibition of autophagy.205
Influenza Virus Transcription and Replication
Overview
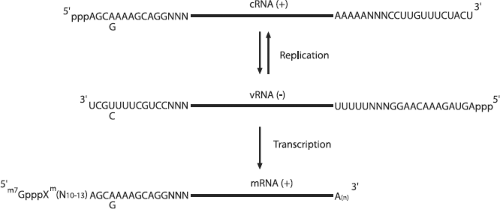
After uncoating, the viral ribonucleoproteins (vRNPs) are transported into the nucleus and the incoming negative-sense viral RNAs (vRNAs) are transcribed into mRNA by a primer-dependent mechanism. These mRNA products are incomplete copies of the vRNA templates and are capped and polyadenylated, unlike vRNA. Replication occurs via a two-step process. A full-length, positive-sense copy of the vRNA is first made, which is referred to as complementary RNA (cRNA), and is in turn used as a template to produce more vRNA (Fig. 40.12).
Nuclear Import of Ribonucleoproteins
One of the characteristics of the influenza virus life cycle, and one that is unusual for an RNA virus, is its dependence on nuclear functions. All viral RNA synthesis occurs in the nucleus, and the trafficking of the viral genome into and out of the nucleus is a tightly regulated process (reviewed in (54,127)). The eight influenza virus genome segments never exist as naked RNA but are associated with four viral proteins to form vRNP complexes. The major viral protein in the RNP complex is the nucleocapsid protein (NP), which coats the RNA. The remaining proteins are the three polymerase proteins (PB1, PB2, and PA), which bind to the partially complementary ends of the viral RNA, creating the distinctive panhandle structure.290 These RNPs (10 to 20 nm wide)115,419 are considered too large to allow for passive diffusion into the nucleus and therefore, once released from an incoming particle, they must rely on an active nuclear import mechanism. All proteins in the RNP complex possess nuclear localization signals (NLSs), which mediate their interaction with the nuclear import machinery.320,456,470,483,625,702,720 However, it is the signals on NP that have been shown to be both sufficient and necessary for the import of viral RNA73,126,496,503 (e-Fig. 40.11).
NP Interactions with Karyopherin α
The transport of proteins across the nuclear membrane is an energy-driven process that is initiated upon recognition of an NLS-containing cargo protein by members of the karyopherin α (also called importin α) family. Karyopherin α binds directly to the NLS and then recruits karyopherin β into a trimeric complex, which docks at the nuclear pore (e-Fig. 40.12). Prior to the identification of a definitive NLS within NP, the human homolog of yeast SRP-1 was identified as an NP-interacting partner.497 This uncharacterized protein was subsequently identified as karyopherin α1 and NP was also shown to interact with another family member, karyopherin α2.496 The minimal karyopherin α–binding site was used to identify the unconventional NLS in NP, so named because it does not contain a cluster of basic residues.702 As described earlier,126 this NLS has been shown to be essential for RNP import, and this implicates karyopherin α as a critical component of RNP nuclear import. Interestingly, the NP NLS-binding site on karyopherin α is distinct from that of classical NLS-containing proteins.126,437 One could postulate that this serves to avoid competition for karyopherin α binding with host proteins and may explain the use of an unconventional NLS. There is also evidence that differential interactions with human versus avian karyopherin α proteins may determine species specificity of influenza viruses.197,198
The Viral Ribonucleoprotein Template
Each viral RNA segment exists as an RNP complex in which the RNA is coated with NP and forms a helical hairpin that is bound on one end by the heterotrimeric polymerase complex (reviewed in (554)). NP is an arginine-rich protein and has a net positive charge (at pH 6.5), which reflects its RNA-binding activity and its primary role in encapsidation (reviewed in (541)). The RNA/NP interaction is thought to be mediated by the positively charged residues on NP and the negatively charged phosphate backbone of the RNA, and thus there is no apparent sequence specificity to the interaction.30,161 The RNA within the influenza virus RNP also remains sensitive to digestion with RNase,147 supporting the model that the RNA is wrapped around the outside of the NP with its bases exposed so that they can be accessed by the polymerase without disrupting the RNP structure.30 Approximately 24 nucleotides of RNA are bound by each NP monomer,502 and NP also has homo-oligomerization properties,547 which adds a higher-order structure to the RNP complex. This is maintained even in the absence of RNA147,576 and has been shown to be crucial for maintaining the RNP in a transcriptionally active form.83,160 Crystal structures of NP show that it is composed of a head domain and body domain and that a flexible tail loop mediates oligomerization.481,745 A potential RNA-binding groove, which is highly positively charged, has been identified between the head and body domains.745 Structural data based on electron microscopy also provide evidence that NP makes direct contact with the bound polymerase complex on the RNP13,113,419 (e-Fig. 40.13A), which may reflect the previously reported interaction of free NP with both PB1 and PB2.41 Detailed mutagenesis of conserved
residues in NP indicates regions that are involved in genome replication/transcription and also genome packaging.388
residues in NP indicates regions that are involved in genome replication/transcription and also genome packaging.388
The RNA Polymerase Complex
The influenza virus RNA-dependent RNA polymerase is a 250-kD complex of three proteins: PB1, PB2 and PA.59 A three-dimensional image of the complex obtained by electron microscopy indicates that the three subunits are tightly associated to form a compact structure13,669 (e-Fig. 40.13B). Protein interaction studies have shown that PB1 binds to both PA and PB2, through its N- and C-terminal domains, respectively,228,492 and that the N-terminus of PA interacts with PB2.268 The details of how the newly synthesized polymerase is assembled are still under debate. One model proposes that PB1 and PA enter the nucleus as a dimer through interactions with RanBP5 and then bind to PB2, which is imported independently via a karyopherin α interaction.135,183,300,555,661 This is supported by fluorescent spectroscopy data in live cells.295 However, another model suggests that PB1-PB2 dimer is transported into the nucleus (via the chaperone Hsp90) and that PA traffics separately.465 It has also been shown that vRNA can bind to the PB1-PA dimer in vitro prior to PB2 binding as well as to the preformed trimeric complex.136
The PB1 Protein
The PB1 protein catalyzes the sequential addition of nucleotides during RNA chain elongation59 and contains the conserved motifs characteristic of RNA-dependent RNA polymerases.42 The active site for the polymerization activity is an S-D-D motif at positions 444 to 446,42 but we currently lack structural information for this region. In fact, only small regions encompassing the extreme N- and C-termini of PB1 have been crystallized in complex with portions of PA and PB2, respectively. Two studies have provided x-ray structures of either residues 1 to 81 or 1 to 25 of PB1 in complex with the C-terminal portion of PA,265,490 and it has been shown that synthetic peptides corresponding to this region of PB1 can compete with full-length PB1 for PA binding and inhibit virus replication.219 The PB1 C-terminus (residues 678 to 757) has been shown to mediate an interaction with residues 1 through 37 of PB2, and structural analysis of this complex indicates that both peptides form three helices whose folding is dependent on the presence of each partner.646 PB1 is also responsible for binding to the terminal ends of both vRNA226,377 and cRNA227 for initiation of transcription and replication.
The PB2 Protein
The PB2 protein plays a critical role in the initiation of transcription as it is responsible for binding the cap on host pre-mRNA molecules.43,674 Despite earlier discrepancy in the position of the binding site, it has now been shown that a domain encompassing PB2 residues 318 to 483 is sufficient for cap binding236 and confirms the findings of a mutagenesis study that identified two aromatic residues (F363 and F404) as being important for the interaction.171 An x-ray structure of this minimal domain bound to cap analog m7GTP reveals that the cap is sandwiched between phenylalanine 404 and histidine 357 in a mode similar to that described for other cap-binding proteins, although the involvement of a histidine is unique.236 In fact, in influenza B and C virus PB2 proteins, the histidine is replaced with a more traditional tryptophan. Both NMR and x-ray structures of the C-terminal domain (aa 678 to 759) of PB2 have been obtained, the latter in complex with karyopherin α1.661 Within this domain the authors report the presence of a bipartite nuclear localization signal, and in the co-crystal it is shown how this region unfolds to allow for interaction with karyopherin α1. PB2 has also been shown to localize to the mitochondria, and this is determined by an N-terminal mitochondrial-targeting signal.81 However, avian influenza viruses have a polymorphism in this signal, so it appears that mitochondrial localization of PB2 is unique to human influenza viruses.229 Finally, as well as interacting with PB1 via its N-terminus (see PB1 section earlier), PB2 is also reported to interact with PA; however, the region of PB2 involved has not yet been defined.268 PB2 also participates in genome replication as mutations affecting this activity but not transcription have been reported.215
The PA Protein
Until recently, the specific function of PA was unknown, but crystal structures of the N-terminal domain revealed that the endonuclease activity of the polymerase, which is required to generate the capped primer, resides in the PA protein.139,752 Previous work had mistakenly attributed this function to the PB1 protein. In the structure, the fold and position of the active site identifies the PA endonuclease as a member of the PD-(D/E)XK family of nucleases. The catalytic site involves residues His 41, Glu 80, Asp 108, Glu 119, and Lys 134 and harbors two Mn(2+) ions.124,139 Mutation of these residues abolishes the transcriptional activity of the trimeric polymerase but replication activity is unaffected, confirming the specific role of the endonuclease in viral transcription.124,752 PA does, however, participate in genome replication as mutations affecting this process have been described.179,294 In addition to encoding nuclease function, the N-terminus of PA (aa 1 to 100) is also reported to be involved in an interaction with PB2,268 while the C-terminus makes contact with PB1. Structures of PA residues 257 to 716 show this region forming a “dragon-like head” with the N-terminal peptide of PB1 inserted into the mouth.265,490 Another function ascribed to PA is that it possesses proteolytic activity.582 Two residues, S624255 and T157,525 have been reported to be involved in the proteolytic function, although viruses containing mutations at position 157 appear to be more severely affected than those mutated at position 624.294,670 PA has also been shown to be a target for casein kinase II and to be phosphorylated at serine and threonine residues.583
The vRNA Promoter
All influenza virus RNA segments contain noncoding sequences at their 5′ and 3′ ends, which flank the coding region. Some of this sequence is segment specific,766 but the terminal ends are conserved between all segments in all influenza viruses. These conserved 13 nucleotides at the 5′ end and 12 nucleotides at the 3′ end display partial and inverted complementarity, which led to the proposal of a panhandle structure created by base-pairing of the 5′ and 3′ ends.138,565 This is supported by cross-linking experiments that demonstrated a circular configuration for virion RNAs290 as well as by more recent structural analysis.19,419 Studies using in vitro transcription of model RNA templates or reporter gene expression in vivo have shown that both 5′ and 3′ terminal ends are necessary for promoter activity and that base-pairing is required (reviewed in (177,475)). Furthermore,
it has been demonstrated that the polymerase can interact with both 5′ and 3′ ends and that the binding affinity decreases when the duplex is disrupted.182,226,240,373,664 These data define the vRNA promoter as a double-stranded element formed by the conserved 5′ and 3′ terminal ends of the vRNA molecule.487 While the need for base-pairing is clear, several models for the secondary structure of the promoter have been proposed based on mutational analyses (e-Fig. 40.14). The original panhandle model predicts extensive Watson-Crick base-pairing, whereas the RNA-fork model proposes that the extreme termini remain single stranded.181,182,332 The corkscrew model suggests that these single-stranded regions in fact base-pair within themselves to form 5′ and 3′ hairpin loops.176 The presence of both 5′ and 3′ stem-loops has been shown to be critical for endonuclease activity, and the 5′ stem-loop is also required for polyadenylation.364,365,545 This favors a model where the stem-loop structures are involved in binding and stabilizing the polymerase complex.64
it has been demonstrated that the polymerase can interact with both 5′ and 3′ ends and that the binding affinity decreases when the duplex is disrupted.182,226,240,373,664 These data define the vRNA promoter as a double-stranded element formed by the conserved 5′ and 3′ terminal ends of the vRNA molecule.487 While the need for base-pairing is clear, several models for the secondary structure of the promoter have been proposed based on mutational analyses (e-Fig. 40.14). The original panhandle model predicts extensive Watson-Crick base-pairing, whereas the RNA-fork model proposes that the extreme termini remain single stranded.181,182,332 The corkscrew model suggests that these single-stranded regions in fact base-pair within themselves to form 5′ and 3′ hairpin loops.176 The presence of both 5′ and 3′ stem-loops has been shown to be critical for endonuclease activity, and the 5′ stem-loop is also required for polyadenylation.364,365,545 This favors a model where the stem-loop structures are involved in binding and stabilizing the polymerase complex.64
Initiation of Messenger RNA Synthesis
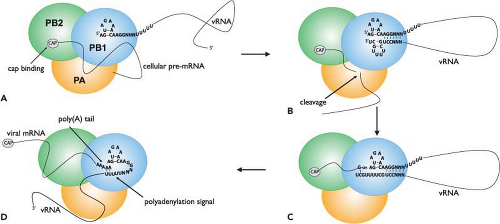
Influenza virus mRNA synthesis is dependent on cellular RNA polymerase II activity. This is because it requires a 5′-capped primer, which it steals from host pre-mRNA transcripts, to initiate its own mRNA synthesis. This process is known as cap snatching and involves the cap-binding function of the PB2 protein and endonuclease function of the PA protein. The initiation of transcription commences with binding of the 5′ end of the vRNA to the PB1 subunit (Fig. 40.13). This induces an allosteric change in the polymerase, which allows the PB2 protein to recognize and bind the cap structure on host pre-mRNAs106,377 (reviewed in (170)). The change in the polymerase also increases its affinity for the 3′ vRNA end, which is bound by PB1. Binding of the 3′ terminus stabilizes the polymerase complex64 and also serves to activate the endonuclease function.106,240,364,377 However, in contrast to this model, one report states that endonuclease activation only requires a bound 5′ end,551 the difference being that this study used capped RNA fragments with “CA” 3′ termini as primers. Primers with this specific end have previously been shown to be used preferentially for transcription initiation in infected cells.32,604 Another study indicates that both primer-binding and endonuclease activities are greatly enhanced when the polymerase binds simultaneously to the 5′ and 3′ ends (i.e., in a preformed duplex).373 Endonuclease activation leads to cleavage of the bound pre-mRNAs. This occurs approximately 10 to 13 nucleotides from their 5′ caps, usually after a purine residue.32,537 Transcription is then initiated by the addition of a “G” residue to the primer, directed by the penultimate “C” nucleotide at the 3′ end of the vRNA template,32 although in some instances the incorporation of a “C” that is directed by the “G” at position 3 in the vRNA has also been observed.181 Unlike influenza viruses, Thogoto viruses lack host-derived sequences at the 5′ end of their capped mRNAs.719 RNA chain elongation is
catalyzed by the polymerase function of PB1 and continues until a stretch of uridine residues is encountered approximately 16 nucleotides before the 5′ end of the vRNA.383,407,566 This is the signal for polyadenylation (Fig. 40.13).
catalyzed by the polymerase function of PB1 and continues until a stretch of uridine residues is encountered approximately 16 nucleotides before the 5′ end of the vRNA.383,407,566 This is the signal for polyadenylation (Fig. 40.13).
Polyadenylation
Unlike host cells, which use a specific poly(A) polymerase for generating the poly(A) tail on mRNA transcripts, polyadenylation of influenza virus mRNAs is catalyzed by the same polymerase that is used for transcription. This activity is dependent on an uninterrupted stretch of five to seven “U” residues and the adjacent double-stranded region of the vRNA promoter.383,407,566 The current model proposes that the 5′ end of the vRNA remains bound to the polymerase during elongation while the template is threaded through in a 3′ to 5′ direction (Fig. 40.13). When the polymerase nears the 5′ end to which it is bound, it is blocked by steric hindrance and consequently it stutters on the preceding stretch of uridines, which it repeatedly copies to produce a poly(A) tail.182,240,539,765 In support of this model, mutations introduced into the 5′ end of the vRNA that prevent or weaken polymerase binding have been shown to also inhibit polyadenylation.180,540,545,546 The polyadenylation signal is vital for gene expression as replacement of the uridines with adenosines has been shown to result in transcripts with poly(U) tails, which fail to be properly exported from the nucleus.538
Splicing
Members of the Orthomyxovirus family can extend the coding capacity of their genomes by producing two proteins from one gene via an alternative splicing mechanism. Genome segments that encode proteins from both spliced and unspliced mRNA transcripts are segments 7 and 8 of influenza A virus,358,360 segment 8 of influenza B virus,63 segments 6 and 7 of influenza C virus,468,741 segment 6 of Thogoto virus,339 and segment 7 of isavirus40 (see Genome Structure and Organization section). The primary transcripts from these segments have 5′ and 3′ splice sites, which (more or less) fit the consensus sequence for the exon/intron boundaries of cellular transcripts. This, combined with the fact that splicing can be demonstrated in the absence of any viral proteins,357,359 indicates that the virus is using the cellular splicing machinery. However, unlike cellular splicing, which is extremely efficient, splicing of viral mRNA has to be relatively inefficient because proteins must be expressed from both spliced and unspliced mRNAs. In influenza virus–infected cells, splicing is tightly regulated such that the steady-state level of spliced viral transcripts is only 10% that of the unspliced viral transcripts.356,360 These control mechanisms may act on several different levels. The rate of nuclear export of the unspliced transcript is certainly crucial as this determines its availability for splicing. It has been proposed that the NS1 protein inhibits both the splicing and nuclear export of NS1 transcripts via negative feedback,8,209 but contradictory reports559 suggest that alternative mechanisms may exist for regulating splicing of viral transcripts. Potentially these may involve cis-acting sequences in the NS1 transcript that negatively control the rate of splicing.7,474 Strangely, the influenza C virus NS1 protein has been reported to up-regulate viral mRNA splicing.458 Splicing of the influenza A virus M1 transcript is controlled by the aforementioned rate of nuclear export675 as well as by the viral polymerase and a cellular splicing factor, SF2/ASF. The polymerase determines the time at which splicing (and hence production of M2) occurs, and SF2/ASF is required to activate splicing.605,606
Replication Products: cRNA and vRNA
Full-length copies of the incoming vRNA have to be made, and these positive-sense cRNAs serve as templates for the synthesis of new negative-sense genomic vRNA. In vitro evidence suggests that the de novo initiation mode for vRNA synthesis may occur via terminal initiation and elongation, whereas for cRNA synthesis it involves internal initiation and realignment.137 This is a primer-independent model; however, a primer-dependent mode of vRNA initiation has also been proposed.528 The cRNA promoter is complementary to the vRNA promoter and has also been reported to assume a corkscrew configuration, albeit with subtle differences.18,129,519,766 This variation has been implicated in determining whether or not the endonuclease function of the polymerase is activated and therefore may play an important regulatory role.366
The Switch from Transcription to Replication
The vRNA serves as a template for both mRNA and cRNA synthesis, and yet the means of initiation and termination for the generation of these two molecules are quite different. In contrast to the primer-dependent mechanism of initiation of mRNA synthesis, initiation of cRNA synthesis occurs without a capped primer and cRNA molecules are full-length copies of the vRNA and thus are not prematurely terminated and polyadenylated as are mRNAs. The different initiation and termination reactions therefore have to be coordinated, but exactly how the polymerase switches between these two modes is not well understood. It has been proposed that the transcription-competent polymerase is structurally different from the replication-competent polymerase, and support for this theory comes from evidence that different domains of PB1 are involved in binding vRNA versus cRNA and that PA is more critical for binding the cRNA than the vRNA promoter.227,412 One obvious difference is that the cap-binding and endonuclease functions of PB2 and PA are not required when the polymerase is in replication mode.
In contrast to mRNAs, newly synthesized cRNAs and vRNAs are encapsidated, and it has been proposed that the availability of soluble NP (i.e., not associated with RNPs) controls the switch between mRNA and cRNA synthesis. This hypothesis arose from the observation that replication is dependent on de novo protein synthesis, which means that the incoming RNPs are only capable of transcription.263 Indeed, free NP has been shown to be required for production of full-length cRNA (antitermination),33 and this is consistent with data from temperature-sensitive (ts) NP mutants,345,435,598 which show that cRNA but not mRNA synthesis is affected at the nonpermissive temperature. However, this model has been challenged by a report demonstrating that overexpressed NP does not promote replication.457 Another study disputes the existence of a switch, rather suggesting a stabilization role for NP and the polymerase.694 It claims that the incoming polymerase is able to synthesize both mRNA and cRNA,690 but until there is a sufficient pool of polymerase and NP to encapsidate the cRNA, it is degraded and therefore at early times postinfection there is a bias toward mRNA accumulation. Also, requirements for higher nucleotide concentrations
to initiate cRNA synthesis may determine the timing of transcription versus replication.693 Furthermore, the accumulation of NEP/NS2 is associated with a decrease in transcription and an increase in replication, suggesting a regulatory role.74,560 Interestingly, NEP/NS2 is also required for the generation of small viral RNAs (svRNAs), which have been implicated in the initiation of vRNA synthesis.528 The svRNAs are 22 to 27 nt in length and correspond to the 5′ end of each viral RNA segment. These segment-specific svRNAs are needed for vRNA but not cRNA synthesis, so according to this model the polymerase is in replication mode when these svRNAs are present. It has also been proposed that the switch from transcription to replication is the result of accumulation of a newly synthesized free polymerase complex, which enhances cRNA to vRNA synthesis (and vice versa) over mRNA synthesis.322 The role of host factors in regulating influenza virus replication, including posttranslational modification of viral proteins, should also not be excluded.51,323,392,427,449,450
to initiate cRNA synthesis may determine the timing of transcription versus replication.693 Furthermore, the accumulation of NEP/NS2 is associated with a decrease in transcription and an increase in replication, suggesting a regulatory role.74,560 Interestingly, NEP/NS2 is also required for the generation of small viral RNAs (svRNAs), which have been implicated in the initiation of vRNA synthesis.528 The svRNAs are 22 to 27 nt in length and correspond to the 5′ end of each viral RNA segment. These segment-specific svRNAs are needed for vRNA but not cRNA synthesis, so according to this model the polymerase is in replication mode when these svRNAs are present. It has also been proposed that the switch from transcription to replication is the result of accumulation of a newly synthesized free polymerase complex, which enhances cRNA to vRNA synthesis (and vice versa) over mRNA synthesis.322 The role of host factors in regulating influenza virus replication, including posttranslational modification of viral proteins, should also not be excluded.51,323,392,427,449,450
Regulation of Viral Gene Expression
Early studies have provided evidence for temporal regulation of viral gene expression,263,624 but the mechanism(s) is still unresolved. Disproportionate accumulation of mRNAs from the eight segments has been observed, but whether this represents specific up-regulation of transcription for these segments164,260 or reflects different rates of vRNA synthesis597,624 or RNA stability is unclear. Suffice to say that the synthesis of NP and NS1 mRNAs and protein is favored at early stages, whereas the synthesis of HA, NA, and particularly M1 mRNAs and proteins is delayed.260,263,597,624 This differential expression is mirrored by the roles these proteins play at different points in the virus life cycle. As discussed earlier, NP is required for replication, and NS1 plays a crucial role in combating the host immune response; thus, both these proteins are needed early in the virus life cycle. M1 has been found to inhibit viral transcription,527,713 which demands its delayed expression, and at later stages M1 accumulation probably dictates the arrest of viral mRNA synthesis. M1 is also involved in the export of RNPs from the nucleus,420 which must only occur once replication is complete.
Another control mechanism for differential gene expression resides in the vRNA promoter. A natural variation is found at position 4 from the 3′ vRNA end in an otherwise totally conserved region. The PB1, PB2, and PA RNA segments have a “C” at this position, while the remaining segments usually have a “U.” The C4-containing promoter is associated with a down-regulation in transcription and an up-regulation in replication compared to the U4 promoter,370 which correlates with the lower amounts of polymerase mRNAs and proteins found in infected cells.263,624 A structural analysis of the C4 and U4 promoters has revealed differences that may alter their interaction with the polymerase and thereby regulate gene expression.372
As observed with many other viruses, influenza virus gene expression is also controlled at the level of translation. This is achieved via numerous mechanisms and results in the selective translation of viral genes and suppression of host protein synthesis (reviewed in (200,692,743)). These mechanisms include (a) degradation of host pre-mRNAs following cleavage (due to cap-snatching), (b) inhibition of host mRNA processing, (c) degradation of cellular RNA polymerase II, and (d) preferential translation of viral mRNA transcripts. Several of these processes involve the influenza virus NS1 protein. The NS1-mediated effect on mRNA processing is discussed in a later section (see The Actions of Influenza Virus Nonstructural Proteins on the Host Cell). NS1 is also involved in the specific translational enhancement of viral mRNAs through its association with the 5′ noncoding region of viral mRNA transcripts and with cellular proteins involved in translation initiation.11,76,520 These include the translation initiation factor eIF4GI and poly(A)-binding protein 1, and it has been proposed that this protein complex acts to specifically recruit ribosomes to the 5′ end of viral mRNA transcript. Another cellular protein that may play a role is GRSF-1, an RNA-binding protein that has been reported to interact with the 5′ end of the NP transcript and to stimulate the specific translation of a template driven by the NP 5′ noncoding region in a cell-free translation system.326,521 Whether this interaction is relevant in vivo remains to be determined.
An interesting model explaining the selective translation of viral mRNAs has been proposed suggesting that the viral polymerase complex remains associated with the viral mRNA transcript in the cytoplasm. It is thought that this interaction eliminates the need for complex formation with eIF4E. As eIF4E is inactivated in influenza virus–infected cells,172 this would explain the selective translation of viral transcripts over cellular transcripts. Other components of the translation machinery, such as eIF4A and eIF4G, are required for influenza viral protein translation.742 Although this model is compelling in its simplicity, this mechanism is questioned by the finding that viral mRNAs in the cytoplasm are devoid of viral polymerase but are associated with cellular cap-binding proteins, including eIF4E.39 Clearly, further investigation is required to explain the selective translation of viral transcripts in infected cells.
An additional host shut-off mechanism is controlled by the viral polymerase at the level of host transcription. It has been shown that the influenza virus polymerase interacts with the C-terminal domain of the large subunit of cellular RNA polymerase II167 and that this interaction mediates the degradation of RNA polymerase II at late times postinfection.568,691 This may play a role in viral pathogenicity because attenuated influenza viruses have been shown not to induce RNA polymerase II degradation.567
Virus Assembly and Release
Nuclear Export of Ribonucleoproteins
Association of RNP with M1
Following virus replication, newly formed RNP complexes are assembled in the nucleus from where they are exported into the cytoplasm. Two viral proteins, the matrix protein (M1) and the nuclear export protein (NEP/NS2), are involved in directing the nuclear export of RNPs (reviewed in (54,127)). Our present understanding of this process indicates that M1 associates with RNPs in the nucleus and may actually promote the formation of RNP complexes.293 M1 makes contact with both the vRNA and NP31,746 (reviewed in (127,472)), and evidence that M1 also binds to nucleosomes210,769 has led to the hypothesis that M1 interactions cause the dissociation of RNP from the nuclear matrix. This agrees with the significant finding that nuclear import of M1 is required for subsequent export of RNP complexes.72,420 Furthermore, at high temperatures, heat shock protein 70 is found bound to RNP, which prevents association with M1 and results in a block in RNP export.273,580 Recently it has been reported that sumoylation of M1 is essential for its nuclear export function.732
NEP/NS2 Interacts with the Cellular Export Machinery
Initially these data suggested that M1 alone could control RNP export, but as is the case for import, nuclear export of large molecules involves direct interactions with the cellular export machinery and as yet no such interactions with M1 have been demonstrated. However, NEP/NS2 has been found to interact with the export receptor Crm1477 and several nucleoporins.91,498 NEP/NS2 also associates with M1,4,608,744 so the current model is that an RNP–M1–NEP/NS2 complex is formed in the nucleus and that NEP/NS2 is responsible for recruiting the export machinery and directing export of the complex (e-Fig. 40.15). In support of this role, it has been shown that injection of anti-NEP/NS2 antibodies into the nucleus of infected cells inhibits RNP export,498 which is also seen in a system using recombinant virus-like particles lacking NEP/NS2.477 A methionine/leucine-rich nuclear export signal (NES) has been identified in the N-terminus of NEP/NS2498 and shown to be critical for RNP export and virus growth309,477; however, this NES does not appear to mediate the interaction of NEP/NS2 with Crm1.477 Yet treatment with leptomycin B, a Crm1 inhibitor, completely inhibits RNP export in infected cells, which indicates that export does occur in a Crm1-dependent manner.162,309,714
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree