Orbiviruses
Polly Roy
History
Two important orbivirus diseases are African horse sickness (AHS) and Bluetongue (BT) disease of sheep and cattle, both recognized first on the African continent.47,124 The first major outbreak of AHS was observed in 1719, when approximately 1,700 animals died and the most significant outbreak occurred in 1854 to 1855, affecting some 70,000 horses. AHS mainly appeared in years of unusually high rainfall and warm weather, and it was speculated that insect vectors were involved in spreading of the disease. Only in 1900 was it demonstrated that AHS was caused by a virus, when M’Fadyean succeeded in transmitting the disease by a bacteria-free filtrate from an infected animal to a healthy one.88 The major vertebrate reservoir for the virus was believed to be zebra.3 To date, AHS virus occurs in multiple serotypes (AHSV-1 to 9) and is endemic in eastern and central Africa and many parts of sub-Saharan Africa. From 1943 onward, AHS outbreaks have been reported in different parts of the world, including Palestine, Turkey, Israel, Pakistan, and the Indian subcontinent.27,66
In the late 18th century, BT disease was observed in domestic animals as well as in wild ruminants (e.g., Blesbuck, white-tailed deer, elk, and pronghorn antelope). As with AHS, BT disease was confined to Africa for many decades. Distinctive lesions in the mouths of the infected animals with a dark blue tongue were the characteristic symptoms. A detailed description of the disease was first published in 1905,112 when it was named malarial catarrhal fever,26 and Theiler demonstrated that the agent was a filterable virus.125 The first confirmed outbreak outside of Africa occurred in Cyprus in 1924, followed by outbreaks in 1943 to 1944 when about 2,500 sheep died with a mortality rate reaching 70%. In the United States, BT disease in sheep was first recognized in 1948, and in 1956 a major epizootic began in Portugal and extended into Spain. Outbreaks of BT disease in the Middle East, Southeast Asia, southern Europe, and the United States in the early 1940s and 1950s led to its description as an emerging disease.52 Virus isolates, of 26 different serotypes, have been made in tropical, semitropical, and temperate zones of the world, including North and South America, Australia, southern Europe, Israel, Africa, and Southeast Asia. In 1998, significant outbreaks of BT occurred in Europe, initially in Greece and then in many other Mediterranean countries, causing the death of over a million sheep by 2005.96 In 2006 and 2007, BTV emerged in Northwestern Europe eventually expanding into the United Kingdom, Denmark, and the Czech Republic. An important factor in the distribution of BTV worldwide is the availability of suitable vectors, usually biting midges (gnats) of the Culicoides species.
Classification
The Orbivirus genus is one of 15 genera within the Reoviridae family, which includes vertebrate, arthropod, and plant pathogens. Despite a basic similarity, orbiviruses differ greatly in their structure, physicochemical properties, replication cycle, pathogenesis, and epidemiology. Unlike the reoviruses and rotaviruses, orbiviruses are arthropod-borne, and 22 serogroups or virus species of orbiviruses are distinguished in addition to several unclassified isolates.87 Within each serogroup, multiple serotypes are differentiated by neutralization tests. Different orbiviruses infect a wide range of vertebrate hosts, including ruminants (domesticated and wild), equids (domesticated and wild), rodents, bats, marsupials, birds, sloths, and primates, including humans. The orbivirus serogroups, serotypes, principal vectors, and main hosts are listed in e-Table 46.1 with their assigned abbreviations.87
Virion Structure
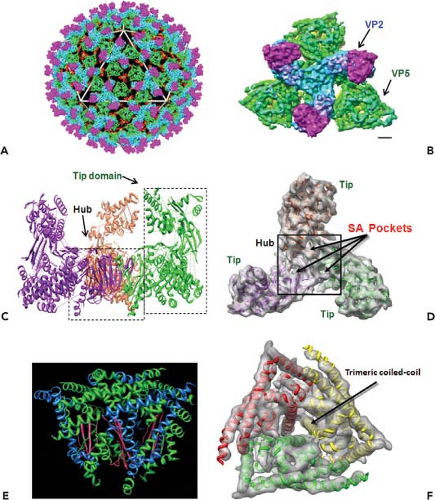
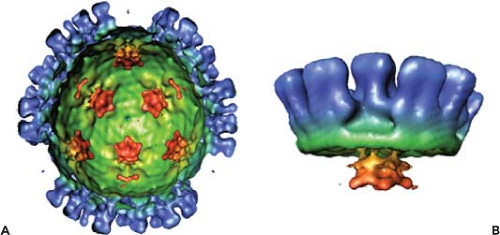
Orbiviruses are nonenveloped, icosahedral particles containing segmented, double-stranded RNA (dsRNA) genomes. BTV, the prototype of the genus, is the main subject of this chapter. BTV virions (550S) are architecturally complex structures composed of seven discrete proteins that are organized into two concentric shells, the inner and outer capsids138 (Fig. 46.1). The virion proteins encapsidate a genome of 10 dsRNA segments that can be divided into three different size classes (large, medium, and small). Unlike reovirus and rotavirus particles, mature BTV particles are relatively fragile, and the infectivity of BTV is easily lost in mildly acidic conditions. The outer capsid of BTV consists of two major proteins, VP2 and VP5, which are removed shortly after infection, to yield a transcriptionally active core (470S) particle (Fig. 46.1) which, in contrast to virions, is fairly robust. Cores contain two major proteins (VP7 and VP3), three minor proteins (VP1, VP4, and VP6), and the dsRNA genome.138 They may be further uncoated to form subcore particles (390S) that lack VP7. Cores can also be derived from virions in vitro by removal of the outer capsid.56,134
Virion Particle and Outer Capsid
Early electron microscopy (EM) visualizations were of characteristically fuzzy particles, suggesting BTV is highly sensitive to preparation conditions.24 However, cryo-electron microscopy (cryo-EM) and image reconstruction of BTV particles revealed an icosahedral symmetry of the virions with a diameter of ∼880 Å and an accurate positioning of VP2 and VP5 proteins in relation to each other and to the underlying core surface.49,91 The images revealed a total of 60 triskelion spike-like structures formed by VP2 trimers and 120 globular structures of VP5 trimers, both of which attach to the underlying core surface layer independently of each other. A recent 7 Å resolution cryo-EM structure of virion (Fig. 46.2) further identified the secondary structural elements and the topological arrangement of these two proteins.144 Each VP2 monomer has two distinct domains, a tip domain and a lower region, three of which form a hub domain at the base of the triskelion spike. The top of the VP2 tip domain is rich in β sheets and projects upward from the surface of the virion. The hub has a distinct β-barrel fold (Fig. 46.2C) and each monomer contains a sialic acid (SA) binding pocket (Fig. 46.2D).
In contrast to VP2, the VP5 trimer is globular, very rich in helices, and possesses only one β sheet (Fig. 46.2E). Five distinct amphipathic α-helical regions lie on the exterior surface of the protein, including an N-terminal α helical region that is positioned on the exposed surface of VP5.144 A cluster of three copies of one amphipathic helix are well positioned to expose their hydrophobic undersides to the endosomal membrane (Fig. 46.2E). Other amphipathic helices are similarly located on the exposed surface, where they may mimic the arrangement seen in other fusion proteins such as the fusion protein of the enveloped HIV (Fig. 46.2F). The gaps among the VP2 triskelions are filled by three VP5 trimers. Density maps suggest that VP5 trimers interact very weakly with the VP2 hub domains and with the underlying core surface, but both the VP2 tip and hub domains connect to their underlying VP7 trimers by stronger forces.
Core Particle and Proteins
Core particles derived from purified virus in vitro are stable and have been studied extensively at the structural level, initially by cryo-EM (at various resolutions between 22 Å and 40 Å) and subsequently by establishing a 3.6-Å atomic structure from crystals, which has revealed four distinct features.
Surface Layer of the Core and VP7 Trimers
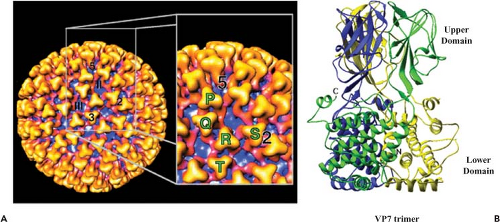
The core surface has a diameter of 73 nm with an icosahedral symmetry triangulation number of 13 (T = 13) in a left-handed configuration. The surface layer of VP7 trimers (appear as prominent triangular shapes) occupies the space between radii of 260 Å and 345 Å. The trimers are arranged around 132 distinctive channels as six-member rings, with five-member rings at the vertices (Fig. 46.3A). A total of 780 VP7 (38 kD) molecules per particle form 260 quasi-equivalent trimers the protomeric unit (P, Q, R, S, T), which can be seen clearly. Aqueous channels surrounded by these trimers are about 8 nm wide and 8 nm deep. Based on their location with respect to the icosahedral symmetry axes, they can be grouped into three types; type I channels run along the icosahedral fivefold axes, type II channels surround the fivefold axes, and type III channels are located around the icosahedral threefold axes. The arrangement of each monomer within the VP7 trimer was shown by x-ray crystallography.4,41 Each monomer (349 amino acids for BTV-10) consists of two distinct domains, “upper” and “lower,” which are twisted in such a way that the top domain of one monomer rests upon the lower domain of another (Fig. 46.3B). The smaller upper domain of VP7 forms the “head
region” of the trimer, which appears as a “knobbly” projection on the surface of the core and contains the central one-third of the polypeptide chain of the molecule (amino acids [aa] 121–249) folded into an anti-parallel β-sandwich. Two other x-ray structures of upper VP7 domains, one from BTV-10 (aa 123–253) and one from AHSV type 4, also revealed a similar structural arrangement. The lower, broader domain of the monomer is composed of nine α-helices and long extended loops, and is formed by both the N (aa 1–120) and the C (aa 250–349) termini of the molecule. A single lysine residue, prone to proteolysis, is situated at the junction between the two domains, and its mutation has a deleterious effect on core assembly.69 A low resolution (5.4 Å) VP7 structure indicated a very different orientation of the helical lower domain from that of the high-resolution structure, suggesting an “unwound” or “open” form compared to the tightly packed helices found in the high-resolution form.4 These two very different structures indicate that VP7 has the potential for internal flexibility in the lower domain and suggest that substantial conformational change in VP7 occurs at some stage in the viral life cycle.
region” of the trimer, which appears as a “knobbly” projection on the surface of the core and contains the central one-third of the polypeptide chain of the molecule (amino acids [aa] 121–249) folded into an anti-parallel β-sandwich. Two other x-ray structures of upper VP7 domains, one from BTV-10 (aa 123–253) and one from AHSV type 4, also revealed a similar structural arrangement. The lower, broader domain of the monomer is composed of nine α-helices and long extended loops, and is formed by both the N (aa 1–120) and the C (aa 250–349) termini of the molecule. A single lysine residue, prone to proteolysis, is situated at the junction between the two domains, and its mutation has a deleterious effect on core assembly.69 A low resolution (5.4 Å) VP7 structure indicated a very different orientation of the helical lower domain from that of the high-resolution structure, suggesting an “unwound” or “open” form compared to the tightly packed helices found in the high-resolution form.4 These two very different structures indicate that VP7 has the potential for internal flexibility in the lower domain and suggest that substantial conformational change in VP7 occurs at some stage in the viral life cycle.
X-ray crystallographic structures for core particles of two different BTV serotypes (BTV-1 and BTV-10) have confirmed the precise organization of the 260 VP7 trimers42 (Fig. 46.4A). The channels, or “pores,” are small (7 Å diameter) at the icosahedral threefold axes (class III) and slightly larger (9 Å diameter) at the fivefold axes (class I), where they act as “portals” to release the newly synthesized transcripts. Although the “T” trimers of VP7 sit tightly on the pores at the threefold axes, the “P” trimers are located loosely above the pores at the fivefold axes. At the interface between VP7 and VP3, the VP7 trimer layer is principally formed by side-to-side packing of short hydrophobic side chains, whereas interaction with the VP3 underneath layer is extensive and relatively tight. The lower domains of VP7 pack down against six corners of each VP3 molecule such that 12 monomers of VP7 (4 trimers) are in direct association with VP3 molecules. The thirteenth monomer of VP7 (organized in the icosahedron T = 13) is not directly in contact with a VP3 molecule, but is slightly raised and is in contact with monomers of adjacent trimers.
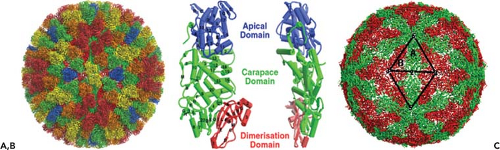
Arrangement of VP3 Molecules in the Inner Layer
The inner core layer, beneath the VP7 layer, is made up solely of VP3 proteins and occupies the space between radii 230 and 260 Å. Unlike the VP7 layer, the VP3 layer appears as an almost spherical structure as revealed by high-resolution x-ray structure of the cores.42,43,48,94
Sixty dimers of VP3 (103 kD) serve as the unit building block to form the disc-shaped VP3 shell of 59 nm diameter and with an internal diameter of 38 nm that contains both the genome, as well as the minor core proteins.42 The 120 molecules of the VP3 shell are organized as an icosahedral lattice with T = 2 symmetry (Fig. 46.4B) and are arranged as two sets (A and B) of 60 subunits (Fig. 46.4C). Five of the “A” group of molecules are arranged as a five-pointed star around each fivefold axis, and five “B” group molecules are positioned, one between each of the points of the star, but at a further distance from the fivefold axis to produce a decamer. Such icosahedral organization of the inner shell is shared by all members of Reoviridae and other viruses with segmented dsRNA genomes. The inner surface of the VP3 shell has relatively few charged residues and has a series of shallow grooves.
Structural Arrangement of Internal Minor Proteins
A clear density internal to the VP3 layer, in both the virion and the core cryo-EM reconstructions and in the x-ray structure, was assigned to the internal minor proteins (VP1, VP4, and VP6). In the absence of the viral genome, cryo-EM reconstruction shows the complex as a flower-shaped density directly beneath the icosahedral fivefold axes and attached to the underside of the VP3 layer91 (Fig. 46.5). It is not clear whether the VP6 protein is complexed with VP1 and VP4 at the fivefold vertices of the core; from in vitro studies, it can be deduced that the VP6 is tightly associated with the genomic RNA.
A modeled structure of VP1 has revealed a typical RNA dependent RNA polymerase (RdRp) organization with a distinct central polymerase domain (PD), an N-terminal domain (NTD), and a C-terminal (CTD) domain similar to that of Reovirus and rotavirus.140 The model was supported by mutagenesis combined with dissection of biological activity of each domain, and it is likely that all Reoviridae polymerases share similar structural organization.
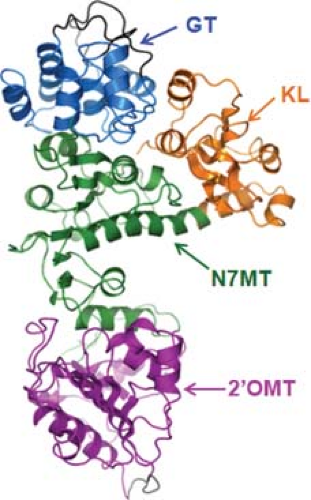
An atomic structure is available for VP4, the capping enzyme of the virus.80,97,119 It shows an elongated morphology composed of four domains, which includes an N-terminal kinase-like domain (KL); a center methyltransferase-1 domain (N7MT) and O2-methyltransferase-2 domain (O2MT), and a C-terminal Guanylyltransferase (GTase) domain (GT) (Fig. 46.6). The 2′-O-MT structure is similar to the catalytic domains of class I S-Adenosyl L-methionine (SAM) SAM-dependent methyltransferases and in particular to vaccinia virus VP39. A Lysine Aspartic Lysine Glutamic (KDKE) tetrad in the active site seems to be characteristic of RNA2′-O-MTases. The N-terminal 108 residues of VP4 show a kinase-like fold (KL domain), and is the least conserved domain among the capping enzymes of different orbivirus species, whereas the GT domain is the most highly conserved region of VP4 between different orbivirus species. Functional analyses indicate that
catalytic residues of the GT lie with the 98 aa of the C-terminus where a number of conserved lysine and histidine residues could form the guanine adduct required for GTase activity.
catalytic residues of the GT lie with the 98 aa of the C-terminus where a number of conserved lysine and histidine residues could form the guanine adduct required for GTase activity.
RNA Genome Seen in X-ray Structure of the Core
The electron density in the x-ray structure of the core exhibited features consistent with layers of highly ordered RNA.40 Approximately 80% of the genome can be modeled as four concentric layers that have center-to-center spacing between RNA strands of 26 to 30 Å. Grooves in the inner surface of the VP3 shell form a spiral around the fivefold axis with which the dsRNA layers appear to interact. The topography of the RNA molecules is uncertain, as the density detected in the inner layers of RNA gets progressively weaker, although each layer maintains an overall spiral organization.40
Table 46.1 Bluetongue Virus (BTV-10) Coding Assignments | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Genome Structure and Organization
The complete sequences of all 10 dsRNA segments were initially determined for BTV-1037 and subsequently for a number of BTV serotypes as well as for many other orbiviruses (see review, 87,108). For BTV-10, the sizes of the RNA segments range from 3,954 (segment 1) to 822 base pairs (bp) (segment 10) (e-Fig. 46.1) and the total genome is 19,220 bp long37 (Table 46.1). The 10 dsRNA segments of BTV have conserved terminal sequences with conserved 5′ (8 nucleotides) and 3′ (6 nucleotides) termini.99 The 5′ terminus of the coding strand of each duplex is capped and methylated, as are the BTV messenger RNA (mRNA) transcripts.
Apart from segment 1, the first AUG initiation codon in the mRNA strand of each segment initiates a single open reading frame (ORF). There are two methionine codons in the same reading frame in the S10 RNA sequence, coding for two related proteins: NS3 and NS3A.34,85 Recently an additional small non-structural protein has been identified, NSP4 encoded by S9, which also encodes the structured protein VP6.145 The encoded structural proteins are numbered VP1 to VP7 in the order of their migration on sodium dodecyl sulfate (SDS)–polyacrylamide gels. In addition to the seven structural proteins in virion particles, four nonstructural proteins (NS1, NS2, NS3, and NS3A) are synthesized in infected cells.108
Molecular Genetics
As early as 1981, the genetic diversity of the different BTV genome segments was identified by RNA oligonucleotide fingerprint analyses of field samples, and both genetic drift and shift were shown to contribute to BTV evolution.108 In some segments, variability was clearly linked to serotype (e.g., segments 2 and 5 encoding outer capsid proteins VP2 and VP5). In addition, oligonucleotide fingerprinting revealed that genome segment reassortment occurs readily between different BTV serotypes and within other orbivirus serogroups, but not between viruses belonging to different groups.39,107 Partly as a result of these data, individual orbivirus serogroups are now recognized as distinct virus species.87
Early molecular hybridization studies confirmed that genome segments 2 and 5 exhibited little or no cross-hybridization between serotypes, whereas the other eight RNA segments consistently showed some level of relatedness. Segments 1, 3, 4, 6, and 8 are relatively conserved, whereas segments 7 and 10 vary somewhat. Apart from segment 10 of AHSV, which appears to be even more variable than that of BTV, similar results have been obtained for AHSV.18,135
Recently, an extensive phylogenetic analysis of segments 2 and 5 from >300 different BTV isolates has shown that the nucleotide sequence of segment 2 varies from 29% to 59% between different serotypes.76,101 Based on this analysis, 26 serotypes were identified forming 10 distinct groups based on the fact that some serotypes are more related than others74,76 (e-Fig. 46.2A). Isolates within each serotype can also be separated into two major geographical groups, group E, from the East (India, Indonesia, China, Australia), and group W, from the West (Africa and North and South America).74 Far less variation occurs in segment 5, which encodes VP5, although it too shows some correlation with serotypes.76 A close genetic relationship has also been demonstrated among BTV, EHDV, and AHSV, three Culicoides transmitted Orbiviruses. Segments encoding the core proteins, for example, VP3, are the most conserved, whereas VP2 is the most variable60,102,141 (e-Fig. 46.2B). Despite as little as 17% to 23% identity, however, the VP2 protein of the three orbiviruses has a similar predicted secondary structure. Unlike VP2, the other outer capsid protein VP5 is more conserved among the three viruses, clearly indicating these viruses share strong phylogenetic relationships.
Stages of Virus Replication
The basic features of replication cycle of orbiviruses are similar to those of reoviruses and rotaviruses. Unlike reoviruses or rotaviruses, however, orbiviruses multiply in arthropod as well as in vertebrate hosts, and thus some stages of orbivirus replication and morphogenesis are unique. The overall information available on the BTV replication cycle and assembly is summarized in a schematic diagram (Fig. 46.7).
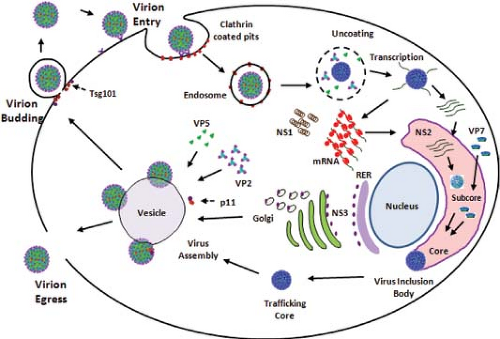 Figure 46.7. Replication cycle of bluetongue virus (BTV). Schematic diagram of an orbivirus replication cycle (deduced from bluetongue virus data). |
Virus Entry
Of the two outer capsid proteins, the larger VP2 (110 kD) is the receptor binding protein responsible for eliciting neutralizing antibodies and also has hemagglutination activity. Furthermore, recent structural data revealed that the VP2 trimer has two putative ligand-binding domains, one at the tip of the monomer and the other, an SA-binding pocket, at the base
of each trimer that binds type 2 oligosaccharide structures (Galâ14GlcNAcâ13) in glycan arrays.9,144 In BTV VP2 the SA binding domain is about 40 Å inward from the outer end of the tip domain, a distance well within the range of lengths of chains of sugars on glycoproteins.144 Because it is known that BTV particles agglutinate erythrocytes of ruminants20 and VP2 alone is responsible for this activity,46 it can be postulated that SA binding may promote adsorption of the virus onto the surface of erythrocytes, thereby increasing the probability of ingestion by the blood-feeding Culicoides vector.
of each trimer that binds type 2 oligosaccharide structures (Galâ14GlcNAcâ13) in glycan arrays.9,144 In BTV VP2 the SA binding domain is about 40 Å inward from the outer end of the tip domain, a distance well within the range of lengths of chains of sugars on glycoproteins.144 Because it is known that BTV particles agglutinate erythrocytes of ruminants20 and VP2 alone is responsible for this activity,46 it can be postulated that SA binding may promote adsorption of the virus onto the surface of erythrocytes, thereby increasing the probability of ingestion by the blood-feeding Culicoides vector.
The attachment of VP2 leads to receptor-mediated endocytosis of virions in clathrin-coated vesicles (Fig. 46.7). Clathrin is subsequently lost, and large endocytic vesicles are formed. Both VP2 and VP5 proteins can be identified within the early endosome where either one or both proteins are degraded.22,31,45 Compounds that raise the lysosomal or endosomal pH (e.g., ammonium chloride) prevent endocytosis of virus particle and the subsequent release of the uncoated cores into the cytoplasm.30
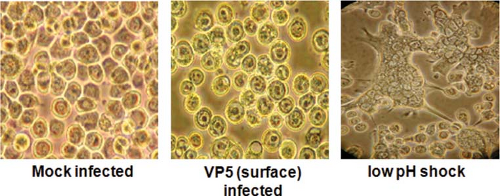
As discussed above, VP5 shares certain structural features with the fusion proteins of enveloped viruses, consistent with membrane penetration activity.144 Peptides representing the two amino terminal amphipathic helices of VP5 cause leakiness of the membrane, and the full-length protein triggers strong syncytia and multinuclear cells when localized to the plasma membrane and treated with low pH31,46 (Fig. 46.8). Therefore, VP5 functionally substitutes for a typical viral fusion protein, although it does not require proteolytic activation to render it fully functional as in the case of reovirus protein.
Combining the available structural and biochemical data, a plausible hypothesis would be that upon receptor binding and entry VP2 trimers undergo rearrangement or degradation, allowing the exposure of VP5 trimers and thereby allowing the VP5 amphipathic helices to freely interact with membranes and initiate the permeabilization process.
In summary, VP2 makes an initial contact with the mammalian host cell and VP5 mediates the penetration of the host cell membrane by destabilizing the endosomal membrane. Core particles lacking both VP2 and VP5 proteins are released into the cytoplasm from the endosomal vesicles. In contrast to mammalian host, BTV cores lacking VP2 and VP5 are infectious for invertebrate cells86 with the Arginine Glycine Aspartic (RGD) motif of VP7 involved in entry into cells of Culicoides species.122
Transcription and Replication
After cell entry, the outer capsid is removed to release a transcriptionally active core particle, which provides a compartment within which the 10 genome segments are repeatedly transcribed by the core-associated enzymes VP1, VP4, and VP6.134,139 Ten capped nonpolyadenylated mRNAs are synthesized from the 10 genome segments and are released from the core particle into the host cell cytoplasm where they act as templates both for translation and for negative-strand viral RNA synthesis to generate genomic dsRNAs.85,133,134 Transfection of mammalian cells with in vitro BTV transcripts also leads to generation of viral proteins and infectious virus particles, as demonstrated by the passage of infectivity in Baby Hampster Kidney cells (BHK) cells.12 These findings have allowed the development of a helper virus-independent reverse genetics system for BTV and other orbiviruses and for the generation of targeted mutant viruses.12,81
When intact cores isolated from virus particles are activated in vitro in the presence of magnesium ions and NTP substrates, distinct conformational changes can be seen around the fivefold axis.40 This is interpreted as an outward movement of VP3 and VP7, allowing opening of a pore in the VP3 layer at the fivefold axes through which the mRNAs are extruded. The structural integrity of the core particle appears to be essential for efficient transcriptional activity in vivo, but it has been possible to use in vitro assays with the three minor proteins of the core to delineate the specific roles of each (see review in 103; Fig. 46.9).
The Largest Protein VP1 Is the RNA-Dependent RNA Polymerase
The first indication that the largest BTV protein, VP1 (149.5 kD) is the virus polymerase protein came from sequence comparison with other DNA and RNA polymerases and from an in vitro polymerase assay that used a crude extract of insect cells infected with a recombinant baculovirus expressing only BTV VP1.37,131 Subsequent in vitro studies have used purified recombinant protein and have shown that VP1 exhibits a processive replicase activity in the absence of any other viral proteins and initiates BTV minus-strand synthesis de novo.13 Recombinant VP1 copies each of the plus-strand RNA segments fully, from the smallest (822 nt) to the largest BTV plus-strand RNA of 3,954 nt, either individually or simultaneously in a single in vitro reaction, to produce a dsRNA duplex that is RNase I resistant, and RNase III sensitive. Synthesis of dsRNAs from capped single stranded RNA (ssRNA) templates is significantly higher than from uncapped ssRNA templates and could be further enhanced by priming with complementary terminal dinucleotide. Some evidence for template specificity was suggested by the fact dsRNAs were not synthesized from nonviral ssRNA templates unless they were fused to specific BTV sequences. These data contrast those of rotavirus and reovirus; in rotavirus the assigned polymerase protein (VP1) is only active in the context of the inner shell protein VP2,128 and recombinant reovirus polymerase λ3 is generally incapable of synthesizing de novo reovirus RNA, although short RNA transcripts were observed.113,123
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree