Henipaviruses
Lin-Fa Wang
John S. Mackenzie
Christopher C. Broder
History
Hendra virus (HeV), the first known member of the genus Henipavirus in the family Paramyxoviridae, came to light in September 1994 as the causative agent of a sudden outbreak of acute respiratory disease in thoroughbred horses at a stable in Brisbane, Australia. A total of 21 horses and 2 humans (a horse trainer and a stable hand) became infected. The horse trainer and 14 horses died, and 7 horses with mild or subclinical infection were killed.130 A virus was isolated, called equine morbillivirus but later named Hendra virus after the Brisbane suburb where the outbreak occurred. A second person died from HeV infection 13 months after the Brisbane outbreak, a farmer from Mackay, nearly 1000 km north of Brisbane. Unlike the first case, however, the man succumbed to encephalitis caused by HeV infection.107 A forensic investigation found that the farmer had suffered a mild meningitic illness 14 months earlier after assisting at the necropsy of two horses that had died of severe respiratory distress—later found to have been caused by HeV125—and that he became infected at that time. After initial serologic evidence suggested that fruit bats (flying foxes) of the genus Pteropus in the suborder Megachiroptera were the reservoir hosts,165 HeV was isolated from two species of flying fox.56 In total, there have been 14 recognized occurrences of HeV in Australia between 1994–2010, with at least one occurrence per year since 2006.5,10 Every occurrence of HeV has involved horses as the initial infected host, causing lethal respiratory disease and encephalitis, along with a total of seven human cases arising from exposure to infected horses, among which four have been fatal and the most recent in 2009.6,10,115
Nipah virus (NiV), the second known member of the genus Henipavirus, emerged as the cause of an outbreak of disease in pigs and humans in Peninsular Malaysia in 1998 through 1999. The epidemic started in Perak State as clusters of cases of encephalitis among pig farmers. It was initially believed to be caused by Japanese encephalitis virus; however, various features of the outbreak, including a high proportion of cases in direct contact with pigs and illness and deaths in pigs, differed from those expected with Japanese encephalitis.30 Indeed, respiratory illness and encephalitis in pigs preceded human cases in the same district.97 The epidemic spread south to the intensive pig-farming areas of Negeri Sembilan in December 1998 and subsequently peaked between February and April 1999. More than 1 million pigs were destroyed to halt the spread of the epidemic, and by late May, 265 human cases of acute encephalitis with 105 deaths were recorded.29,30 A cluster of 11 cases with 1 death occurred among abattoir workers in Singapore.114 In early March 1999, a virus was isolated from the cerebrospinal fluid (CSF) of a patient with encephalitis and identified as the etiologic agent.29,30 Named Nipah virus after the village from which the patient had come, it was shown to be closely related to HeV. NiV was subsequently isolated from the urine of Malaysian flying foxes.33 The virus re-emerged in Bangladesh in 2001,68 and outbreaks of NiV-related encephalitis have occurred in people from that country almost every year since, along with two reports of NiV encephalitis in India23,60 and the most recent occurrence in early 2011 in Bangladesh.7 The human case fatality rate of these occurrences of NiV has approached 75%.68,85
Infectious Agent
Classification
When HeV was first isolated in 1994, partial sequencing of the matrix gene (M) revealed that it most closely resembled members of the genus Morbillivirus in the subfamily Paramyxovirinae.104 Subsequent characterization of the full-length genome, however, revealed that many of the genetic features of HeV were unique among paramyxoviruses and that the virus did not fit within any of the three genera existing at that time, Morbillivirus, Respirovirus, and Rubulavirus.147,148 When NiV was isolated in 1999, it was initially described as Hendra-like on the basis of its strong reactivity with anti-HeV antibodies. Later it was shown that sera raised against HeV were able to neutralize NiV and vice versa, and that both viruses shared a high degree of similarity in genome organization and protein size and sequence.29,58,59,146 In 2002, the genus Henipavirus was created to accommodate these novel paramyxoviruses, and HeV was designated the type species.75 Currently, the genus Henipavirus contains two virus species and several strains isolated from humans, bats, horses, and pigs over a wide geographic area and spanning a period of 15 years (Table 37.1). The susceptibility of humans, the virulence of the viruses, and absence of therapeutics and vaccines led to classification of HeV and NiV as biosafety level 4 (BSL4) pathogens.
Table 37.1 Summary of Henipaviruses Isolated from Different Species and Geographic Locations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Propagation in Cell Culture and Cytopathic Effect
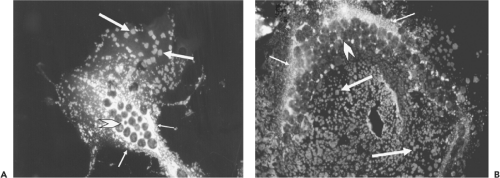
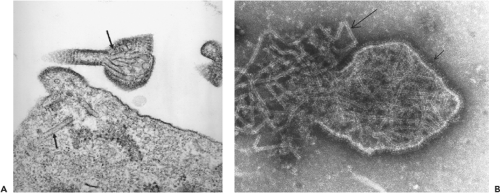
The ultrastructural characteristics of henipavirus-infected cells resemble those found in cells infected by other members of the Paramyxovirinae. Shared features include generation of large syncytia and the presence of viral nucleocapsids in cytoplasmic inclusion bodies and underlying electron-dense areas of the plasma membrane.49,69 In Vero cells, NiV-induced syncytia are significantly larger than those generated by HeV and nuclei and nucleocapsids are frequently located at the cell periphery, compared with HeV-induced syncytia, where they tend to be more centrally located or distributed randomly throughout the cytoplasm (Fig. 37.1). Henipavirus-infected cells also contain structures that are not seen with other paramyxoviruses—specifically a network of membrane-like reticular structures in the cytoplasm and long tubules that appear to be continuous with the plasma membrane in NiV-infected cells. Tubules can also be observed in NiV virions (Fig. 37.2A). In situ hybridization suggests that
these reticular structures contain viral RNA and may play a role in viral transcription.49
these reticular structures contain viral RNA and may play a role in viral transcription.49
Virus Morphology
Henipavirus particles are pleomorphic, varying from spherical to filamentous and ranging in size from 40 to 1,900 nm.49,69,104 Nucleocapsids have a diameter of 18 to 19 nm with an average pitch of 5 nm. When examined by electron microscopy (EM), HeV has a unique double-fringed appearance, caused by the presence of surface projections 15 ± 1 nm and 8 ± 1 nm in length (see Fig. 37.2B). Approximately 95% of virions contain the double fringe, and the remaining 5% display a uniform fringe length of 15 ± 1 nm. Unlike HeV, NiV possesses a single layer of surface projections with an average length of 17 ± 1 nm, and NiV particles released into the culture medium are difficult to image because they are routinely penetrated by negative stains. This suggests that the viruses may differ in the physical nature of their envelope.69
Genome Length and Organization
In the subfamily Paramyxovirinae, the genome length of all characterized viruses is divisible by six, an observation caused by the requirement of each N protein in the viral ribonucleoprotein to bind 6 nucleotide (nt) residues.76 This is also true
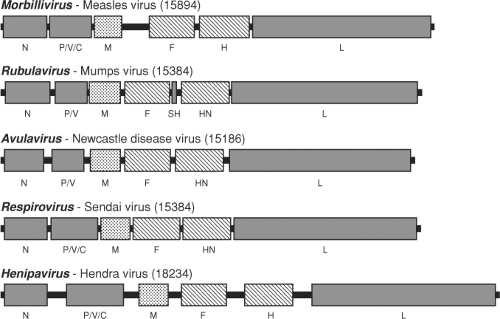
for HeV and NiV despite their much larger genome sizes.146 The genomes of the Malaysian and Bangladesh strains of NiV differ by 6 nt because of a 6-nt increase in the 3′ untranslated region of the F gene.57 A minigenome replicon study confirms that NiV complies with the rule of six.55 When the complete genome sequence of HeV was determined, its length (18,234 nt) was more than 2,700 nt, or 15% longer than the genomes of all other paramyxoviruses known at that time.148 The size of the NiV genome at 18,246 nt to 18,252 nt is slightly larger than that of HeV.57,58,59 The extra length of the henipavirus genome is in the form of unique, long untranslated sequences at the 3′ end of five of the six genes. A comparison of genome length and untranslated regions of representative members of the Paramyxovirinae is shown in Figure 37.3.
for HeV and NiV despite their much larger genome sizes.146 The genomes of the Malaysian and Bangladesh strains of NiV differ by 6 nt because of a 6-nt increase in the 3′ untranslated region of the F gene.57 A minigenome replicon study confirms that NiV complies with the rule of six.55 When the complete genome sequence of HeV was determined, its length (18,234 nt) was more than 2,700 nt, or 15% longer than the genomes of all other paramyxoviruses known at that time.148 The size of the NiV genome at 18,246 nt to 18,252 nt is slightly larger than that of HeV.57,58,59 The extra length of the henipavirus genome is in the form of unique, long untranslated sequences at the 3′ end of five of the six genes. A comparison of genome length and untranslated regions of representative members of the Paramyxovirinae is shown in Figure 37.3.
The genome organization of henipaviruses resembles that in the genera Respirovirus and Morbillivirus. The first 12 nt of the 3′ and 5′ genomic terminal sequences of paramyxoviruses are highly conserved and complementary, containing promoter elements for replication and transcription.76 The first 3 nt of the henipavirus genome termini are 5′-ACC-3′—a sequence that is absolutely conserved in members of the subfamily Paramyxovirinae but different from that found in the Pneumovirinae.
Genetic Diversity
Partial genome sequencing revealed that HeV isolated from equine and human sources during the outbreak in Brisbane appears identical and differs little from HeV isolated from flying foxes 2 years later.56,104 Sequencing of five additional horse isolates from five different locations from the 2006 to 2008 HeV occurrences has demonstrated a very high genetic similarity.90 Similar observations were made in Malaysia, where it was demonstrated that NiV isolated from pigs at the height of the outbreak and at its geographic focus were essentially identical to human isolates made at that time and isolates obtained from flying foxes several years later.2,24,33,58,59
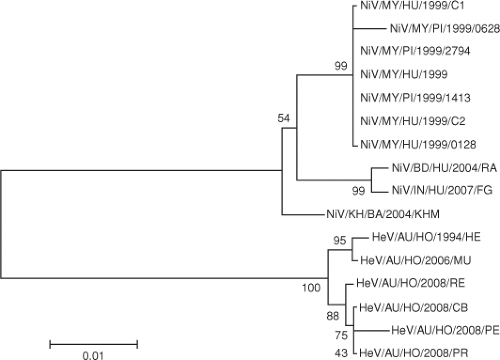
In Bangladesh, four human isolates obtained in 2004 demonstrate significant genetic heterogeneity and suggest multiple spillovers of NiV from flying foxes into the human population.57 The NiV sequence detected from lung tissue of human patients in India was more related to NiV from Bangladesh than NiV from Malaysia.9,23 On the other hand, NiV isolated from the flying fox Pteropus lylei in Cambodia120 represents an evolutionary lineage that is separated from the Malaysia or Bangladesh/India cluster (Fig. 37.4). Partial N gene sequences detected in Pteropus lylei in Thailand indicate the circulation of at least two lineages of NiV—one related to the Bangladesh NiV and the other more related to the Malaysian NiV.144
The presence of henipavirus-reactive (but not neutralizing) antibodies in bats from other regions of the world (see Epidemiology section) predicts that a much greater genetic diversity of henipaviruses exists in different bat populations. This also suggests the existence of henipaviruses with different transmissibility and pathogenicity in nonbat hosts.
Virus Proteins and Their Properties
Analysis of purified viruses by polyacrylamide gel electrophoresis reveals L, P, G, F0, N, F1, and F2 proteins,146,147 where F0 is the uncleaved and F1 and F2 the cleaved products of the F gene. Interestingly, F0 is more readily detected in HeV compared with other paramyxoviruses, including NiV.93,146 Overall, the proteins
of henipaviruses are typical of those of the subfamily Paramyxovirinae, with the exception of the P protein, which is significantly larger than cognate proteins in the subfamily.147 The P protein is translated from messenger RNA (mRNA) that is co-linear with genomic RNA. The P gene also encodes V and W proteins, produced from mRNA in which one and two nontemplated G residues, respectively, are inserted at the RNA editing site during transcription. The P, V, and W proteins, therefore, are identical for the first 405 amino acid residues. A C protein is encoded by the 5′ end of the gene in an overlapping reading frame and is produced by an internal translational initiation mechanism, which is common to other members of Paramyxovirinae, except for rubulaviruses.76 The P, V, and the C proteins predicted from the coding regions in the P gene are present in HeV-infected cells.146 The functional expression of W in NiV-infected cells has recently been demonstrated.84
of henipaviruses are typical of those of the subfamily Paramyxovirinae, with the exception of the P protein, which is significantly larger than cognate proteins in the subfamily.147 The P protein is translated from messenger RNA (mRNA) that is co-linear with genomic RNA. The P gene also encodes V and W proteins, produced from mRNA in which one and two nontemplated G residues, respectively, are inserted at the RNA editing site during transcription. The P, V, and W proteins, therefore, are identical for the first 405 amino acid residues. A C protein is encoded by the 5′ end of the gene in an overlapping reading frame and is produced by an internal translational initiation mechanism, which is common to other members of Paramyxovirinae, except for rubulaviruses.76 The P, V, and the C proteins predicted from the coding regions in the P gene are present in HeV-infected cells.146 The functional expression of W in NiV-infected cells has recently been demonstrated.84
Paramyxovirus N, P, and L proteins are necessary and sufficient for replication of viral RNA both in vitro and in vivo,76 and this has been confirmed for henipaviruses by reverse genetics. Using a minigenome replicon containing leader and trailer sequences of the NiV genome with its entire coding region replaced with a reporter gene, it was shown that efficient genome replication was achieved only when all three proteins were expressed in the same cell.55 NiV N, P, and L proteins were also able to rescue a minigenome constructed from the leader and trailer sequences of the HeV genome, further demonstrating the close genetic relationship between the viruses.
The L protein of nonsegmented, negative-stranded RNA (NNR) viruses in the order Mononegavirales contains a highly conserved GDNQ motif, believed to be important for polymerase activity.116 Henipaviruses were the first NNR viruses in which GDNQ was replaced by GDNE. It was speculated that this motif might be unique to paramyxoviruses with relatively large genomes146,148; however, the GDNE motif has since been found in the L protein of Mossman virus that has a genome length of 16,650 nt.96
The cell attachment proteins of the Paramyxovirinae display hemagglutination (H) and neuraminidase (N) activities in a predominantly genus-specific manner. Viruses in the genera Respirovirus, Avulavirus, and Rubulavirus possess both activities,76 whereas viruses in the genus Morbillivirus do not behave uniformly and only some possess hemagglutination activity.77 In contrast, henipavirus attachment proteins have neither of these activities146,166; rather, they utilize the host cell expressed ephrin-B2 and ephrin-B3 molecules as attachment and entry receptors.11,13,105,106 Recent solution structures of the henipavirus G glycoprotein alone and in complex with the ephrin-B2 and ephrin-B3 receptors have revealed the details of the virus–host cell binding process, distinguishing it from other paramyxovirus receptor strategies.20,21,163
Proteolytic processing of paramyxovirus F proteins is essential for the generation of a fusogenic form of the protein. For most paramyxoviruses that generate systemic infections, cleavage is catalyzed by the cellular protease furin at a multibasic cleavage site.76 Surprisingly, henipavirus F proteins are cleaved without the involvement of furin and, although cleavage occurs at a single basic residue—lysine for HeV and arginine for NiV98—activation of the NiV F protein does not require a basic amino acid at the cleavage site.93 It has been shown that the lysosomal cysteine protease cathepsin L is responsible for the cleavage of the HeV F protein.108 The cytoplasmic tails of henipavirus F proteins contain the endocytosis consensus motif YXX ø.92,143 Endocytosis is required for cleavage activation of the F protein,35,92 consistent with identification of cathepsin L as the enzyme responsible for cleavage.
Host Range
For most paramyxoviruses, host range is limited and interspecies transmission is rare. In contrast, henipaviruses display a broad species tropism. In addition to at least three flying fox species, two nonpteropid fruit bat species, and an insectivorous species, NiV has naturally infected pigs, humans, dogs,
horses, and cats,29,63,97,120,164 whereas HeV infects four Australian flying fox species and has naturally infected humans and horses.42,104 Confirmation of the wide host range of henipaviruses and the identical cell tropism of HeV and NiV were obtained early using an in vitro cell fusion system that relies on vaccinia virus–mediated cell surface expression of G and F glycoproteins.17,18,136 Guinea pigs, hamsters, ferrets, squirrel monkeys, and African green monkeys are also susceptible to experimental NiV infection.19,46,88,94,157 Laboratory studies have added cats, guinea pigs, hamsters, and African green monkeys to the list of HeV-susceptible species,53,121,151,156 along with ferrets (J. Pallister and L.-F. Wang, unpublished results).
horses, and cats,29,63,97,120,164 whereas HeV infects four Australian flying fox species and has naturally infected humans and horses.42,104 Confirmation of the wide host range of henipaviruses and the identical cell tropism of HeV and NiV were obtained early using an in vitro cell fusion system that relies on vaccinia virus–mediated cell surface expression of G and F glycoproteins.17,18,136 Guinea pigs, hamsters, ferrets, squirrel monkeys, and African green monkeys are also susceptible to experimental NiV infection.19,46,88,94,157 Laboratory studies have added cats, guinea pigs, hamsters, and African green monkeys to the list of HeV-susceptible species,53,121,151,156 along with ferrets (J. Pallister and L.-F. Wang, unpublished results).
Pathogenesis and Pathology
Cell and Tissue Tropism
The primary site of NiV replication and the dynamics of virus spread in humans are unknown. However, the distribution and time of appearance of lesions throughout the vasculature and in the brain and lung in NiV encephalitis suggest that secondary infection probably arises via hematogenous spread of the virus, with secondary replication occurring in vascular endothelium.159 Inflammation of blood vessels occurs in most organs but is particularly prominent in the brain, lung, heart, and kidney.30,159,160 Vasculitis is limited to small arteries, arterioles, and capillaries where NiV antigen is found in both endothelial cells and the smooth muscle of the tunica media. The pattern and time of appearance of vasculitis and viral antigen distribution are consistent with endothelial cell infection occurring before infection of the smooth muscle. Syncytial endothelial cells are present in blood vessels of various organs and represent a pathognomonic but insensitive feature of henipavirus infections found in only 25% of human NiV infections.159 Syncytial endothelial cells were also observed in the single human case of encephalitis caused by HeV.107 NiV antigen is also found in neurons and, less frequently, in bronchiolar and renal epithelial cells. The 5-day interval between maximal vasculitis in the brain and parenchymal infection in acute NiV encephalitis suggests that primary virus replication occurs in endothelial cells, with infection of neurons occurring as a result of vascular damage. The presence of inclusion bodies and viral antigen in neurons suggests that neurologic impairment in encephalitis may be caused by both the effects of ischemia and infarction and viral infection of neurons.30,114,159 Human HeV encephalitis has been described as widespread cortical, subcortical, and deep white matter involvement in two cases in 2008115 and similar to those described in a previous HeV encephalitis case82,107 and NiV encephalitis cases.82,127
Although NiV antigen is found less frequently in bronchiolar and renal epithelial cells compared with vascular and neuronal locations, replication in epithelial locations may play a role in virus dissemination, because NiV is found in urine and in tracheal and nasopharyngeal secretions of infected patients in the early phase of their illness.31,48 Despite this, human-to-human transmission in Malaysia was extremely rare.100
The identification of ephrin-B2 and ephrin-B3 as functional receptors for henipaviruses in cultured cells provides an explanation for the observed distribution of viral antigen in arterial endothelial cells, smooth muscle, neurons, and some epithelial cells.11,13,105,106 Ephrin-B2 is found in arteries, arterioles, capillaries in multiple organs, and tissues including arterial smooth muscle and human bronchiolar epithelial cells135 but is absent from venous components of the vasculature.45 Ephrin-B3 is found predominantly in the nervous system as well as the vasculature.113 The ephrins engage Eph receptors and mediate bidirectional cell–cell signaling events and are modulators of cell remodeling events, especially within the nervous and vascular systems.113
Immune Response
In patients with encephalitis, anti-NiV antibodies were observed more frequently in the serum than the CSF. Immunoglobulin M (IgM) antibodies occurred more frequently than immunoglobulin G (IgG) antibodies in both locations.30,114,159 The appearance of specific IgM antibodies in serum preceded their appearance in the CSF, a sequence consistent with viremia preceding central nervous system (CNS) infection. Anti-NiV antibodies were present in most patients with clinical NiV encephalitis; however, no difference was observed in clinical features, laboratory results, or mortality between seropositive and seronegative patients.27,48 Seroconversion of IgG against HeV was seen in two human cases of HeV infection, one fatal, in 2008,115 following an influenza-like illnesses and before progressing to encephalitis.
Inhibition of the Interferon Response
In henipaviruses, as in other paramyxoviruses,66 anti-interferon (IFN) activities are encoded by the P gene and in vitro studies indicate that P gene products inhibit both IFN induction and signaling.37 In IFN induction, viral double-stranded RNA (dsRNA) is detected by intracellular RNA helicase enzymes3,79 and by toll-like receptor 3 (TLR-3).12 Both of these dsRNA signaling pathways lead to activation of the pre-existing transcription factors interferon regulatory factor 3 (IRF-3) and nuclear factor kappa B (NF-kB),50 and the synthesis of IFN-α and a subset of IFN-β proteins.89 HeV inhibits dsRNA signaling by using a strategy adopted by other paramyxoviruses in which the V protein binds to the intracellular RNA helicase sensor and prevents downstream signaling3 but does not abrogate dsRNA signaling through TLR-3.132 In an additional strategy unique to henipaviruses, the W protein, by virtue of a nuclear localization signal located in the unique carboxy terminal, inhibits dsRNA signaling in the nucleus by targeting a process that is part of both helicase-dependent and TLR-3–dependent signaling pathways.132
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree