1 Introduction
Microorganisms differ enormously in terms of their shape, size and appearance and in their genetic and meta-bolic characteristics. All these properties are used in classifying microorganisms into the major groups with which many people are familiar, e.g. bacteria, fungi, protozoa and viruses, and into the less well known categories such as chlamydia, rickettsia and mycoplasmas. The major groups are the subject of individual chapters immediately following this, so the purpose here is not to describe any of them in great detail but to summarize their features so that the reader may better understand the distinctions between them. A further aim of this chapter is to avoid undue repetition of information in the early part of the book by considering such aspects of microbiology as cultivation, enumeration and genetics that are common to some, or all, of the various types of microorganism.
1.1 Viruses, viroids and prions
Viruses do not have a cellular structure. They are particles composed of nucleic acid surrounded by protein; some possess a lipid envelope and associated glycoproteins, but recognizable chromosomes, cytoplasm and cell membranes are invariably absent. Viruses are incapable of independent replication as they do not contain the enzymes necessary to copy their own nucleic acids; as a consequence, all viruses are intracellular parasites and are reproduced using the metabolic capabilities of the host cell. A great deal of variation is observed in shape (helical, linear or spherical), size (20–400 nm) and nucleic acid composition (single-or double-stranded, linear or circular RNA or DNA), but almost all viruses are smaller than bacteria and they cannot be seen with a normal light microscope; instead they may be viewed using an electron microscope which affords much greater magnification.
Viroids (virusoids) are even simpler than viruses,being infectious particles comprising single-stranded RNA without any associated protein. Those that have been described are plant pathogens, and, so far, there are no known human pathogens in this category, although human hepatitis D virus shares some features in common with viroids, and may have originated from them.
Prions are unique as infectious agents in that they contain no nucleic acid. A prion is an atypical form of a mammalian protein that can interact with a normal protein molecule and cause it to undergo a conformational change so that it, in turn, becomes a prion and ceases its normal function. Prions are the agents responsible for transmissible spongiform encephalopathies, e.g. Creutzfeldt–Jakob disease (CJD) and bovine spongiform encephalopathy (BSE). They are the simplest and most recently recognized agents of infectious disease, and are important in a pharmaceutical context owing to their extreme resistance to conventional sterilizing agents like steam, gamma radiation and disinfectants (Chapter 21).
1.2 Prokaryotes and eukaryotes
The most fundamental distinction between the various microorganisms having a cellular structure (i.e. all except those described in section 1.1 above) is their classification into two groups—the prokaryotes and eukaryotes— based primarily on their structural characteristics and mode of reproduction. Expressed in the simplest possible terms, prokaryotes are the bacteria and archaea (section 1.2.1), and eukaryotes are all other cellular microorganisms, e.g. fungi, protozoa and algae. The crucial difference between these two types of cell is the possession by the eukaryotes of a true cell nucleus in which the chromosomes are separated from the cytoplasm by a nuclear membrane. The prokaryotes have no true nucleus; they normally possess just a single chromosome that is not separated from the other cell contents by a membrane. Other major distinguishing features of the two groups are that prokaryotes are normally haploid (possess only one copy of the set of genes in the cell) and reproduce asexually; eukaroyotes, by contrast, are usually diploid (possess two copies of their genes) and normally have the potential to reproduce sexually. The capacity for sexual reproduction confers the major advantage of creating new combinations of genes, which increases the scope for selection and evolutionary development. The restriction to an asexual mode of reproduction means that the organism in question is heavily reliant on mutation as a means of creating genetic variety and new strains with advantageous characteristics, although many bacteria are able to receive new genes from other strains species (see section 6.1 and Chapter 3). Table 2.1 lists some distinguishing features of the prokaryotes and eukaryotes.
Table 2.1 Distinguishing features of prokaryotes and eukaryotes
| Characteristic | Eukaryotes | Prokaryotes |
| Size | Normally > 10 µ m | Typically 1 – 5 µ m |
| Location of chromosomes | Within a true nucleus separated from the cytoplasm by a nuclear membrane | In the cytoplasm, usually attached to the cell membrane |
| Nuclear division | Exhibit mitosis and meiosis | Mitosis and meiosis are absent |
| Nucleolus | Present | Absent |
| Reproduction | Asexual or sexual reproduction | Normally asexual reproduction |
| Chromosome number | >1 | 1 |
| Mitochondria and chloroplasts | May be present | Absent |
| Cell membrane composition | Sterols present | Sterols absent |
| Cell wall composition | Cell walls (when present) usually contain cellulose or chitin but not peptidoglycan | Walls usually contain peptidoglycan |
| Ribosomes | Cytoplasmic ribosomes are 80S | Ribosomes are smaller, usually 70S |
| Flagella | Structurally complex | Structurally simple |
| Pili | Absent | Present |
| Fimbriae | Cilia | Present |
| Storage compounds | Poly-β-hydroxybutyrate absent | Poly-β-hydroxybutyrate often present |
1.2.1 Bacteria and archaea
Bacteria are essentially unicellular, although some species arise as sheathed chains of cells. They possess the properties listed under prokaryotes in Table 2.1 but, like viruses and other categories of microorganisms, exhibit great diversity of form, habitat, metabolism, pathogenicity and other characteristics. The bacteria of interest in pharmacy and medicine belong to the group known as the eubacteria. The other subdivision of prokaryotes, the archaea, have no pharmaceutical importance, and although formerly considered largely to comprise organisms capable of living in extreme environments (e.g. high temperatures, extreme salinity or pH) or organisms exhibiting specialized modes of metabolism (e.g. by deriving energy from sulphur or iron oxidation or the production of methane) they are now known to occur in a wide variety of habitats.
The eubacteria are typically rod-shaped (bacillus),spherical (cocci), curved or spiral cells of approximately 0.5–5.0 mm (longest dimension) and are divided into two groups designated Gram-positive and Gramnegative according to their reaction to a staining procedure developed in 1884 by Christian Gram (see Chapter 3). Although all the pathogenic species are included within this category, there are very many other eubacteria that are harmless or positively beneficial. Some of the bacteria that contaminate or cause spoilage of pharmaceutical materials are saprophytes, i.e. they obtain their energy by decomposition of animal and vegetable material, while many could also be described as parasites (benefiting from growth on or in other living organisms without causing detrimental effects) or pathogens (parasites damaging the host). Rickettsia and chlamydia are types of bacteria that are obligate intracellular parasites, i.e. they are incapable of growing outside a host cell and so cannot easily be cultivated in the laboratory. Most bacteria of pharmaceutical and medical importance possess cell walls (and are therefore relatively resistant to osmotic stress), grow well at temperatures between ambient and human body temperature, and exhibit wide variations in their requirement for, or tolerance of, oxygen. Strict aerobes require atmospheric oxygen, but for strict anaerobes oxygen is toxic. Many other bacteria would be described as facultative anaerobes (normally growing best in air but can grow without it) or microaerophils(preferring oxygen concentrations lower than those i normal air).
1.2.2 Fungi
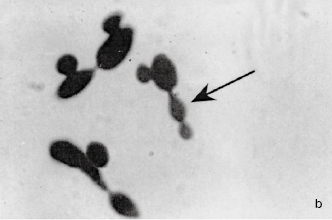
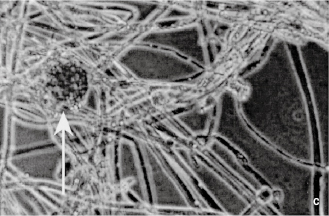
Fungi are structurally more complex and varied in appearance than bacteria and, being eukaryotes, differ from them in the ways described in Table 2.1. Fungi are considered to be non-photosynthesizing plants, and the term fungus covers both yeasts and moulds, although the distinction between these two groups is not always clear. Yeasts are normally unicellular organisms that are larger than bacteria (typically 5–10 μm) and divide either by a process of binary fission (see section 4.2 and Figure 2.1a) or budding (whereby a daughter cell arises as a swelling or protrusion from the parent that eventually separates to lead an independent existence, (Figure 2.1b). Mould is an imprecise term used to describe fungi that do not form fruiting bodies visible to the naked eye, thus excluding toadstools and mushrooms. Most moulds consist of a tangled mas (mycelium) of filaments or threads (hyphae) which vary between 1 and over 50 µ m wide (Figure 2.1c); they may be differentiated for specialized functions, e.g. absorption of nutrients or reproduction. Some fungi may exhibit a unicellular (yeast-like) or mycelial (mould-like) appearance depending upon cultivation conditions. Although fungi are eukaryotes that should, in theory, be capable of sexual reproduction, there are some species in which this has never been observed. Most fungi are saprophytes with relatively few having pathogenic potential, but their ability to form spores that are resistant to drying makes them important as contaminants of pharmaceutical raw materials, particularly materials of vegetable origin
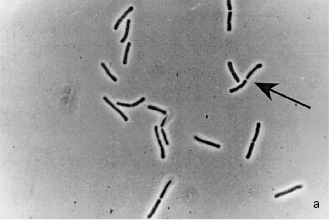
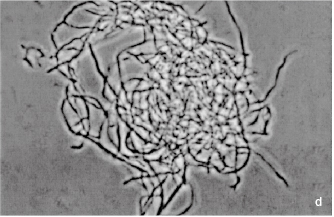
Figure 2.1 (a) A growing culture of Bacillus megaterium in which cells about to divide by binary fission display constrictions (arrowed) prior to separation. (b) A growing culture of the yeast Saccharomyces cerevisiae displaying budding (arrowed). (c) The mould Mucor plumbeus exhibiting the typical appearance of a mycelium in which masses of asexual zygospores (arrowed) are formed on specialized hyphae. (d) The bacterium Streptomyces rimosus displaying the branched network of filaments that superficially resembles a mould mycelium. (e) The typical appearance of an overnight agar culture of Micrococcus luteus inoculated to produce isolated colonies (arrowed). (f) A single colony of the mould Aspergillus niger in which the actively growing periphery of the colony (arrowed) contrasts with the mature central region where pigmented asexual spores have developed.
1.2.3 Protozoa
Protozoa are eukaryotic, predominantly unicellular microorganisms that are regarded as animals rather than plants, although the distinction between protozoa and fungi is not always clear and there are some organisms whose taxonomic status is uncertain. Many protozoa are free-living motile organisms that occur in water and soil, although some are parasites of plants and animals, including humans, e.g. the organisms responsible for malaria and amoebic dysentery. Protozoa are not normally found as contaminants of raw materials or manufactured medicines and the relatively few that are of pharmaceutical interest owe that status primarily to their potential to cause disease.
Microorganisms, just like other organisms, are normally known by two names: that of the genus (plural = genera) and that of the species. The former is normally written with an upper case initial letter and the latter with a lower case initial letter, e.g. Staphylococcus aureus or Escherichia coli. These may be abbreviated by shortening the name of the genus provided that the shortened form is unambiguous, e.g. Staph. aureus, E. coli. Both the full and the shortened names are printed in italics to designate their status as proper names (in old books, theses or manuscripts they might be in roman type but underlined). The species within a genus are sometimes referred to by a collective name, e.g. staphylococci or pseudomonads, and neither these names, nor names describing groups of organisms from different genera, e.g. coliforms, are italicized or spelt with an upper case initial letter.
As in most other aspects of their physiology, microorganisms exhibit marked differences in their metabolism. While some species can obtain carbon from carbon dioxide and energy from sunlight or the oxidation of inorganic materials like sulphides, the vast majority of organisms of interest in pharmacy and medicine are described as chemoheterotrophs—they obtain carbon, nitrogen and energy by breaking down organic compounds. The chemical reactions by which energy is liberated by digestion of food materials are termed catabolic reactions, while those that use the liberated energy to make complex cellular polymers, proteins, carbohydrates and nucleic acids, are called anabolic reactions.
Food materials are oxidized in order to break them down and release energy from them. The term oxidation is defined as the removal or loss of electrons, but oxidation does not invariably involve oxygen, as a wide variet of other molecules can accept electrons and thus act as oxidizing agents. As the oxidizing molecule accepts the electrons, the other molecule in the reaction that provides them is simultaneously reduced. Consequently, oxidation and reduction are invariably linked and such reactions are often termed redox reactions. The term redox potential is also used, and this indicates whether oxidizing or reducing conditions prevail in a particular situation, e.g. in a body fluid or a culture medium. Anaerobic organisms prefer low redox potentials (typically zero to −200 mV or less) while aerobes thrive in high redox potential environments (e.g. zero to +200mV or more).
There are marked similarities in the metabolic pathways used by pathogenic bacteria and by mammals. Many bacteria use the same process of glycolysis that is used by humans to begin the breakdown of glucose and the release of energy from it. Glycolysis describes the conversion of glucose, through a series of reactions, to pyruvic acid, and it is a process for which oxygen is not required, although glycolysis is undertaken by both aerobic and anaerobic organisms. The process releases only a relatively small amount of the energy stored in a sugar molecule, and aerobic microorganisms, in common with mammals, release much more of the energy by aerobic respiration. Oxygen is the molecule at the end of the sequence of respiratory reactions that finally accepts the electrons and allows the whole process to proceed, but it is worth noting that many organisms can also undertake anaerobic respiration, which uses other final electron acceptors, e.g. nitrate or fumarate.
As an alternative to respiration many microorganisms use fermentation as a means of releasing more energy from sugar; fermentation is, by definition, a process in which the final electron acceptor is an organic molecule. The term is widely understood to mean the production by yeast of ethanol and carbon dioxide from sugar, but in fact many organisms apart from yeasts can undertake fermentation and the process is not restricted to common sugar (sucrose) as a starting material or to ethanol and carbon dioxide as metabolic products. Many pathogenic bacteria are capable of fermenting several different sugars and other organic materials to give a range of metabolic products that includes acids (e.g. lactic, acetic and propionic), alcohols (e.g. ethanol, propanol, butanediol) and other commercially important materials like the solvents acetone and butanol. Fermentation is, like glycolysis, an anaerobic process, although the term is commonly used in the pharmaceutical and biotechnology industries to describe the manufacture of a wide rang of substances by microorganisms where the biochemical process is neither fermentative nor even anaerobic, e.g. many textbooks refer to antibiotic fermentation, but the production vessels are usually vigorously aerated.
Microorganisms are far more versatile than mammals with respect to the materials that they can use as foods and the means by which those foods are broken down. Some pathogenic organisms can grow on dilute solutions of mineral salts and sugar (or other simple molecules like glycerol, lactic or pyruvic acids), while others can obtain energy from rarely encountered carbohydrates or by the digestion of proteins or other non-carbohydrate foods. In addition to accepting a wide variety of food materials, many microorganisms can use alternative metabolic pathways to break the food down depending on the environmental conditions, e.g. facultative anaerobes can switch from respiration to fermentation if oxygen supplies are depleted. It is partly this ability to switch to different metabolic pathways that explains why none of the major antibiotics work by interfering with the chemical reactions microorganisms use to metabolize their food. It is a fundamental principle of antibiotic action that the drug must exploit a difference in metabolism between the organism to be killed and the human host; without such a difference the antibiotic would be very toxic to the patient too. However, not only do bacteria use metabolic pathways for food digestion that are similar to our own, many of them would have the ability to switch to an alternative energy-producing pathway if an antibiotic were developed that interfered with a reaction that is unique to bacteria.
The metabolic products that arise during the period when a microbial culture is actually growing are termed primary metabolites, while those that are produced after cell multiplication has slowed or stopped, i.e. in the ‘stationary phase’ (see Chapter 3), are termed secondary metabolites. Ethanol is a primary metabolite of major commercial importance although it is produced in large quantities only by some species of yeast. More common than ethanol as primary metabolites are organic acids, so it is a common observation that the pH of a culture progressively falls during growth, and many organisms further metabolize the acids so the pH often rises after cell growth has ceased. The metabolites that are found during secondary metabolism are diverse, and many of them have commercial or therapeutic importance. They include antibiotics, enzymes (e.g. amylases that digest starch and proteolytic enzymes used in biological washing powders), toxins (responsible for many of the symptoms of infection but some also of therapeutic value, e.g. botox—the toxin of Clostridium botulinum) and carbohydrates (e.g. dextran, used as a plasma expander and for molecular separations by gel filtration).
The vast majority of microorganisms of interest in pharmacy and medicine can be cultivated in the laboratory and most of them require relatively simple techniques and facilities. Some organisms are parasites and so can only be grown inside the cells of a host species—which often necessitates mammalian cell culture facilities—and there are a few (e.g. the organism responsible for leprosy) that are not cultivated outside the living animal.
4.1 Culture media
A significant number of common microorganisms are capable of synthesizing all the materials they need for growth (e.g. amino acids, nucleotides and vitamins) from simple carbon and nitrogen sources and mineral salts. Such organisms can grow on truly synthetic (chemically defined) media, but many organisms do not have this capability and need a medium that already contains these biochemicals. Such media are far more commonly used than synthetic ones, and several terms have been used to describe them, e.g. routine laboratory media, general purpose media and complex media. They are complex in the sense that their precise chemical composition is unknown and likely to vary slightly from batch to batch. In general, they are aqueous solutions of animal or plant extracts that contain hydrolysed proteins, B-group vitamins and carbohydrates.
Readily available and relatively inexpensive sources of protein include meat extracts (from those parts of animal carcasses that are not used for human or domestic animal consumption), milk and soya. The protein is hydrolysed to varying degrees to give peptones (by definition not coagulable by heat or ammonium sulphate) or amino acids. Trypsin or other proteolytic enzymes are preferred to acids as a means of hydrolysis because acids cause more amino acid destruction; the term ‘tryptic’ denotes the use of the enzyme. Many microorganisms require B-group vitamins (but not the other water-or fat-soluble vitamins required by mammals) and this require-ment is satisfied by yeast extract. Carbohydrates are used in the form of starch or sugars, but glucose (dextrose) is the only sugar regularly employed as a nutrient.
Microorganisms differ in terms of their ability to ferment various sugars, and their fermentation patterns may be used as an aid in identification. Thus, other sugars included in culture media are normally present for these diagnostic purposes rather than as carbon and energy sources. Sodium chloride may be incorporated in culture media to adjust osmotic pressure, and occasionally buffers are added to neutralize acids that result from sugar metabolism. Routine culture media may be enriched by the addition of materials like milk, blood or serum, and organisms that need such supplements in order to grow are described as ‘exacting’ in their nutritional requirements.
Culture media may be either liquid or solid; the latter term describes liquid media that have been gelled by the addition of agar, which is a carbohydrate extracted from certain seaweeds. Agar at a concentration of about 1–1.5% w/v will provide a firm gel that cannot be liquefied by the enzymes normally produced during bacterial growth (which is one reason it is used in preference to gelatin). Agar is unusual in that the melting and setting temperatures for its gels are quite dissimilar. Fluid agar solutions set at approximately 40 °C, but do not re-liquefy on heating until the temperature is in excess of 90 °C. Thus agar forms a firm gel at 37°C which is the normal incubation temperature for many pathogenic organisms (whereas gelatin does not) and when used as a liquid at 45 °C is at a sufficiently low temperature to avoid killing microorganisms—this property is important in pour plate counting methods (see section 5).
In contrast to medium ingredients designed to support microbial growth, there are many materials commonly added to selective or diagnostic media whose function is to restrict the growth of certain types of microorganism while permitting or enhancing the growth of others. Examples include antibacterial antibiotics added to fungal media to suppress bacterial contaminants, and bile to suppress organisms from anatomical sites other than the gastrointestinal tract. Many such additives are used in media for organism identification purposes, and these are considered further in subsequent chapters. The term enrichment sometimes causes confusion in this context. It is occasionally used in the sense of making a medium nutritionally richer to achieve more rapid or profuse growth. Alternatively, and more commonly, an enrichment medium is one designed to permit a particular type of organism to grow while restricting others, so the one that grows increases in relative numbers and is ‘enriched’ in a mixed culture.
Solid media designed for the growth of anaerobic organisms usually contain non-toxic reducing agents, e.g. sodium thioglycollate or sulphur-containing amino acids; these compounds create redox potentials of −200 m V or less and so diminish or eliminate the inhibitory effects of oxygen or oxidizing molecules on anaerobic growth. The inclusion of such compounds is less important in liquid media where a sufficiently low redox potential may be achieved simply by boiling; this expels dissolved oxygen, which in unstirred liquids only slowly resaturates the upper few millimetres of liquid. Redox indicators like methylene blue or resazurin may be incorporated in anaerobic media to confirm that a sufficiently low redox potential has been achieved.
Media for yeasts and moulds often have a lower pH (5.5–6.0) than bacterial culture media (7.0–7.4). Lactic acid may be used to impart a low pH because it is not, itself, inhibitory to fungi at the concentrations used. Some fungal media that are intended for use with specimens that may also contain bacteria may be supplemented with antibacterial antibiotics, e.g. chloramphenicol or tetracyclines.
4.2 Cultivation methods
Most bacteria and some yeasts divide by a process of binary fission whereby the cell enlarges or elongates, then forms a cross-wall (septum) that separates the cell into two more or less equal compartments each containing a copy of the genetic material. Septum formation is often followed by constriction such that the connection between the two cell compartments is progressively reduced (see Figure 2.1a) until finally it is broken and the daughter cells separate. In bacteria this pattern of division may take place every 25–30 minutes under optimal conditions of laboratory cultivation, although growth at infection sites in the body is normally much slower owing to the effects of the immune system and scarcity of essential nutrients, particularly iron. Growth continues until one or more nutrients is exhausted, or toxic metabolites (often organic acids) accumulate and inhibit enzyme systems. Starting from a single cell many bacteria can achieve concentrations of the order of 109 cellsml−1 or more following overnight incubation in common liquid media. At concentrations below about 107 cells ml−1 culture media are clear, but the liquid becomes progressively more cloudy (turbid) as the concentration increases above this value; turbidity is, therefore, an indirect means of monitoring culture growth. Some bacteria produce chains of cells, and some produce elongated cells (filaments) that may exhibit branching to create a tangle mass resembling a mould mycelium (Figure 2.1d). Many yeasts divide by budding (see section 1.2.3 and Figure 2.1b) but they, too, would normally grow in liquid media to produce a turbid culture. Moulds, however, grow by extension and branching of hyphae to produce a mycelium (Figure 2.1c) or, in agitated liquid cultures, pellet growth may arise.
When growing on solid media in Petri dishes (often referred to as ‘plates’) individual bacterial cells can give rise to colonies following overnight incubation under optimal conditions. A colony is simply a collection of cells arising by multiplication of a single original cell or a small cluster of them (called a colony-forming unit or CFU). The term ‘colony ’ does not, strictly speaking, imply any particular number of cells, but it is usually taken to mean a number sufficiently large to be visible by eye. Thus, macroscopic bacterial colonies usually comprise hundreds of thousands, millions or tens of millions of cells in an area on a Petri dish that is typically 1–10 mm in diameter (Figure 2.1e). Colony size is limited by nutrient availability and/or waste product accumulation in just the same way as cell concentration in liquid media. Colonies vary between bacterial species, and their shapes, sizes, opacities, surface markings and pigmentation may all be characteristic of the species in question, so these properties may be an aid in identification procedures (see Chapter 3).
Anaerobic organisms may be grown on Petri dishes provided that they are incubated in an anaerobic jar. Such jars are usually made of rigid plastic with airtight lids, and Petri dishes are placed in them together with a low-temperature catalyst. The catalyst, consisting of palladium-coated pellets or wire, causes the oxygen inside the jar to be combined with hydrogen that is generated by the addition of water to sodium borohydride; this is usually contained in a foil sachet that is also placed in the jar; alternatively, oxygen may be removed by combination with ascorbic acid. After its removal, an anaerobic atmosphere is achieved and this is monitored by an oxidation-reduction (redox) indicator; resazurin is frequently used as a solution soaking a fabric strip.
Yeast colonies often look similar to those of bacteria, although they may be larger and more frequently coloured. The appearance of moulds growing on solid microbiological media is similar to their appearance when growing on common foods. The mould colony consists of a mycelium that may be loosely or densely entangled depending on the species, often with the central area (the oldest, most mature region of the colony) showing pigmentation associated with spor production (Figure 2.1f). The periphery of the colony is that part which is actively growing and it is usually non-pigmented.
4.3 Planktonic and sessile (biofilm) growth
Bacteria growing in liquid culture in the laboratory usually exist as individual cells or small aggregates of cells suspended in the culture medium; the term planktonic is used to describe such freely suspended cells. In recent years, however, it has become recognized that planktonic growth is not the normal situation for bacteria growing in their natural habitats. In fact, bacteria in their natural state far more commonly grow attached to a surface which, for many species, may be solid, e.g. soil particles, stone, metal or glass, or for pathogens, an epithelial surface in the body, e.g. lung or intestinal mucosa. Bacteria attached to a substrate in this way are described as sessile, and are said to exhibit the biofilm or microcolony mode of growth.
Planktonic cells are routinely used for almost all the testing procedures that have been designed to assess the activity of antimicrobial chemicals and processes, but the recognition that planktonic growth is not the natural state for many organisms prompted investigations of the relative susceptibilities of planktonic-and biofilm-grown cells to antibiotics, disinfectants and decontamination or sterilization procedures. In many cases it has been found that planktonic and sessile bacteria exhibit markedly different susceptibilities to these lethal agents, and this has prompted a reappraisal of the appropriateness of some of the procedures used (see Chapters 8, 13 and 18).
5 Enumeration of microorganisms
In a pharmaceutical context there are several situations where it is necessary to measure the number of microbial cells in a culture, sample or specimen:
• when measuring the levels of microbial contamination in a raw material or manufactured medicine
• when evaluating the effects of an antimicrobial chemical or decontamination process
• when using microorganisms in the manufacture of therapeutic agents
• when assessing the nutrient capability of a growth medium.
In some cases it is necessary to know the total number of microbial cells present, i.e. both living and dead: e.g. in vaccine manufacture dead and living cells may both produce an immune response, and in pyrogen testin both dead and living cells induce fever when injected into the body. However, in many cases it is the number or concentration of living cells that is required. The terminology in microbial counting sometimes causes confusion. A total count is a counting procedure enumerating both living and dead cells, whereas a viable count, which is far more common, records the living cells alone. However, the term total viable count (TVC) is used in most pharmacopoeias and by many regulatory agencies to mean a viable count that records all the different species or types of microorganism that might be present in a sample (e.g. bacteria plus fungi).
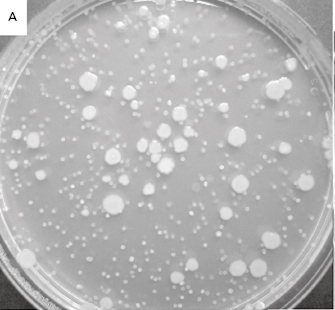
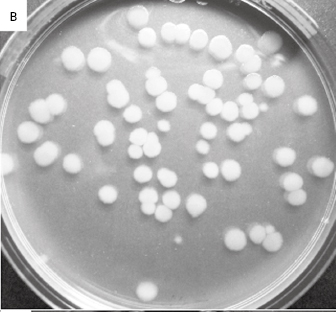
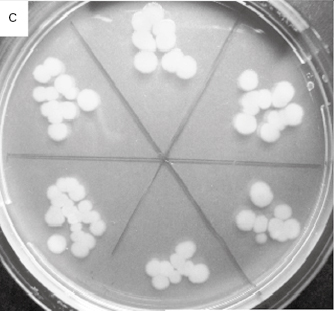
Table 2.2 lists the more common counting methods available. The first three traditional methods of viable counting all operate on the basis that a living cell (or a CFU) will give rise to a visible colony when introduced into or onto the surface of a suitable medium and incubated. Thus, the procedure for pour plating (Figure 2.2A) usually involves the addition of a small volume (typically 1.0 ml) of sample (or a suitable dilution thereof) into molten agar at 45 °C which is then poured into empty sterile Petri dishes. After incubation the resultant colonies are counted and the total is multiplied by the dilution factor (if any) to give the concentration in the original sample. In a surface spread technique (Figure 2.2B) the sample (usually 0.1–0.25 ml) is spread over the surface of agar which has previously been dried to permit absorption of the added liquid. The Miles Misra (surface drop) method (Figure 2.2C) is similar in principle, but several individual drops of culture are allowed to spread over discrete areas of about 1 cm diameter on the agar surface. These procedures are suitable for samples that are expected to contain concentrations exceeding approximately 100 CFU ml−1 so that the number of colonies arising on the plate is sufficiently large to be statistically reliable. If there are no clear indi-cations of the order of magnitude of the concentration in the sample, it is necessary to plate out the sample at each of two, three or more (decimal, i.e. 10-fold) dilutions so as to obtain Petri dishes with conveniently countable numbers of colonies (usually taken to be 30–300 colonies).
Table 2.2 Traditional and rapid methods of enumerating cells
| Traditional methods | ||
| Viable counts | Total counts | Rapid methods (indirect viable counts) |
| 1 Pour plate (counting colonies in agar) | 1 Direct microscopic counting (using Helber or haemocytometer counting chambers) | 1 Epifl uorescence (uses dyes that give characteristic fl uorescence only in living cells) often coupled to image analysis |
| 2 Surface spread or surface drop (Miles Misra) methods (counting colonies on agar surface) | 2 Turbidity methods (measures turbidity (opacity) in suspensions or cultures) | 2 ATP methods (measure ATP production in living cells using bioluminescence) |
| 3 Membrane filter methods (colonies growing on membranes on agar surface) | 3 Dry weight determinations | 3 I mpedance (measures changes in resistance,capacitance or impedance in growing cultures) |
| 4 MPN (counts based on the proportion of liquid cultures growing after receiving low inocula) | 4 Nitrogen, protein or nucleic acid determinations | 4 Manometric methods (measure oxygen consumption or CO 2 production by growing cultures) |
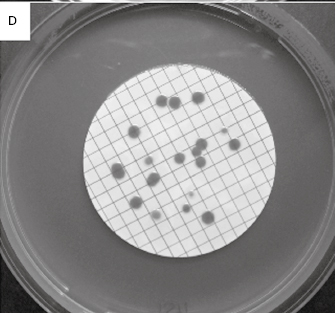
Figure 2.2 Viable counts of bacteria: (A) Pour plate method using Bacillus subtilis; the colonies on the surface of the agar are growing larger than those within the agar due to greater oxygen availability. (B) Surface spread, and (C) surface drop (Miles Misra) methods using Bacillus subtilis. (D) Membrane filtration method showing Serratia marcescens colonies growing on a 47 mm diameter membrane.