Filoviridae: Marburg and Ebola Viruses
Heinz Feldmann
Anthony Sanchez
Thomas W. Geisbert
Classification
Taxonomy
Filoviruses are taxonomically classified within the order Mononegavirales, a large group of enveloped viruses whose genomes are composed of a nonsegmented, negative-strand (NNS) RNA molecule.98 Following their discovery, filoviruses were initially grouped with rhabdoviruses, based primarily on the appearance of virus particles. However, subsequent morphologic, genetic, physiochemical, and virologic studies of Marburg virus (MARV) and Ebola virus (EBOV) isolates revealed unique properties and led to their placement into a separate family, the Filoviridae.224 Further characterization of these agents demonstrated that EBOV and MARV represent divergent lineages of filoviruses; their differences were significant enough to warrant the formation of the genera Marburgvirus and Ebolavirus. According to the International Committee on Taxonomy of Viruses (ICTV) (http://www.ictvonline.org/virusTaxonomy.asp?version=2009&bhcp=1), the Marburgvirus genus contains a single species, Lake Victoria marburgvirus, because strains exhibit only limited genetic variation. However, the appearance of distinct MARV lineages may lead to further speciation.401 There is a greater divergence within the Ebolavirus genus, five species having been recognized: Zaire ebolavirus (type species; ZEBOV), Sudan ebolavirus (SEBOV), Reston ebolavirus (REBOV), Tai Forest ebolavirus (formerly Ivory Coast or Cote d’Ivoire ebolavirus, ICEBOV or CIEBOV), and Bundibugyo ebolavirus (BEBOV), of which BEBOV has still to be approved.403 A distinct filovirus sequence has recently been obtained from bats in Spain, but no virus has yet been isolated. This new “putative virus” (designated Lloviu virus) is proposed to represent the single species Lloviu cuevavirus in the new genus Cuevavirus of the Filoviridae family.299
Biosafety and Biosecurity
Because of their high mortality rate, their potential for person-to-person transmission, and a lack of an approved vaccine or antiviral therapy, MARV and EBOV are classified as biosafety level 4 (BSL-4; risk group 4) pathogens, for which maximum containment facilities are required when handling the infectious agent (http://www.cdc.gov/biosafety/publications/bmbl5/BMBL.pdf). Filovirus infectivity is quite stable at room temperature (20°C), but is largely inactivated in 30 minutes at 60°C; MARV is somewhat resistant to desiccation. Infectivity is greatly reduced or destroyed by high doses of ultraviolet light and gamma irradiation, lipid solvents, β-propiolactone, photo-induced alkylating probe 1,5-iodonaphthylazide, guanidinium isothiocyanate, and commercial hypochlorite and phenolic disinfectants.
The threat of bioterrorism in the aftermath of the September 11, 2001 and anthrax attacks against the United States has
prompted governments to implement countermeasures. This has led to greater restrictions on the acquisition and use of a variety of agents that pose serious threats to public health. Filoviruses have been classified as Centers for Disease Control and Prevention (CDC) Category A Agents (http://www.bt.cdc.gov/agent/agentlist-category.asp#a), as part of a system for prioritizing initial public health preparedness efforts and grading the potential of agents for large-scale dissemination. Filoviruses are also classified as “select agents” by the CDC Select Agent Program. This program is mandated by federal law to regulate activities involving these agents within the United States and to register laboratories and entities handling one or more select agents (http://www.cdc.gov/od/sap/).
prompted governments to implement countermeasures. This has led to greater restrictions on the acquisition and use of a variety of agents that pose serious threats to public health. Filoviruses have been classified as Centers for Disease Control and Prevention (CDC) Category A Agents (http://www.bt.cdc.gov/agent/agentlist-category.asp#a), as part of a system for prioritizing initial public health preparedness efforts and grading the potential of agents for large-scale dissemination. Filoviruses are also classified as “select agents” by the CDC Select Agent Program. This program is mandated by federal law to regulate activities involving these agents within the United States and to register laboratories and entities handling one or more select agents (http://www.cdc.gov/od/sap/).
Table 32.1 Outbreaks of Filovirus Disease | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
History
Table 32.1 lists the documented occurrences of MARV and EBOV disease, along with information regarding these outbreaks. Additional details of these episodes are described in the following sections.
Marburg Hemorrhagic Fever (MHF)
The first identified instance of filovirus disease occurred in 1967, when MARV caused severe cases of hemorrhagic fever in Europe.274,275,276 The epidemic started in mid-August in Marburg, Germany with three laboratory workers who contracted the disease after processing organs from African green monkeys (Cercopithecus aethiops) imported from Uganda. Seventeen more patients were hospitalized and two medical personnel contracted the disease while attending to patients. The last patient, who apparently had been infected by her husband during the convalescent period, was admitted in November 1967.276 Six cases (including two secondary infections) occurred in Frankfurt, Germany concomitant with the Marburg infections.372 In September, two cases were identified in Belgrade, former Yugoslavia, in which a veterinarian was infected while performing a necropsy on a dead monkey and transmitted the virus to his wife, who nursed him early in his illness.276 A total of 31 cases (including six secondary infections) were identified, with seven fatalities in primary infections (23%). Subsequent serologic investigations have suggested that there was one additional primary case in Marburg.370
MHF remained an obscure medical curiosity until 1975, when three cases were reported in Johannesburg, South Africa.122 The index case was a white male who just prior to his infection had traveled in Zimbabwe. Seven days after onset, his travel companion also became ill and transmitted the disease to an attending nurse; the index case was the only fatality. An investigation was conducted along the travel route of the index case, but the source of the virus was not determined.66 Two further episodes of MHF were reported from Kenya in 1980 and 1987. The index case in 1980 became ill in western Kenya and died in Nairobi, where an attending physician was infected but survived the disease.371 In 1987, a fatal case occurred in the same region of western Kenya.218 The 1980 and 1987 index cases both traveled to the Mt. Elgon region, which is located close to Lake Victoria and was the source of the monkeys that initiated the original 1967 outbreak (trapped near Lake Kyogo, Uganda).
The first outbreak of MHF in a community setting of central Africa started in October 1998 in Durba/Watsa, located in the northeastern region of the Democratic Republic of Congo (DRC). Its remote location and the hazards of an ongoing armed conflict hampered efforts to study this outbreak, but an investigation was initiated after the death of an attending physician in 1999. Sporadic cases continued and were directly or indirectly linked to activity in the vicinity of an underground gold mine. Primary cases were mainly gold miners who started multiple, usually short chains of human-to-human transmission within their families. Overall, 154 cases were reported with a case fatality rate (CFR) of 83%. Analysis of viral sequences derived from clinical specimens and virus isolates showed nucleotide diversity up to approximately 20%.22,23,65,400,401 The largest outbreak of MHF (252 cases with 227 deaths; CFR of 90%) took place in northern Angola, Uige province. The first cases date back to October/November 2004, but initial diagnostic tests were negative for filoviruses. The main outbreak started in February/March 2005 and the last confirmed case died in July. Initial infections were linked to a Uige hospital and included a high number of pediatric cases. Sequence analysis of virus isolates suggested a single introduction into the community.151,211,401 The latest MHF episode dates to 2007, with four documented cases associated with a single mine in western Uganda.400 In addition, two imported cases were reported from the United States (nonlethal) and the Netherlands (lethal), who independently visited the same cave in Uganda in 2008.6,396
Ebola Hemorrhagic Fever (EHF)
EHF was first reported in 1976, when EBOV appeared simultaneously in the DRC (at that time Zaire) and Sudan with 318 (CFR of 88%) and 284 cases (CFR of 53%), respectively. These epidemics were determined to have been caused by two distinct species (ZEBOV and SEBOV), a fact not recognized until years later. Viruses were isolated from patients of both outbreaks and named after a small river in northwestern DRC.36,449,450
No index case was clearly identified in the Sudan outbreak in 1976, although initial cases originating in Nzara, Sudan, involved six cotton factory workers and their close relatives. The epidemic was augmented by the spread of cases to neighboring areas (Maridi, Tembura, and Juba). High levels of transmission occurred in the hospital of Maridi (a teaching center for student nurses), primarily through the use of contaminated needles and a lack of barrier nursing practices. At the same time, a larger outbreak in the DRC, centered around a Belgian mission hospital in Yambuku, Equateur Region, was being fueled by similar circumstances. During a 7-week period of the outbreak, the single most significant factor in the spread of infection in the hospital was the reuse of contaminated syringes and needles, although secondary transmission to family members caused 45% of all recorded infections. The outbreak ended with closure of the hospital and quarantining of infected patients.
In 1977, a single fatal ZEBOV case was reported from Tandala, DRC, about 325 km from the original focus of the 1976 Yambuku outbreak.179 SEBOV reemerged in 1979 in Nzara and Yambio, Sudan. The index case worked in the same textile factory cited as the potential source of infection in the 1976 Sudan outbreak. Hospitalization of the patient led to four nosocomial infections and further transmission to five families (34 cases with 22 fatalities).451
No further cases of EHF were reported until 1994, when a novel EBOV (ICEBOV [CIEBOV]) was isolated from an ethnologist who had become ill while working in the Tai Forest reserve of Ivory Coast. The infection was determined to have occurred while performing a necropsy on a dead chimpanzee (whose troop had lost several members to EHF).114,243 Later, a single seroconversion suggested a second nonfatal human case in nearby Liberia. This episode extended the geographic distribution of known EBOV cases to include most of the African rain forest and was the first case in West Africa.
In 1995, a strain of ZEBOV very similar to the original 1976 virus reemerged in the DRC, causing a large hospital and community outbreak of EHF in and around Kikwit.223,453 The presumed index case was a charcoal worker, but transmission escalated following two consecutive laparotomies performed on an infected male laboratory worker at Kikwit General Hospital. About three-quarters of the first 70 patients within the subsequent developing epidemic were health care workers. In total, there were 315 cases and 250 deaths (CFR of 81%). Major risk factors for contracting disease were involvement in patient care in hospitals and households and preparations of bodies for burial.
Beginning in 1994, ZEBOV became active in or adjacent to the central African rain forest on both sites of the border between Gabon and the Republic of Congo (RC).5,148 Almost all outbreaks in this region described in this section were associated with hunting and butchering of wildlife, often
great apes. The first epidemic was reported in 1994 from the Ogooué-Ivindo Province in northeast Gabon with a total of 52 cases (CFR of 60%).147 In 1996, two more outbreaks were reported from the same province.147 The first epidemic started in early February and included 37 cases (CFR of 57%); the second episode began in July/August and resulted in 60 cases (CFR of 75%). The latter epidemic included an imported case in South Africa where an ill Gabonese physician infected a nurse who died with EHF (2 cases; CFR of 50%). The first reported epidemic that crossed the border into the RC began in late November of 2001 with the index case again reported from Ogooué-Ivindo Province in northeast Gabon. The epidemic spread to Mekambo and Makokou and from there into the RC by ill Gabonese who sought medical care by traditional healers. In total, there were 65 (CFR of 82%) and 59 (75%) cases from Gabon and the RC, respectively.251 The next occurrence of EHF was a large epidemic reported from the districts of Mbomo and Kelle in Cuvette Ouest Region, RC, in late 2002 to May 2003 with 143 cases (CFR of 90%), followed in late 2003 by a smaller episode in the district of Mbomo with 35 cases (CFR of 83%).115 A neighboring area (Etoumbi) was affected in 2005 by a small outbreak of EHF with 12 cases (CFR of 75%) (http://www.who.int.easyaccess1.lib.cuhk.edu.hk/csr/don/2005_06_16/en/index.html) (Table 32.1). This has so far been the last reported outbreak in this region.
great apes. The first epidemic was reported in 1994 from the Ogooué-Ivindo Province in northeast Gabon with a total of 52 cases (CFR of 60%).147 In 1996, two more outbreaks were reported from the same province.147 The first epidemic started in early February and included 37 cases (CFR of 57%); the second episode began in July/August and resulted in 60 cases (CFR of 75%). The latter epidemic included an imported case in South Africa where an ill Gabonese physician infected a nurse who died with EHF (2 cases; CFR of 50%). The first reported epidemic that crossed the border into the RC began in late November of 2001 with the index case again reported from Ogooué-Ivindo Province in northeast Gabon. The epidemic spread to Mekambo and Makokou and from there into the RC by ill Gabonese who sought medical care by traditional healers. In total, there were 65 (CFR of 82%) and 59 (75%) cases from Gabon and the RC, respectively.251 The next occurrence of EHF was a large epidemic reported from the districts of Mbomo and Kelle in Cuvette Ouest Region, RC, in late 2002 to May 2003 with 143 cases (CFR of 90%), followed in late 2003 by a smaller episode in the district of Mbomo with 35 cases (CFR of 83%).115 A neighboring area (Etoumbi) was affected in 2005 by a small outbreak of EHF with 12 cases (CFR of 75%) (http://www.who.int.easyaccess1.lib.cuhk.edu.hk/csr/don/2005_06_16/en/index.html) (Table 32.1). This has so far been the last reported outbreak in this region.
In 2000 to 2001, the largest known epidemic of filovirus disease occurred in Uganda, with 425 cases and 224 deaths. The causative agent was closely related to SEBOV from the Sudan 1976 and 1979 outbreaks, and marked the first appearance of EBOV in Uganda. The CFR of 53% was in line with the generally lower mortality associated with the SEBOV species.454 The epidemic was mainly concentrated in the Gulu district, a savannah area located in the north of the country close to the Sudanese border, with person-to-person transmission including nosocomial infections. The index case was never identified. During the epidemic, the virus spread to the neighboring Masindi district and more distantly to the town of Mbarara in southwestern Uganda.31 During this outbreak a high number of health care workers were infected after barrier nursing procedures were instituted. It was also the first time that laboratory diagnostics were performed in the field to assist in outbreak management.454 In 2004, southern Sudan was again affected by a small SEBOV outbreak with 17 cases, of which 7 died (CFR of 41%).455 The index case had butchered a monkey and human-to-human transmission was mainly by contact.
The last reported ZEBOV outbreaks occurred in the Kasai Occidental province of the DRC in 2007 and 2008/09. The first larger outbreak included 264 reported cases with a CFR of 71%457; the second smaller outbreak had 32 cases, of which 15 died (CFR of 47%).458 Both outbreaks affected rural communities in the vicinity of the city of Luebo and are thought to be related to hunting and handling of migratory fruit bats.248
A new EBOV species, designated Bundibugyo ebolavirus (BEBOV), has been identified as the causative agent for an outbreak that occurred in the Bundibugyo district in western Uganda in 2007.403,423 In total, there were 149 reported cases, with 37 deaths (CFR of 25%); of these, 56 cases were laboratory confirmed. This single outbreak had the lowest reported CFR among all EBOV that have caused outbreaks in central Africa so far.
Most recently, a single case of SEBOV has been reported from Central Uganda.7 No further cases have been reported.
EHF in Nonhuman Primates
In November 1989, an EBOV with low or no apparent pathogenicity for humans was recognized in a shipment of cynomolgus monkeys (Macaca fascicularis) housed at a quarantine facility in Reston, Virginia. These monkeys were imported from a single supplier in the Philippines, and an unusually high mortality was observed in animals during transportation and quarantine. Simian hemorrhagic fever virus was also circulating in the facility; efforts to culture this virus led to the detection of a new species of EBOV that was named Reston ebolavirus (REBOV).72,203 The actual origin of this novel EBOV was never determined. Resumption of importation of monkeys led to new outbreaks of monkey disease in the United States in 1990 and 1996336 and in Italy in 1992.452 Subsequent investigations have traced all shipments except one to a single supplier in the Philippines. The mode of contamination of this exporter’s holding compound has never been ascertained, but whether the virus persisted in the facility or was reintroduced from wild-caught animals, the result was a continued movement of infected macaques. A few infected handlers were also identified by serologic methods without reports of severe illness or suspicious deaths among this cohort.285 Improved shipping, housing, and quarantine regulations regarding importation of monkeys have been implemented to protect the United States from future episodes of EBOV introductions.74 Recently, REBOV emerged in pigs in the Philippines.14 The pigs were co-infected with porcine respiratory and reproductive virus (PRRS) and the actual pathogenic potential of REBOV in pigs remains unclear. This discovery certainly raises issues for food production. Six workers from pig farms and slaughterhouses developed antibodies to REBOV, indicating that they became infected but did not develop disease. The potential for REBOV as a human pathogen remains unanswered but should not be totally dismissed. As of yet, REBOV infections/exposures have never resulted in clinical disease in humans.
Note, in 2012, Uganda has reported two EHF and one MHF outbreak; another EHF outbreak was reported from DRC. The EHF outbreaks were caused by SEBOV and BEBOV. The border region between Uganda and DRC seems to be the new “hot spot” for filovirus infections. For more information, please see the CDC and WHO websites.
Laboratory Infections/Exposures
A single laboratory infection of EHF occurred in the United Kingdom in 1976. Treatment with human leukocyte interferon and human convalescent plasma was initiated and the patient survived.87 In the past two decades there have been at least three laboratory infections with MARV (1 fatal) in Russia.282 In 2004, accidental ZEBOV exposures via needlesticks while working with animals occurred in the United States and Russia, but only the latter became infected (fatally).196,233 In 2009, a German researcher had an accidental ZEBOV exposure via needlestick while working with animals. The person was treated with a recombinant vesicular stomatitis virus (VSV)–based vaccine expressing the ZEBOV glycoprotein. It could not be determined if the exposure resulted in infection.158
Virion Structure
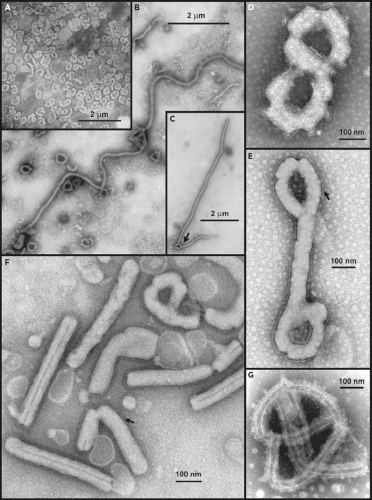
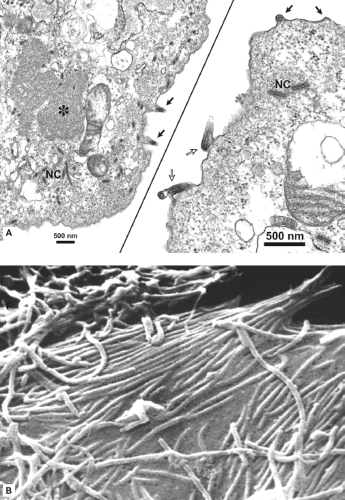
Initial electron microscopic (EM) observations of filoviruses revealed distinctive bacilliform to filamentous virus particles; it was this highly characteristic morphology that inspired their name (Latin filum, thread).294,316 The virions of MARV and EBOV produced in tissue culture are pleomorphic, appearing as either U-shaped, 6-shaped, or circular (torus) configurations, or as elongated filamentous forms of varying length (up to 14,000 nm), all from the same culture fluid (Figs. 32.1A–C). The filamentous forms can also be seen to form branched structures (Fig. 32.1C, arrow). The unit length associated with peak infectivity for MARV and EBOV was measured to be 860 and 1,200 nm, respectively.139 Virions have a uniform diameter of 80 nm, contain a helical ribonucleoprotein complex or nucleocapsid (NC) roughly 50 nm in diameter (Figs. 32.1 and 32.2), and have a central axial space (∼20 nm in diameter) running the length of the particle. The NC has a helical periodicity of ∼5 nm (Fig. 32.1G), and is surrounded by a matrix protein and a closely apposed outer envelope derived from the host-cell plasma membrane. The virion surface is studded with membrane-anchored peplomers projecting ∼10 nm from the surface (Figs. 32.1E and 32.1F). Virions can often appear ragged or “moth-eaten” (Fig. 32.1D) (especially late in the infection). The density of virions has been determined to be 1.14 g/mL by centrifugation in a potassium tartrate gradient.
Genome Structure and Organization
The single-stranded, negative-sense RNA molecule that makes up a filovirus genome constitutes ∼1% of the virion mass.330 The genomes of filoviruses are very similar in their organization, which generally conform to those of paramyxoviruses and rhabdoviruses, but their complexity is more akin to those of paramyxoviruses. Filovirus genomes are approximately 19,000 bases in length, making them the largest in the order Mononegavirales (Fig. 32.2), and contain seven sequentially arranged genes in the order nucleoprotein (NP)–viron protein (VP) 35–VP40—glycoprotein (GP)–VP30–VP24—polymerase (L). Genes are delineated by conserved transcriptional signals, and begin close to the 3′ end of the genomic sequence with a start site and end with a stop (polyadenylation) site. For rhabdo- and paramyxoviruses, genes are usually separated by short intergenic regions of one or more nucleotides, which are also seen in filovirus genomes. An unusual feature of all filovirus genomes is the presence of gene overlaps, which have been identified in the genomes of some paramyxo- and rhabdoviruses, but do not resemble those of filoviruses. As seen in Figure 32.2, the stop site of an upstream (3′) gene overlaps the start of the downstream gene, and overlapping sequences are limited to the conserved transcriptional signals and are centered on a 3′-UAAUU pentanucleotide sequence common to start and stop sites.49,98,103,157,192,347 There is one overlap in the MARV genome (VP30–VP24), but the characterized EBOV genomes contain at least two overlaps (VP35–VP40, GP–VP30, and VP24–L; REBOV lacks the GP–VP30 overlap). Intergenic regions of filovirus genomes are generally short, although all genomes have a single lengthy sequence (>120 bases) separating the GP and VP30 genes of MARV and the VP30 and VP24 genes of EBOV. The positioning of the MARV overlap and a long intergenic region (that precedes the VP30 gene) appears to be shifted one gene (in the 3′ direction) with respect to the genome of the EBOV. The significance of this arrangement and how it may have been generated are unknown. The extragenic sequences at the 3′ end of all filovirus genomes (leader) are short, ranging in length from 50 to 70 bases, while the length of the 5′ end (trailer) sequences are variable. The extreme 3′ and 5′ ends of the filovirus genomes are conserved, show a high degree of complementarity, and potentially form stem-loop structures.67,290,352,414 Filovirus trailer sequences are more variable in length, the longest being that of ZEBOV (677 bases), followed by BEBOV (475 bases), ICEBOV (474 bases), SEBOV (381 bases), MARV (76–95 bases), and REBOV (25 bases).
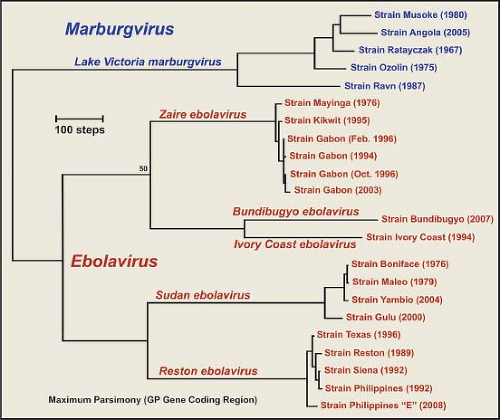
The evolutionary profile of the family Filoviridae (Fig. 32.3) indicates that EBOV and MARV represent distinct filovirus lineages, the five species of EBOV also represent distinct lineages, and there is an extraordinary level of genetic stasis within the lineages of EBOV. Nucleotide and amino acid differences between MARV and EBOV are both approximately 55%, whereas EBOV species show 32% to 41% differences in nucleotide and amino acid sequences.98,353,400 These same levels of sequence variation are also seen when other genes are compared. Within species of EBOV, however, there is a remarkable degree of genetic stability, indicating that these viruses have most likely reached a high degree of fitness as they have adapted to their respective niches. MARV isolates have not shown the degree of variation seen among EBOV species, but two lineages of MARV have been described that are genetically distinct by more than 20% genetic diversity.98,354,400
Virus Proteins
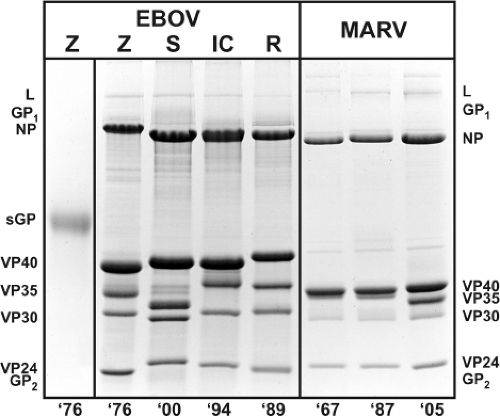
Filovirus structural proteins can be subdivided into two categories, those that form the NC and those that are associated with the envelope (Fig. 32.2 and Table 32.2). The NC-associated proteins are involved in the transcription and replication of the genome, whereas the envelope-associated proteins have a role in either the assembly of the virion or virus entry. Shown in Figure 32.4 are the characteristic migration patterns of purified filovirus proteins separated by SDS-PAGE.
Nucleoproteins
The NP and VP30 proteins of filoviruses are the major and minor nucleoproteins, respectively, are phosphorylated, and interact strongly with the genomic RNA molecule to form the viral NC (along with VP35 and L).27,261 Expression of recombinant NP alone in mammalian cells results in the formation of inclusions and nonspecific association with cellular RNA to form helical structures.306,435 Analysis of NP amino acid sequences has identified a conserved, hydrophobic N-terminal half that contains all the cysteine residues, and a divergent, hydrophilic C-terminal half that contains most of the proline residues and is extremely acidic.348,349 The N-terminal 450 amino acids of the ZEBOV NP have been linked with self-assembly of NP into tube-like structures that may function as a platform for NC formation.435 Predicted mass values for NP molecules are approximately 20 kd smaller than estimated sizes derived from SDS-PAGE migration, possibly as a result of reduced binding of SDS molecules to the negatively charged NP. This hypothesis is supported by a study of recombinant
ZEBOV NP363 that mapped this property to two C-terminal domains (aa 439–492 and 589–739). In the central region of the NPs of filoviruses is a highly conserved region that shows some homology with nucleoprotein sequences of paramyxoviruses, and to a lesser extent rhabdoviruses, and likely has a similar structure and function.183,349
ZEBOV NP363 that mapped this property to two C-terminal domains (aa 439–492 and 589–739). In the central region of the NPs of filoviruses is a highly conserved region that shows some homology with nucleoprotein sequences of paramyxoviruses, and to a lesser extent rhabdoviruses, and likely has a similar structure and function.183,349
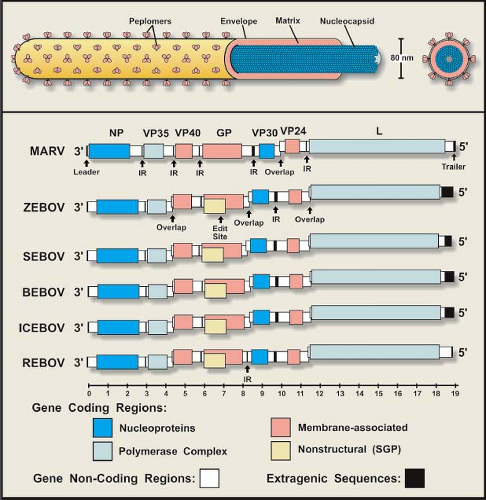 Figure 32.2. Schematic representation of a filovirus particle (top) and the organization of filovirus genomes (bottom). |
The VP30 protein of ZEBOV is also capable of binding RNA, particularly a stem-loop structure located near the leader sequence; this property mapped to residues 26 to 40 that is arginine rich.214 This region may have an additional role of binding to the acidic C-terminal half of NP. VP30 contains a zinc-finger motif ∼70 to 80 residues from the N-terminus that is highly conserved in filoviruses (consensus = CX8CX4CX3HX2D/E),286 and RNA binding activity is increased by Zn2+.214 Immediately C-terminal to this sequence (separated by six residues) is a conserved tetraleucine sequence linked to co-translational homo-oligomerization of VP30.168 Additional studies have shown that the C-terminal half of VP30 also contains a homo-oligomerization domain and that hexamerization occurs via an N-terminal domain.169 A functional study of recombinant ZEBOV VP30 has revealed that it behaves as a transcription activator169,440 regulated by its phosphorylation state,287 but this property appears to be absent from the VP30 of MARV.289 The ZEBOV VP30 also interacts with L; a role of bridging NP and L in the NC complex has been postulated.154 Recently, the ZEBOV VP30 (along with VP35 and VP40) has been implicated in suppressing antiviral immunity through its antagonistic effect on the host cellular RNA interference (RNAi) pathway.91
Polymerase Complex Proteins
The L and VP35 proteins form the polymerase complex, which transcribes and replicates the filovirus genome. The
L protein provides the RNA-dependent RNA polymerase activity of the complex; motifs linked to RNA (template) binding, phosphodiester bonding (catalytic site), and a ribonucleotide triphosphate binding have been described.290,414 Conserved regions or “blocks” of sequences have been identified in filovirus L proteins, but there are also areas of divergence (particularly within the C-terminal quarter of the molecule) and sequences that are unique to the larger MARV L protein.
L protein provides the RNA-dependent RNA polymerase activity of the complex; motifs linked to RNA (template) binding, phosphodiester bonding (catalytic site), and a ribonucleotide triphosphate binding have been described.290,414 Conserved regions or “blocks” of sequences have been identified in filovirus L proteins, but there are also areas of divergence (particularly within the C-terminal quarter of the molecule) and sequences that are unique to the larger MARV L protein.
Table 32.2 Filovirus Genes and the Functions and Relative Molecular Weights of Their Gene Products | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||
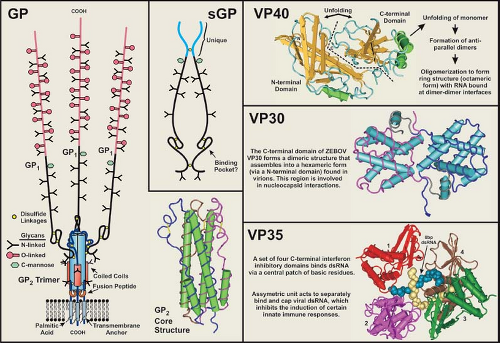
VP35 has an essential role as a cofactor that affects the mode of RNA synthesis (transcription or replication), similar to that of the P proteins of other NNS viruses290,292,321,415; VP35 acts as a link between L and NP.28 Basic residues in the C-terminal region have been identified as critical to viral RNA synthesis.321 The ZEBOV VP35 has also been shown to be a virulence factor through its inhibitory effect on the host innate immune system.255 VP35 has an antagonistic effect on the interferon type I pathway (see Host Immune Response section) by binding virus-generated double-stranded RNA (dsRNA) and by directly interfering with pathway kinases19,20,21,54,90,170,171,253,322,323; as noted previously, it also acts as a suppressor of RNA silencing.91,162 A C-terminal domain in VP35 confers its dsRNA binding property171 and X-ray crystallography studies of ZEBOV and REBOV VP35 have shown that it forms an asymmetrical dimer, the units of which separately bind and “end-cap” dsRNA226,253,254,255 (Fig. 32.5). Additionally, the ZEBOV VP35 interacts with the 8kDa dynein light chain, a component of the microtubule transport system,237 which may have an effect on the virus life cycle.
Structural (Surface) Glycoprotein
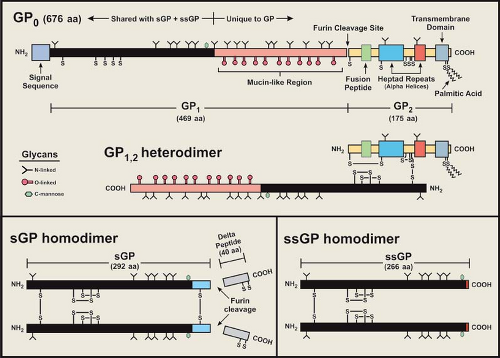
As noted earlier, the surface of the filovirus virion is covered with peplomers (spike structures) composed of the structural glycoprotein, GP, and is anchored in the envelope in a type I orientation (Figs. 32.1 and 32.2). GP has been the most studied of the filovirus proteins, due in large part to its role in virus entry, its influence on pathogenesis, its antigenicity, and its attractiveness as an immunogen in vaccine development. The GP of EBOV species is encoded in two reading frames and expressed through transcriptional editing (353,410; see Stages of Replication, next section), while the GP of MARV is encoded in a single ORF.49,103 Despite this difference, the features of their amino acid sequences are very similar. A schematic representation of the EBOV GP is depicted in Figure 32.6 and illustrates the general characteristics of a filovirus GP molecule.
The glycoproteins of filoviruses are translocated into the endoplasmic reticulum (ER) by a signal sequence at the N-terminus of GP0 (precursor molecule) and are anchored by a membrane-spanning sequence at the C-terminus (Fig. 32.6); the cytoplasmic tail is extremely short (3 residues for EBOV and 7 for MARV). As GP0 is transported through the ER and Golgi apparatus, it is glycosylated with both N-linked glycans (hybrid and complex) and O-linked glycans.104,107,149,409 An extremely divergent, mucin-like region (rich in threonine, serine, and proline residues) is located in the middle of GP0 and is heavily glycosylated; all O-linked glycans are located in this region. Analysis of the carbohydrate composition of GP has shown that MARV isolates lack terminal sialic acid when grown in Vero E6 or MA-104 cells, unlike the GP of EBOV species, which contain abundant α(2–6) and/or α(2–3) linked sialic acids.104,149 Differences in sialic acid addition may be caused by differences in targeting as they are directed though the trans-Golgi apparatus. In addition, no neuraminidase activity has been found with any filovirus. The MARV GP (Musoke strain) is also phosphorylated by Golgi protein
kinases, putatively at serine residues near the center of GP1 (260SSDDEDLATSGSGS273)357 the C-terminal set of 3 serines is conserved in MARV.354 The implications of this processing are unknown, but could influence the trafficking of MARV GP.
kinases, putatively at serine residues near the center of GP1 (260SSDDEDLATSGSGS273)357 the C-terminal set of 3 serines is conserved in MARV.354 The implications of this processing are unknown, but could influence the trafficking of MARV GP.
GP0 is cleaved by furin, a subtilisin/kexin-like convertase localized in the trans Golgi, or a furin-like endoprotease at a site just C-terminal to a long, variable, mucin-like region.356,413,417 Cleavage leads to the formation of a GP1,2 heterodimer that is held together by a single disulfide bond formed between the most N-terminal cysteine of GP1 (cysteine at position 53 in ZEBOV) and the fifth cysteine from the N-terminus GP2 (predicted) (Fig. 32.6). The MARV cleavage site is located ∼70 residues N-terminal to that of the EBOV site, and a second conserved furin/furin-like cleavage sequence is located immediately after the second cysteine from the N-terminus of GP2 (just within the first heptad repeat), but there is no evidence that this sequence is cleaved. It should be noted that cleavage of GP0 to form the GP1,2 heterodimer is not required for virus entry in tissue culture, as mutation of the furin cleavage site does not prevent entry by pseudotyped virus,199,448 nor does it significantly affect infection and subsequent spread by a recombinant ZEBOV or virulence in nonhuman primates.301,302 Nevertheless, cleavage may be required for efficient maintenance in the natural host.
Peplomers are composed of trimerized GP1,2 heterodimers, and X-ray crystallography studies of a recombinant-expressed portion of GP2 have shown that trimerization occurs when heptad repeat sequences form coiled coils in a rod-shaped structure (Figs. 32.5 and 32.6) similar to those of the HA2 of influenza, the transmembrane (TM) of retroviruses, and SNAREs.269,442,443 Two conserved cysteine residues at the C-terminal end of the membrane spanning sequence are palmitoylated,119,200 which could stabilize the anchorage of the peplomer and may influence virus entry, although GP pseudotyping studies suggests that these cysteines are not essential for infectivity.200,209 The TM region of GP2 has been linked to increased permeability of infected cells with the 667ALF669 sequence particularly important.165 A fusion peptide is internally positioned near the N-terminus of GP2 and is flanked by two conserved cysteines that are predicted to form a disulfide bond; this arrangement is very similar to the TM of Rous sarcoma virus and avian leukosis virus.120,209 The fusion peptide of ZEBOV inserts efficiently
into synthetic membranes containing phosphatidylinositol and promotes fusion of lipid vesicles.1,200,341,375
into synthetic membranes containing phosphatidylinositol and promotes fusion of lipid vesicles.1,200,341,375
Sequence analysis of the GP gene coding regions indicates that the N-terminal end (∼200 residues) of GP1 and most of GP2 are conserved and have regions of increased hydrophobicity. The N-terminal region of EBOV GP1 contains conserved cysteine residues that are closely positioned and form intramolecular disulfide bonds (C108–C135, C121–C147), which are also found in the sGP molecule15,209,418 and likely form an important structural feature. The MARV GP has conserved cysteines that correspond to the C108–C135 linkage of EBOV, but the other two closely positioned cysteines appear to be shifted towards the center of the molecule (and likely form a disulfide bridge). The abundant O-glycans of the mucin-like region confers an extended structure and its heavy glycosylation makes it very hydrophilic. The mucin-like region is located at the C-terminus of GP1, and was predicted to project away from the virion membrane (toward the aqueous environment) with the N-terminal end (linked to GP2) contributing to the stalk structure of the peplomer (Fig. 32.5). X-ray crystallography of GP1 has verified this prediction.241 In tissue culture, it has been shown that the ectodomain portion of the ZEBOV peplomer is released from cells (separate from virions) through proteolytic cleavage by tumor necrosis factor α-converting enzyme (TACE; zinc-dependent metalloprotease) near the transmembrane anchor (residue D637 of ZEBOV).78 GP can also be released into the medium as peplomers anchored in vesicles extruded from the plasma membrane.416
Nonstructural Glycoproteins
The expression of a nonstructural soluble glycoprotein (sGP) as the primary product of the GP gene of EBOV is unusual and an important distinction from MARV.353,410 The N-terminal ∼300 amino acids of sGP are identical to those of the structural GP, but the C-terminus is unique in sequence (Fig. 32.6). sGP is produced from a precursor molecule that is also cleaved by furin (or a furin-like endoprotease) near the C-terminus to release a short peptide that seems to contain exclusively O-linked glycans and has been named delta peptide.409,419 No biologic activity has been attributed to delta peptide. Biochemical and antigenic analyses of the ZEBOV sGP have shown that it is structurally distinct from GP15,356,418,419 and is secreted from infected cells as a homodimer that is likely formed in
the ER. Initial structural studies indicated an antiparallel orientation for sGP molecules in the dimer by disulfide bonding between cysteine residues C53 and C306.418 However, subsequent MALDI-TOF MS analysis of sGP peptide fragments have unequivocally demonstrated a parallel orientation for the homodimer, which is held together by disulfide bonds between the N-terminal (C53–C53′) and C-terminal (C306–C306′) cysteines that fix the orientation of the molecules.15,95 The intramolecular disulfide bonds are similar in topology and spacing to the fibronectin type II module (binding site for collagen and gelatin), and may form a binding pocket for an as-of-yet unidentified ligand. Biophysical characterization of ZEBOV sGP has also revealed that the tryptophan residue at position 288 is C-mannosylated,16,94 an unusual form of glycosylation, at the specific motif W-X-X-W (first W is C-mannosylated) that is in the shared N-terminal region and is conserved in all EBOV species. GP1 is presumed to have this same type of glycosylation. It is possible that sGP could contribute to disease progression, because large amounts circulate through the blood of acutely infected humans,350 but there has been no evidence linking sGP to a role in pathogenesis.
the ER. Initial structural studies indicated an antiparallel orientation for sGP molecules in the dimer by disulfide bonding between cysteine residues C53 and C306.418 However, subsequent MALDI-TOF MS analysis of sGP peptide fragments have unequivocally demonstrated a parallel orientation for the homodimer, which is held together by disulfide bonds between the N-terminal (C53–C53′) and C-terminal (C306–C306′) cysteines that fix the orientation of the molecules.15,95 The intramolecular disulfide bonds are similar in topology and spacing to the fibronectin type II module (binding site for collagen and gelatin), and may form a binding pocket for an as-of-yet unidentified ligand. Biophysical characterization of ZEBOV sGP has also revealed that the tryptophan residue at position 288 is C-mannosylated,16,94 an unusual form of glycosylation, at the specific motif W-X-X-W (first W is C-mannosylated) that is in the shared N-terminal region and is conserved in all EBOV species. GP1 is presumed to have this same type of glycosylation. It is possible that sGP could contribute to disease progression, because large amounts circulate through the blood of acutely infected humans,350 but there has been no evidence linking sGP to a role in pathogenesis.
Recently, another ZEBOV nonstructural glycoprotein, termed small soluble glycoprotein (ssGP), has been identified and partially characterized,281 and outwardly appears to be a truncated version of sGP. As with GP, ssGP is expressed through transcriptional editing. This glycoprotein is expressed at a low level (∼1/20 that of sGP+GP) and has structural properties similar to that of sGP, in that it has N-linked glycans (no O-linked) and exists as a homodimer (disulfide bond between cysteines at position 53). As with sGP, the function of ssGP has yet to be adequately defined, but ssGP appears to lack an anti-inflammatory property reported for sGP.281
Matrix Proteins
The VP40 protein functions as the matrix protein and the VP24 protein may have a secondary/minor matrix protein function.164 VP40 is the most abundant protein in the virion, while only small amounts of VP24 are incorporated into virus particles (Fig. 32.4). Both proteins have an affinity for membranes and are associated with the virion envelope (no membrane-spanning regions),208,340,362 and are easily released from virions by nonionic detergents under low-salt conditions.85,225 VP40 is critical to the budding process, as it initiates and drives the envelopment of the NC by the plasma membrane.207 In addition, it has been reported that both VP40 and VP24 of EBOV contribute to regulation of genome replication and transcription.180
VP24 has a decidedly hydrophobic profile, and a study of a recombinant-expressed form (ZEBOV) indicates that it has an affinity for the plasma membrane and perinuclear region of infected cells.164 VP24 is capable of forming homotetramers, which is influenced by pH and divalent cation changes. Because disulfide-bonded oligomers of VP24 are not evident in the virion,352 the formation of multimers is likely due to ionic and/or hydrophobic interactions. The precise role of VP24 in the replication of filoviruses is still unclear and direct interactions with other virus proteins have not been described, but a role in formation of nucleocapsid-like structures has been described.164 The VP24 of EBOV has also been reported to antagonize the interferon type I signalling pathway, similar to that of VP35 (see Host Immune Response section).20
Stages of Replication
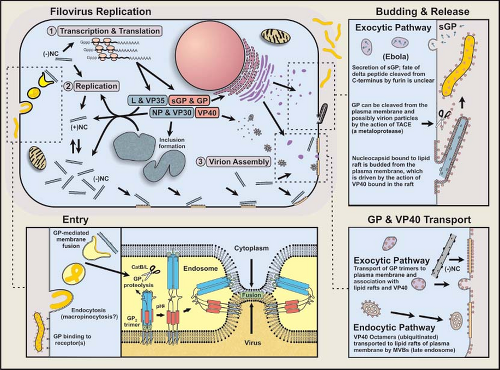
Recent studies have provided valuable insights into filovirus entry into host cells and the mechanisms leading to the production and release of infectious progeny. Although very much incomplete, the details of this complicated series of molecular events are slowly being revealed. The current understanding of this process is illustrated in Figure 32.7 and described in the following sections.
Mechanism of Attachment
In filovirus infections a variety of host organs and cell types are involved; this broad tropism is related in large part to the binding properties of the peplomers that populate the surface of the virion. Because GP is the only filovirus protein involved in initiating infection, it has been intensely studied for its ability to bind cellular receptors. Much of the work directed at receptor binding (and subsequent entry processes) has utilized recombinant pseudotyping systems, which provide a safer and easier approach to characterizing these properties and events.56,58,200,390,447,448,467 However, these results need to be verified using infectious filoviruses, as interactions of GP with VP24 and/or VP40 need to be considered along with other properties that may be peculiar to filovirus virions.
Identification of attachment molecules (receptors?) involved in filovirus entry is complicated by the ability of GP trimers to specifically or nonspecifically bind a variety of host-cell surface molecules. The asialoglycoprotein receptor found on hepatocytes binds MARV,29 yet EBOV also infects hepatocytes despite its GP having sialylated glycans. The β1 group of integrins has been suggested to interact with ZEBOV GP on the cell surface and during intracellular trafficking (when co-expressed),393 although cells that express this molecule (such as Jurkat cells) are not easily infectible. The folate receptor alpha has been implicated as a cofactor in filovirus entry,56 but virus entry independent of this molecule has been shown to take place.366,369 C-type lectins (DC-SIGN and DC-SIGNR; bind oligosaccharide ligands)—present on certain forms of dendritic cells, macrophages, and endothelial cells—are also capable of binding filovirus peplomers,3,12,279,365 especially when N-linked glycans contain high mannose carbohydrates.213,258 However, there are indications that DC-SIGN may not act as an EBOV receptor, but instead acts to promote attachment and other host factors are involved in entry.280 One study has shown that macrophages are more susceptible to virus entry by ZEBOV GP-pseudotyped HIV-1 particles than are monocytes, and that HUVEC cultures pretreated with TNF-α showed increased entry over untreated cells.467 These results imply that changes in cellular gene expression can alter the makeup of surface attachment molecules (and cofactors?). Antibody binding to peplomers might also enhance infectivity through its interaction with the Fc portion of the complement protein C1q bound to the surface of host cells.391 Very recently, T-cell Ig and mucin domain 1 (TIM-1) has been described as a potential binding protein for EBOV GP and enhances virus entry into cells.232
Results of site-directed mutagenesis studies of ZEBOV GP have shown that individual glycosylation sites are not critical to virus entry,209,270 and deletion of the entire mucin-like region can actually increase virus entry in vitro.209 A role in receptor binding or increased binding has yet to be attributed to the mucin-like region. Further deletion of the GP1 C-terminal
sequences (past the mucin-like region), together with mutagenesis of N-terminal residues, has localized the entry function to ∼150 residues at the N-terminus (residues 33–185).270 It should be noted that this region also contains the same intramolecular disulfide bonds as sGP, which potentially forms a binding pocket that is involved in cell attachment and/or receptor binding. If this is the case, then the corresponding region of MARV GP1 would likely have distinctive binding characteristics due to a differing disulfide bonding pattern. In contrast, another study identified approximately the same region as the potential receptor binding site for both viruses, indicating that EBOV and MARV utilize a common receptor.239
sequences (past the mucin-like region), together with mutagenesis of N-terminal residues, has localized the entry function to ∼150 residues at the N-terminus (residues 33–185).270 It should be noted that this region also contains the same intramolecular disulfide bonds as sGP, which potentially forms a binding pocket that is involved in cell attachment and/or receptor binding. If this is the case, then the corresponding region of MARV GP1 would likely have distinctive binding characteristics due to a differing disulfide bonding pattern. In contrast, another study identified approximately the same region as the potential receptor binding site for both viruses, indicating that EBOV and MARV utilize a common receptor.239
Mechanism of Entry and Intracellular Trafficking
Following attachment, virions are presumed to enter the cell by a process of endocytosis, acidification of the endocytic vesicle, and fusion of virus and host membranes resulting in the release of the NC into the cytoplasm. ZEBOV GP-mediated entry and fusion are affected by the treatment of host cells with agents that disrupt microtubules or inhibit the function of microfilaments.467 These cytoskeletal components are key to clathrin-dependent and caveolae-mediated internalization, and support the theory that filoviruses enter the cell through endocytosis. However, studies examining the type of endocytic pathway utilized by filoviruses are conflicting. One study has demonstrated that disruption of the caveola vesicular system (via cholesterol binding compounds) inhibited ZEBOV and MARV entry and that filovirus GP-pseudotyped virus co-localized with the caveolin-1 (cholesterol binding protein) marker.88 However, cells lacking caveolae are infectible and co-expression of folate receptor alpha and caveolin-1 in a T-cell line did not increase infectivity.366 Clathrin-mediated endocytosis and GP-dependent macropinocytosis or a macropinocytosis-like mechanism for EBOV internalization and an involvement of lipid rafts.2,298,344 It should be noted that inhibitors of macropinocytosis (amiloride) and the lipid raft-caveolae endocytosis pathway did not significantly affect ZEBOV entry.345 Latest, it was reported that EBOV entry was dependent on Niemann-Pick CI (NPC1), a protein known to function in cholesterol transport.53 It thus appears that the entry of ZEBOV (and possibly other filoviruses) occurs through multiple routes.
A filovirus virion internalized in a vesicle at the plasma membrane traffics through the endosomal pathway, and at some point in time the NC is released into the cytoplasm by GP2-mediated fusion of the virus envelope and endosomal membrane. Membrane fusion is dependent on endosomal acidification,392,447 and endosomal proteolysis of the GP1 subunit peplomer by the cysteine proteases CatL and CatB (active in acidic pH environments) can enhance ZEBOV entry.44,59,222,345 CatL removes the glycan cap and mucin-like domain, exposing core residues of a recombinant peplomer and increasing infectivity.182,222 Removal of GP1 is believed to set off a conformational change in the GP2 trimer that triggers the deployment of the fusion machinery, resulting in the insertion of the GP2 fusion peptides into the endosomal membrane. This event would link and draw viral and host membranes together to induce fusion and the release of the NC into the cytoplasm.436 The minimum number of peplomers needed to induce fusion has not been determined.
A filovirus virion internalized in a vesicle at the plasma membrane traffics through the endosomal pathway, and at some point in time the NC is released into the cytoplasm by GP2-mediated fusion of the virus envelope and endosomal membrane. Membrane fusion is dependent on endosomal acidification,392,447 and endosomal proteolysis of the GP1 subunit peplomer by the cysteine proteases CatL and CatB (active in acidic pH environments) can enhance ZEBOV entry.44,59,222,345 CatL removes the glycan cap and mucin-like domain, exposing core residues of a recombinant peplomer and increasing infectivity.182,222 Removal of GP1 is believed to set off a conformational change in the GP2 trimer that triggers the deployment of the fusion machinery, resulting in the insertion of the GP2 fusion peptides into the endosomal membrane. This event would link and draw viral and host membranes together to induce fusion and the release of the NC into the cytoplasm.436 The minimum number of peplomers needed to induce fusion has not been determined.
Transcription and Translation
Following filovirus entry, negative-strand RNA genetics dictates that transcription is the first (and obligatory) viral process, similar to paramyxoviruses and rhabdoviruses. Once the nucleocapsid is released into the cytoplasm, polyadenylated monocistronic messenger RNAs (mRNAs) are synthesized from virus genes in a 3′ to 5′ direction (with polar attenuation) from the encapsidated genomic RNA template. Transcription seems to involve a process of starting and stopping as the polymerase complex encounters conserved start (initiation) and stop (termination/polyadenylation) sites along the genome. Synthesis of the “leader” sequence is postulated to occur, but intergenic sequences and the “trailer” sequence seem to be ignored, although this has not been shown experimentally. NP mRNA can be detected as early as 7 hours postinfection, and peaks around 18 hours.346 It is assumed that transcripts are capped at the 5′ end (7MeG5′-ppp5′-R) by the L protein, as it contains conserved motifs associated with this enzymatic activity.108
Analyses of defective interfering particles of ZEBOV have shown that promoters for initiating RNA synthesis are contained within 156 and 177 nucleotide regions of the genomic and antigenomic RNA 3′ termini, respectively.52 Subsequent to these studies, it was shown that a bipartite promoter is located within the first 128 nucleotides of the 3′ end of the ZEBOV genome.441 One element is located at the extreme 3′ end and the other within the nontranslated region of the NP gene. These elements are separated by a nonspecific sequence (nucleotides 56–80) that acts to provide proper spacing and also contains the NP gene transcription start site. This bipartite promoter is similar to that of various paramyxoviruses (i.e., Sendai virus) and obeys the “rule of 6”.441
Transcriptional start sites are 12 or 14 nucleotides in length and end in the consensus sequence 3′-CUUCUAAUU for EBOV and 3′-CUURUAAUU for MARV, while stop sites are 11 or 12 nucleotides long with the conserved sequence 3′-UAAUUC(U)5/6. Polyadenylation is believed to occur by slippage or stuttering of the polymerase at the 5 to 6 uridines ending the stop site. A characteristic that is unique to the transcriptional signals of filoviruses is a common pentanucleotide sequence, 3′-UAAUU, present at the 5′ end of start sites and at the 3′ end of stop sites103; the stop sites of ZEBOV and REBOV polymerase genes deviate slightly from this sequence (3′-UAAUA).
The mechanism initiating transcription of a downstream (5′) gene involved in a gene overlap and the consequences of this arrangement are unknown. Because the overlaps are short (18–21 nucleotides), the proximity of the polymerase may not affect recognition of the start site as it finishes polyadenylating the upstream gene. The function of these overlaps remains unclear, but attenuation of transcription does not appear to take place, as the transcription of the VP40 and VP30 genes of ZEBOV is substantial and expression of VP40 is very strong.
Filovirus mRNA molecules have characteristics that make them somewhat unique. They contain long noncoding regions at their 3′ and/or 5′ ends, which contribute to the increased length of the genome and may function in the stability of transcripts. In addition, the 5′ ends of filovirus transcripts have the potential to form stable, stem-loop structures, which might affect their stability and ribosome binding capacity/translation.291,347,352
The ZEBOV VP30 has a transcription activation property that is linked to an RNA secondary structure formed at the 5′ end of the NP gene transcript as it is synthesized168,169,286; the presence of VP30 is required for transcription of downstream genes. This property is impaired by phosphorylation at six serines and one threonine at the N-terminus, and restored by the action of cellular phosphatases.286 Because ZEBOV VP30 in the virion is at least partly phosphorylated,85 the action of phosphatases on the NC may be required before transcription proceeds efficiently. Thus, the phosphorylation state of VP30 may be a critical component in regulating EBOV RNA synthesis; a corresponding mechanism has not been shown for MARV.
The organization and transcription of the GP genes of EBOV are unusual and provide an important distinction between MARV and EBOV. The MARV GP gene encodes a single product, GP, in a conventional open reading frame (ORF), whereas all EBOV species encode their GP in two ORFs (−0 and −1 frames). Expression of the EBOV GP requires a transcriptional editing event281,353,410 comparable to the editing described for the phosphoprotein gene of certain paramyxoviruses. Translation of the unedited transcript of the EBOV GP gene results in the production of sGP, a smaller, nonstructural, secreted glycoprotein, the primary gene product (Figs. 32.4 and 32.6). The transcriptional editing event that leads to GP expression occurs at a series of seven uridines on the genomic RNA template and results in the insertion of an additional adenosine, which connects the GP open coding frames; approximately 20% to 25% of the transcripts are edited. The mechanism of insertion most likely evolved out of the polymerase’s ability to polyadenylate by stuttering on a poly(U) template. However, insertion of a single nucleotide at the editing site appears to occur with a high degree of fidelity, but insertion of two adenosines can occur (in ∼5% of GP gene transcripts), which leads to the synthesis of low levels of ssGP.281 The editing of EBOV GP gene transcripts is the only example of a virus glycoprotein that is expressed through this type of mechanism. Sequence analysis of the GP genes of MARV isolates indicates that a nucleotide sequence that corresponds to the editing region of EBOV GP genes is totally absent.49,103,356,445 The difference in filovirus GP gene
organization provides important evidence pointing to a divergent evolution for EBOV and MARV.
organization provides important evidence pointing to a divergent evolution for EBOV and MARV.
Replication of Genomic Nucleic Acid
In addition to transcription, the promoter at the 3′ end of the genomic RNA also drives the synthesis of full-length complementary/antigenomic RNA from the encapsidated template. As with other NNS RNA viruses, the ends of the genome have a high degree of sequence complementarity,67,352 and stem-loop structures are predicted to form at the 3′ and 5′ ends of genomic and antigenomic RNAs. These structures are believed to be essential to the replication of filoviruses.67 The initial expression of virus genes leads to a buildup of viral proteins (especially NP), which is thought to trigger a switch from transcription to replication. This switch results in the synthesis and encapsidation of antigenomic RNA molecules, which in turn serve as templates for genomic RNA that is also rapidly encapsidated. Depletion of capsid proteins is believed to cause a return to transcription, and eventually an equilibrium is established wherein transcription and replication are concurrent processes. As replication progresses in the infected cell, NC particles containing genomic RNA accumulate and are directed to the plasma membrane for virion assembly.
The development of reverse genetics systems based on EBOV and MARV genetics has provided significant advances in understanding filovirus replication32,89,156,289,292,301,395,415 and has allowed the production/reconstitution of recombinant ZEBOV and MARV (Musoke strain) from plasmid DNA.89,301,415 For MARV, the NP, VP35, and L proteins are all that is required to transcribe and replicate minigenomes,298 but systems developed for ZEBOV also required VP30.301,395,415 When components of minigenome reporter gene systems for REBOV and ZEBOV were switched, it was noted that cis-acting signals and nearly all combinations of proteins were exchangeable.32 Rescue of recombinant ZEBOV using NC-associated proteins from REBOV or MARV has also shown that exchanging of these heterologous proteins can lead to recovery of recombinant virus.395
Assembly and Release
When sufficient levels of negative-sense nucleocapsids and envelope-associated proteins are reached, a coalescing of these components occurs at the plasma membrane,207 or to a lesser extent at membranes forming intracellular vacuoles.96 Filovirus-infected cells develop prominent inclusion bodies, easily visualized by light, immunofluorescent, and electron microscopy.183,294 Inclusions are induced by NP, but also contain other proteins that form the NC.28 Inclusions may be a source of components for forming NCs, which can be seen associated with inclusions (Fig. 32.8A). Recombinant-derived, NC-like structures form in cells expressing NP, which may be facilitated by the expression of VP35 and VP24.183,305 NC particles are believed to interact with VP40 molecules in the budding process.
Membrane/lipid rafts have been identified as platforms for the assembly of filovirus virions.24,310 Membrane rafts are rigid microdomains (containing sphingolipids and cholesterol) present in biological membranes and are isolated from the fluid phospholipids surrounding them. GP trimers conveyed to the plasma membrane have an affinity for these lipid rafts, which is associated with palmitoylation of the membrane-spanning anchor sequence.24
Structural and functional studies of VP40 have provided important insights into the assembly of filovirus virions.75,173,198,207,229,230,257,461 Posttranslational processing and intracellular trafficking of VP40 result in the deposition of VP40 at the plasma membrane via the vacuolar protein sorting/endosomal pathway. By itself, ZEBOV VP40 is capable of mediating its own release from mammalian cells to form enveloped virus-like particles (VLPs),208,256,307,399 which are more efficiently produced when GP and NP are present256; VP40 interacts with the C-terminal 50 amino acids of NP.257 ZEBOV VP40 determines VLP morphology and density,219 and likely has the same influence on infectious filovirus particles. A structural study of ZEBOV VP40 demonstrated that it associates with lipid bilayers containing a high level of L-α-phosphatidyl-L-serine (abolished by 1 M NaCl).340 It was also found that this property maps to the C-terminal ∼110 residues, which contains basic and hydrophobic regions that could bind membranes.397 In addition, the N-terminal region is involved in oligomerization, and deletion of the C-terminal region of VP40 allowed it to hexamerize into ring structures. The crystal structure of monomeric ZEBOV VP40 is composed of similar/related β sandwich domains (N-terminal and C-terminal) connected by a hinge region, which unfolds upon interaction with membranes and dimerizes in an antiparallel orientation (Fig. 32.5).362 These dimers form octomeric rings (∼84 Å diameter) with a central pore and RNA binding properties152 that may be essential for replication.181 Late (L) domain motifs are positioned near the N-terminus of filovirus VP40 molecules, and are important in posttranslational processing and tracking events that facilitate virus budding.173,405 The VP40 of EBOV contains overlapping PT/SAP and PPXY motifs (PTAPPE/AY), while MARV contains only the PPXY motif (PPPY). These L domain motifs on the ZEBOV VP40 interact with cellular proteins (with WW domains) associated with the endocytic pathway of mammalian cells229,230,257,272,398,465; for ZEBOV the PPXY domain appears to have a greater role in budding efficiencies.300 Results of in vitro studies have suggested that VP40 is bound as an oligomeric form at its PPXY motif by Nedd4 and ubiquitinated, is subsequently targeted to endosomes or multivesicular bodies (MVB) by Tsg101 and VPS-4 (components of the vacuole sorting pathway), and is recruited to membrane rafts through Tsg101 interactions with VP40 and raft proteins (Fig. 32.7). The finding that small interfering RNA (siRNA)-silencing of Rab9, an enzyme important in late endosome transport, inhibits filovirus replication in Vero cells,60,297 as well as the observation that Rab11 is incorporated into MARV virions,231 support the involvement of the endosomal sorting machinery in filovirus assembly. However, the details of VP40 transport from the late endosome to the plasma membrane have not been defined. In addition, mutation of L domains from VP40 did not prevent recovery of a recombinant ZEBOV, nor did it significantly reduce virus production in cell culture.300 This information suggests that VP40 can be transported to the plasma membrane through a process separate from endosomal trafficking. Raft-associated VP40 is believed to associate with NCs, drawing them tightly to the membrane where they are enveloped and extruded from the host cell as infectious virions (Figs. 32.7 and 32.8). Electron tomography studies of MARV budding indicate that the entire length of nucleocapsids associate laterally with the plasma membrane (much like a rising submarine), which is followed by its protrusion and release of
the mature virion particle by being pinched off at the trailing end.405,444
the mature virion particle by being pinched off at the trailing end.405,444
Effects on Host Cell Cultures
The growth of adapted strains of MARV and EBOV in cultured cells can be striking. Intracytoplasmic vesiculation and mitochondrial swelling are followed by a breakdown of organelles and terminal cytoplasmic rarification or condensation. In African green monkey kidney cell lines infected with filoviruses, cytopathic effects (CPE) are evidenced by a rounding and detachment of cells (without syncytia formation), which can result in a total loss of the monolayer (∼5 days). However, replication of REBOV and ICEBOV is slow in tissue culture, and CPE is usually less evident and generally does not develop until after 7 to 9 days incubation. Persistent REBOV infection with continued production of large amounts of virus particles can be established in Vero E6 cells (A. Sanchez, unpublished observations) and ZEBOV can establish persistent infection under partial immunity.161 Filovirus infection does not lead to the shutdown of host-cell protein synthesis, but expression levels diminish as the infection progresses and virus proteins accumulate. The expression of GP has a cytotoxic effect that is associated with the mucin-like region.117,415,463 Elevated expression would likely impact the function of host-cell adhesion proteins by downregulating and/or displacing them,367,380,393 and could also cause cell detachment without cell death through a phosphorylation-dependent signal cascade.57 Using a reverse genetics system,
it was further demonstrated that cytotoxicity depends on the level of GP expression, with overexpression leading to an early detachment and cytotoxicity of infected cells.415 The effects of filovirus infections on endothelial cells and immunocompetent cells will be discussed in the next section.
it was further demonstrated that cytotoxicity depends on the level of GP expression, with overexpression leading to an early detachment and cytotoxicity of infected cells.415 The effects of filovirus infections on endothelial cells and immunocompetent cells will be discussed in the next section.
Pathology and Pathogenesis
Clinical investigations from episodes and outbreaks of human EBOV and MARV infections have provided important descriptive information on the pathology and pathogenesis of these agents; however, the available data are sparse, often fragmentary and sometimes paradoxical. Comprehensive studies have been carried out to a much greater extent in laboratory animals. Rodents—including guinea pigs, mice, and hamsters—have been employed to study viral hemorrhagic fever (VHF) caused by filoviruses.26,38,343,428,472 Because filovirus isolates derived from primates do not typically produce severe disease in rodents upon initial exposure, serial adaptation is required to produce a uniformly lethal infection. Mice and guinea pigs have served well as early screens for evaluating antiviral drugs and candidate vaccines, and genetically engineered mice clearly have utility for dissecting out specific host–pathogen interactions. However, the disease pathogenesis in rodent models is far less faithful in portraying the human condition than disease observed in nonhuman primates.40,143 As data derived from studies using rodents may not correlate with human disease or may be deficient in identifying certain processes, this section primarily focuses on data obtained from human clinical studies and experimental infections of nonhuman primates (Fig. 32.9).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree