9
Laboratory Diagnosis
CHAPTER CONTENTS
APPROACH TO LABORATORY DIAGNOSIS
The laboratory diagnosis of infectious diseases involves two main approaches: one is the bacteriologic approach, in which the organism is identifixded by staining and culturing the organism, and the other is the immunologic (serologic) approach, in which the organism is identified by detection of antibodies against the organism in the patient’s serum.
In the bacteriologic approach to the diagnosis of infectious diseases, several important steps precede the actual laboratory work, namely, (1) choosing the appropriate specimen to examine, which requires an understanding of the pathogenesis of the infection; (2) obtaining the specimen properly to avoid contamination from the normal flora; (3) transporting the specimen promptly to the laboratory or storing it correctly; and (4) providing essential information to guide the laboratory personnel.
In general, there are three approaches to the bacteriologic laboratory work:
(1) Observing the organism in the microscope after staining.
(2) Obtaining a pure culture of the organism by inoculating it onto a bacteriologic medium.
(3) Identifying the organism by using biochemical reactions, growth on selective media, DNA probes, or specific antibody reactions. Which of these approaches are used and in what sequence depend on the type of specimen and organism. After the organism is grown in pure culture, its sensitivity to various antibiotics is determined by procedures described in Chapter 11.
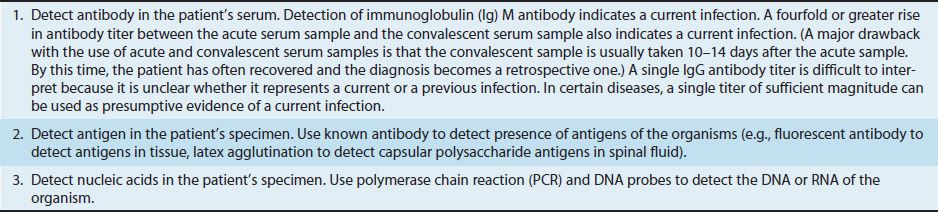
A general approach to the diagnosis of a bacterial infection is described in Table 9–1. This approach emphasizes the importance of performing a Gram stain and obtaining a “pure culture” of the organism. However, sometimes the organism is not recovered by culturing, and other techniques must be used. Table 9–2 describes some approaches to making a diagnosis when the cultures are negative. One approach that is commonly used is serologic testing, which determines the presence of antibodies specific for the organism. In most cases, a fourfold rise in antibody titer between the acute- and convalescent-phase serum samples is considered to be significant.
TABLE 9–1 General Approach to the Diagnosis of a Bacterial Infection
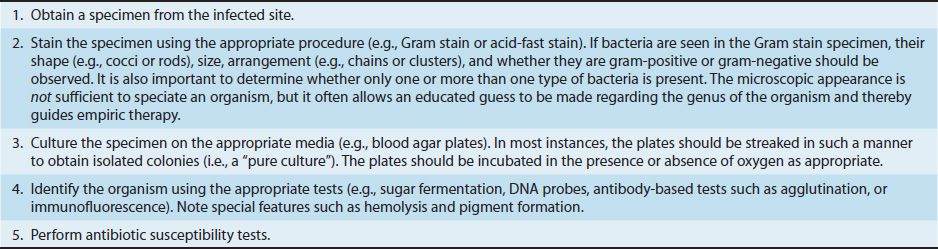
TABLE 9–2 How to Diagnose a Bacterial Infection When the Culture Is Negative
Obtaining a pure culture involves culturing the organism on bacteriologic agar. Initially, blood agar is used because it supports the growth of many bacteria and the type of hemolysis can be observed.
Blood agar contains red blood cells, but it should be noted that viruses and obligate intracellular bacteria, such as Chlamydia and Rickettsia, will not grow on blood agar. Red blood cells do not have a functioning nucleus and therefore are incapable of supporting the growth of either viruses or the obligate intracellular bacteria.
Blood agar contains inhibitors for certain bacteria, such as members of the Neisseria and Haemophilus genera, and the blood must be heated to inactivate these inhibitors. These bacteria therefore are grown on cooked blood agar or chocolate agar (so named because the heated blood turns a chocolate color). Other media contain either specific growth factors required for the bacteria to grow or antibiotics that inhibit normal flora, which allows the pathogenic bacteria to obtain sufficient nutrients to grow.
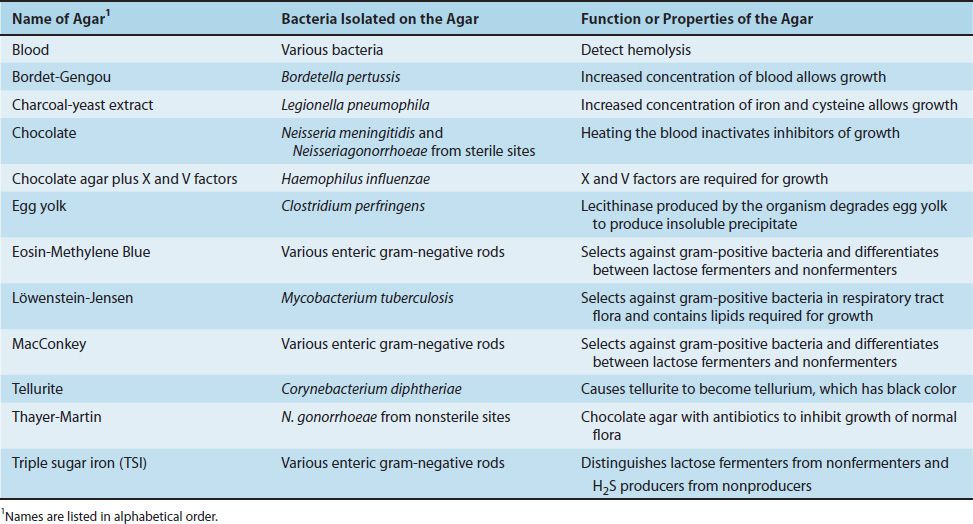
Certain other media, called “selective, differential” media, are often used. These media are selective because they contain compounds that selectively allow certain bacteria to grow and differential because they contain other compounds that allow one type of bacteria to be differentiated from another based on some biochemical reaction. Table 9–3 contains a list of various bacteriologic agars commonly used in the diagnostic laboratory and the function of these agars.
TABLE 9–3 Commonly Used Bacteriologic Agars and Their Function
BACTERIOLOGIC METHODS
Blood Cultures
Blood cultures are performed most often when sepsis, endocarditis, osteomyelitis, meningitis, or pneumonia is suspected. The organisms most frequently isolated from blood cultures are two gram-positive cocci, Staphylococcus aureus and Streptococcus pneumoniae, and three gram-negative rods, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa.
It is important to obtain at least three 10-mL blood samples in a 24-hour period because the number of organisms can be small and their presence intermittent. The site for venipuncture must be cleansed with 2% iodine to prevent contamination by members of the flora of the skin, usually Staphylococcus epidermidis. The blood obtained is added to 100 mL of a rich growth medium such as brain–heart infusion broth. Whether one or two bottles are inoculated varies among hospitals. If two bottles are used, one is kept under anaerobic conditions and the other is not. If one bottle is used, the low oxygen tension at the bottom of the bottle permits anaerobes to grow.
Blood cultures are checked for turbidity or for CO2 production daily for 7 days or longer. If growth occurs, Gram stain, subculture, and antibiotic sensitivity tests are performed. If no growth is observed after 1 or 2 days, blind subculturing onto other media may reveal organisms. Cultures should be held for 14 days when infective endocarditis, fungemia, or infection by slow-growing bacteria (e.g., Brucella) is suspected.
Throat Cultures
Throat cultures are used primarily to detect the presence of group A β-hemolytic streptococci (Streptococcus pyogenes), an important and treatable cause of pharyngitis. They are also used when diphtheria, gonococcal pharyngitis, or thrush (Candida) is suspected.
When the specimen is being obtained, the swab should touch not only the posterior pharynx, but also both tonsils or tonsillar fossae as well. The material on the swab is inoculated onto a blood agar plate and streaked to obtain single colonies. If colonies of β-hemolytic streptococci are found after 24 hours of incubation at 35°C, a bacitracin disk is used to determine whether the organism is likely to be a group A streptococcus. If growth is inhibited around the disk, it is a group A streptococcus; if not, it is a non–group A β-hemolytic streptococcus.
Note that a Gram stain is typically not done on a throat swab because it is impossible to distinguish between the appearance of the normal flora streptococci and S. pyogenes.
Sputum Cultures
Sputum cultures are performed primarily when pneumonia or tuberculosis is suspected. The most frequent cause of community-acquired pneumonia is S. pneumoniae, whereas S. aureus and gram-negative rods, such as K. pneumoniae and P. aeruginosa, are common causes of hospital-acquired pneumonias.
It is important that the specimen for culture really be sputum, not saliva. Examination of a gram-stained smear of the specimen frequently reveals whether the specimen is satisfactory. A reliable specimen has more than 25 leukocytes and fewer than 10 epithelial cells per 100× field. An unreliable sample can be misleading and should be rejected by the laboratory. If the patient cannot cough and the need for a microbiologic diagnosis is strong, induction of sputum, transtracheal aspirate, bronchial lavage, or lung biopsy may be necessary. Because these procedures bypass the normal flora of the upper airway, they are more likely to provide an accurate microbiologic diagnosis. A preliminary assessment of the cause of the pneumonia can be made by Gram stain if large numbers of typical organisms are seen.
Culture of the sputum on blood agar frequently reveals characteristic colonies, and identification is made by various serologic or biochemical tests. Cultures of Mycoplasma are infrequently done; diagnosis is usually confirmed by a rise in antibody titer. If Legionella pneumonia is suspected, the organism can be cultured on charcoal-yeast agar, which contains the high concentrations of iron and sulfur required for growth.
If tuberculosis is suspected, an acid-fast stain should be done immediately and the sputum cultured on special media, which are incubated for at least 6 weeks. In diagnosing aspiration pneumonia and lung abscesses, anaerobic cultures are important.
Spinal Fluid Cultures
Spinal fluid cultures are performed primarily when meningitis is suspected. Spinal fluid specimens from cases of encephalitis, brain abscess, and subdural empyema usually show negative cultures. The most important causes of acute bacterial meningitis are three encapsulated organisms: Neisseria meningitidis, S. pneumoniae, and Haemophilus influenzae.
Because acute meningitis is a medical emergency, the specimen should be taken immediately to the laboratory. The gram-stained smear of the sediment of the centrifuged sample guides the immediate empirical treatment. If organisms resembling N. meningitidis, H. influenzae, or S. pneumoniae are seen, the quellung test or immunofluorescence with specific antisera can identify the organism rapidly. Cultures are done on blood and on chocolate agar and incubated at 35°C in a 5% CO2 atmosphere. Hematin and nicotinamide adenine dinucleotide (NAD) (factors X and V, respectively) are added to enhance the growth of H. influenzae.
In cases of subacute meningitis, Mycobacterium tuberculosis and the fungus Cryptococcus neoformans are the most common organisms isolated. Acid-fast stains of the spinal fluid should be performed, although M. tuberculosis may not be seen, because it can be present in small numbers. The fluid should be cultured and the cultures held for a minimum of 6 weeks. C. neoformans, a budding yeast with a prominent capsule, can be seen in spinal fluid when India ink is used.
Immunologic tests to detect the presence of capsular antigen in the spinal fluid can be used to identify N. meningitidis, S. pneumoniae, H. influenzae, group B streptococci, E. coli, and C. neoformans. The two tests most frequently used are latex particle agglutination and counterimmunoelectrophoresis.
Stool Cultures
Stool cultures are performed primarily for cases of enterocolitis. The most common bacterial pathogens causing diarrhea in the United States are Shigella, Salmonella, and Campylobacter. E. coli O157 strains are also an important cause of diarrhea.
A direct microscopic examination of the stool can be informative from two points of view: (1) a methylene blue stain that reveals many leukocytes indicates that an invasive organism rather than a toxigenic one is involved, and (2) a Gram stain may reveal large numbers of certain organisms, such as staphylococci, clostridia, or campylobacters. Gram stain of the stool is not usually done because the large numbers of bacteria in the normal flora of the colon make the interpretation difficult.
For culture of Salmonella and Shigella, a selective, differential medium such as MacConkey or Eosin–Methylene Blue (EMB) agar is used. These media are selective because they allow gram-negative rods to grow but inhibit many gram-positive organisms. Their differential properties are based on the fact that Salmonella and Shigella do not ferment lactose, whereas many other enteric gram-negative rods do. On EMB agar, colonies of E. coli, a lactose fermenter, appear purple and have a green sheen. In contrast, colonies of non–lactose fermenters, such as Salmonella and Shigella, appear colorless.
If non–lactose-fermenting colonies are found, a triple sugar iron (TSI) agar slant is used to distinguish Salmonella from Shigella. Some species of Proteus resemble Salmonella on TSI agar but can be distinguished because they produce the enzyme urease, whereas Salmonella does not. The organism is further identified as either a Salmonella or a Shigella species by using a specific antisera to the organism’s cell wall O antigen in an agglutination test. This is usually done in hospital laboratories, but precise identification of the species is performed in public health laboratories.
Campylobacter jejuni is cultured on antibiotic-containing media (e.g., Skirrow’s agar) at 42°C in an atmosphere containing 5% O2 and 10% CO2. It grows well under these conditions, unlike many other intestinal pathogens. Although the techniques are available, stool cultures are infrequently performed for organisms such as Yersinia enterocolitica, Vibrio parahaemolyticus, and enteropathic or toxigenic E. coli. Despite the presence of large numbers of anaerobes in feces, they are rarely pathogens in the intestinal tract, and anaerobic cultures of stool specimens are therefore unnecessary.
Stool specimens that are grossly bloody are typically cultured on MacConkey-sorbitol media. E. coli O157 strains do not ferment sorbitol and appear as colorless colonies, whereas typical E. coli strains do ferment sorbitol and appear red.
Urine Cultures
Urine cultures are performed primarily when pyelonephritis or cystitis is suspected. By far the most frequent cause of urinary tract infections is E. coli. Other common agents are Enterobacter, Proteus, and Enterococcus faecalis.
Urine in the bladder of a healthy person is sterile, but it acquires organisms of the normal flora as it passes through the distal portion of the urethra. To avoid these organisms, a midstream specimen, voided after washing the external orifice, is used for urine cultures. In special situations, suprapubic aspiration or catheterization may be required to obtain a specimen. Because urine is a good culture medium, it is essential that the cultures be done within 1 hour after collection or stored in a refrigerator at 4°C for no more than 18 hours.
It is commonly accepted that a bacterial count of at least 100,000/mL must be found to conclude that significant bacteriuria is present (in asymptomatic persons). There is evidence that as few as 100/mL are significant in symptomatic patients. For this determination to be made, quantitative or semiquantitative cultures must be performed. There are several techniques: (1) A calibrated loop that holds 0.001 mL of urine can be used to streak the culture; (2) serial 10-fold dilutions can be made and samples from the dilutions streaked; and (3) a screening procedure suitable for the physician’s office involves an agar-covered “paddle” that is dipped into the urine—after the paddle is incubated, the density of the colonies is compared with standard charts to obtain an estimate of the concentration of bacteria.
Genital Tract Cultures
Genital tract cultures are performed primarily on specimens from individuals with an abnormal discharge or on specimens from asymptomatic contacts of a person with a sexually transmitted disease. One of the most important pathogens in the genital tract is Neisseria gonorrhoeae. The laboratory diagnosis of gonorrhea is made by microscopic examination of a gram-stained smear and by culture of the organism.
Specimens are obtained by swabbing the urethral canal (for men), the cervix (for women), or the anal canal (for men and women). A urethral discharge from the penis is frequently used. Because N. gonorrhoeae is very delicate, the specimen should be inoculated directly onto a Thayer-Martin chocolate agar plate or onto a special transport medium (e.g., Trans-grow).
Gram-negative diplococci found intracellularly within neutrophils on a smear of a urethral discharge from a man have over 90% probability of being N. gonorrhoeae. Because smears are less reliable when made from swabs of the endocervix and anal canal, cultures are necessary. The finding of only extracellular diplococci suggests that these neisseriae may be members of the normal flora and that the patient may have nongonococcal urethritis.
Nongonococcal urethritis and cervicitis are also extremely common infections. The most frequent cause is Chlamydia trachomatis, which cannot grow on artificial medium but must be grown in living cells. For this purpose, cultures of human cells or the yolk sacs of embryonated eggs are used. The finding of typical intracytoplasmic inclusions when using Giemsa stain or fluorescent antibody is diagnostic. Because of the difficulty of culturing C. trachomatis, nonbacteriologic methods, such as enzyme-linked immunosorbent assay (ELISA) to detect chlamydial antigens in exudates or urine or DNA probe assays to detect chlamydial nucleic acids, are now often used to diagnose sexually transmitted diseases caused by this organism.
Because Treponema pallidum, the agent of syphilis, cannot be cultured, diagnosis is made by microscopy and serology. The presence of motile spirochetes with typical morphologic features seen by darkfield microscopy of the fluid from a painless genital lesion is sufficient for the diagnosis. The serologic tests fall into two groups: (1) the nontreponemal antibody tests such as the Venereal Disease Research Laboratory (VDRL) or rapid plasma reagin (RPR) test and (2) the treponemal antibody tests such as the fluorescent treponemal antibody-absorption (FTA-ABS) test. These tests are described on pages 66 and 198.
Wound & Abscess Cultures
A great variety of organisms are involved in wound and abscess infections. The bacteria most frequently isolated differ according to anatomic site and predisposing factors. Abscesses of the brain, lungs, and abdomen are frequently caused by anaerobes such as Bacteroides fragilis and gram-positive cocci such as S. aureus and S. pyogenes. Traumatic open-wound infections are caused primarily by members of the soil flora such as Clostridium perfringens; surgical-wound infections are usually due to S. aureus. Infections of dog or cat bites are commonly due to Pasteurella multocida, whereas human bites primarily involve the mouth anaerobes.
Because anaerobes are frequently involved in these types of infection, it is important to place the specimen in anaerobic collection tubes and transport it promptly to the laboratory. Because many of these infections are due to multiple organisms, including mixtures of anaerobes and nonanaerobes, it is important to culture the specimen on several different media under different atmospheric conditions. The Gram stain can provide valuable information regarding the range of organisms under consideration.
IMMUNOLOGIC METHODS
These methods are described in more detail in Chapter 64. However, it is of interest here to present information on how serologic reactions aid the microbiologic diagnosis. There are essentially two basic approaches: (1) using known antibody to identify the microorganism, and (2) using known antigens to detect antibodies in the patient’s serum.
Identification of an Organism with Known Antiserum
Capsular Swelling (Quellung) Reaction
Several bacteria can be identified directly in clinical specimens by this reaction, which is based on the microscopic observation that the capsule swells in the presence of homologous antiserum. Antisera against the following organisms are available: all serotypes of S. pneumoniae (Omniserum), H. influenzae type b, and N. meningitidis groups A and C.
Slide Agglutination Test
Antisera can be used to identify Salmonella and Shigella by causing agglutination (clumping) of the unknown organism. Antisera directed against the cell wall O antigens of Salmonella and Shigella are commonly used in hospital laboratories. Antisera against the flagellar H antigens and the capsular Vi antigen of Salmonella are used in public health laboratories for epidemiologic purposes.
Latex Agglutination Test
Latex beads coated with specific antibody are agglutinated in the presence of the homologous bacteria or antigen. This test is used to determine the presence of the capsular antigen of H. influenzae, N. meningitidis, several species of streptococci, and the yeast C. neoformans.
Counterimmunoelectrophoresis Test
In this test, the unknown bacterial antigen and a known specific antibody move toward each other in an electrical field. If they are homologous, a precipitate forms within the agar matrix. Because antibodies are positively charged at the pH of the test, only negatively charged antigens, usually capsular polysaccharides, can be assayed. The test can be used to detect the presence in the spinal fluid of the capsular antigens of H. influenzae, N. meningitidis, S. pneumoniae, and group B streptococci.
Enzyme-Linked Immunosorbent Assay
In this test, a specific antibody to which an easily assayed enzyme has been linked is used to detect the presence of the homologous antigen. Because several techniques have been devised to implement this principle, the specific steps used cannot be detailed here (see Chapter 64). This test is useful in detecting a wide variety of bacterial, viral, and fungal infections.
Fluorescent Antibody Tests
A variety of bacteria can be identified by exposure to known antibody labeled with fluorescent dye, which is detected visually in the ultraviolet microscope. Various methods can be used, such as the direct and indirect techniques (see Chapter 64).
Identification of Serum Antibodies with Known Antigens
Slide or Tube Agglutination Test
In this test, serial twofold dilutions of a sample of the patient’s serum are mixed with standard bacterial suspensions. The highest dilution of serum capable of agglutinating the bacteria is the titer of the antibody. As with most tests of a patient’s antibody, at least a fourfold rise in titer between the early and late samples must be demonstrated for a diagnosis to be made. This test is used primarily to aid in the diagnosis of typhoid fever, brucellosis, tularemia, plague, leptospirosis, and rickettsial diseases.
Serologic Tests for Syphilis
The detection of antibody in the patient’s serum is frequently used to diagnose syphilis, because T. pallidum does not grow on laboratory media. There are two kinds of tests.
(1) The nontreponemal tests use a cardiolipin–lecithin–cholesterol mixture as the antigen, not an antigen of the organism. Cardiolipin (diphosphatidylglycerol) is a lipid extracted from normal beef heart. Flocculation (clumping) of the cardiolipin occurs in the presence of antibody to T. pallidum. The VDRL and RPR tests are nontreponemal tests commonly used as screening procedures. They are not specific for syphilis but are inexpensive and easy to perform.
The treponemal tests use T. pallidum as the antigen. The two most widely used treponemal tests are the FTA-ABS and the MHA-TP (microhemagglutination–Treponema pallidum) tests. In the FTA-ABS test, the patient’s serum sample, which has been absorbed with treponemes other than T. pallidum to remove nonspecific antibodies, is reacted with nonviable T. pallidum on a slide. Fluorescein-labeled antibody against human immunoglobulin G (IgG) is then used to determine whether IgG antibody against T. pallidum is bound to the organism. In the MHA-TP test, the patient’s serum sample is reacted with sheep erythrocytes coated with antigens of T. pallidum. If antibodies are present, hemagglutination occurs.
Cold Agglutinin Test
Patients with Mycoplasma pneumoniae infections develop autoimmune antibodies that agglutinate human red blood cells in the cold (4°C) but not at 37°C. These antibodies occur in certain diseases other than Mycoplasma infections; thus false-positive results can occur.
NUCLEIC ACID–BASED METHODS
There are three types of nucleic acid–based tests used in the diagnosis of bacterial diseases: nucleic acid amplification tests, nucleic acid probes, and nucleic acid sequence analysis. Nucleic acid–based tests are highly specific, quite sensitive (especially the amplification tests), and much faster than culturing the organism. These tests are especially useful for those bacteria that are difficult to culture, such as Chlamydia and Mycobacterium species.
Nucleic acid amplification tests utilize the polymerase chain reaction (PCR) or other amplifying process to increase the number of bacteria-specific DNA or RNA molecules so the sensitivity of the test is significantly higher than that of unamplified tests. Many bacteria can be identified using these tests, but they are especially useful in detecting Chlamydia trachomatis and Neisseria gonorrhoeae in urine samples in sexually transmitted disease (STD) clinics.
Tests that use nucleic acid probes are designed to detect bacterial DNA or RNA directly (without amplification) using a labeled DNA or RNA probe that will hybridize specifically to the bacterial nucleic acid. These tests are simpler to perform than the amplification tests but are less sensitive.
Nucleic acid sequence analysis is used to identify bacteria based on the base sequence of the organism’s ribosomal RNA. An organism that has never been cultured, Tropheryma whipplei, was identified using this approach.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


