Fig. 5.1
Molecular subgroups of posterior fossa ependymoma, as described by Witt et al., Wani et al., and Johnson et al. Group A, Group 1, and clusters G, H, and I describe to same biological subgroup (red), as do Group B, Group 2, and clusters E and F (blue). Both subgroups differ significantly in their molecular and clinical variables.
In conclusion, several independent studies have confirmed the presence of at least two, genetically and biologically, different variants of posterior fossa ependymoma. At present, the standard treatment for patients with posterior fossa ependymomas remains maximal safe surgical resection followed by adjuvant radiation therapy. The role of additional adjuvant chemotherapy is being investigated in an ongoing phase III clinical trial by the Children’s Oncology Group (COG), ACNS0831, which includes planned post hoc molecular subgroup analysis. Future studies will be needed to investigate experimental, intensified treatment regimens in prospectively selected high-risk patients.
Subgroup-specific preclinical models are being developed [17, 19] and are expected to help inform the rational selection of novel therapies for testing in future clinical trials.
Prognostic Stratification
The two most widely accepted factors used for patient stratification are extent of resection, metastatic status, and WHO grading. WHO grading, as an important, independent prognostic marker has been described early on [20], and has recently been confirmed by a large meta-analysis investigating 2408 ependymoma patients [21], whereas other studies have suggested that tumor grading is highly dependent upon the experience of individual neuropathologists [22, 23]. Regarding molecular markers, deletion of CDKN2A along with 1q gain was identified as the strongest indicator of poor prognosis in a cohort of 292 intracranial ependymomas [5]. The same study was able to identify reliable cytogenetic markers for standard, intermediate, and high-risk ependymoma, comprising the first molecular staging system for ependymoma that could be validated in a completely non-overlapping patient cohort [5]. This cytogenetic risk stratification model for intracranial ependymoma comprises three cytogenetic subgroups. Group 1 is associated with standard risk, with tumors displaying large aberrations of chromosomes 6, 9, 15, and 18. Group 2 is associated with intermediate risk, and tumors show a balanced genome. Group 3 is associated with high risk, and defined by 1q gain and/or homozygous deletion of CDKN2A/B [5]. Gain of 1q25 as a negative prognostic marker has since been confirmed in three independent clinical cohorts (CCLG/SIOP, BBSFOP, and SIOP) [24]. A separate study confirmed gain of 1q as a negative prognostic marker in posterior fossa ependymoma [25].
In conclusion, copy number gain of chromosome 1q is the most widely published negative prognostic molecular marker and applies to both supra- and infratentorial ependymoma [5, 7, 18, 24–28].
Other prognostic markers that are based on immunohistochemistry, rather than cytogenetics, have also been identified. Tenascin C is an extracellular matrix protein and has been shown to be a negative prognostic marker in ependymoma [11, 12, 18]. The NSC marker Nestin is a negative prognostic marker identifying ependymoma with poor prognosis especially in WHO II tumors [29], possibly indicating a less favorable, undifferentiated phenotype. Conversely, expression of neurofilament light polypeptide 70 (encoded by NEFL) is a positive predictive marker in supratentorial ependymoma [30] and may indicate a more favorable, differentiated phenotype. The immunohistochemical markers LAMA2 and NELL2 delineate the two molecular subgroups in posterior fossa ependymoma described above: Group A tumors (with poor prognosis) are characterized by the pattern LAMA2 positive and NELL2 negative, and Group B tumors (with more favorable prognosis) by the pattern LAMA2 negative and NELL2 positive [18]. The delineation of two molecular subgroups by the expression of LAMA2 and NELL2, and their prognostic values, have since been confirmed in a separate study [16].
Finally, miRNAs associated with prognosis have been described in ependymoma: let-7d, miR-596, and miR-367 are associated with poor survival, and miR-203 is an independent predictor for time to relapse [31].
Molecular Signaling Pathways
Identification of molecular signaling pathways that can be targeted for therapeutic purposes will be crucial for the rational development of novel drug-based treatments. It is important to note that thorough characterization of molecular subgroups and establishment of faithful subgroup-specific models will be needed for successful preclinical testing. It has become evident that different molecular subgroups of posterior fossa ependymoma show distinct activation of molecular signaling pathways (Fig. 5.2): Group A shows activation of epidermal growth factor receptor (EGFR), platelet-derived growth factor (PDGF), vascular endothelial growth factor (VEGF), and mitogen-activated protein kinase (MAPK) among others [18]. Group B shows less activation of classic oncogenic signaling pathways; gene expression profiles, however, indicate activation of ciliogenesis, microtubule assembly and mitochondrial metabolism [18]. These promising findings display novel treatment opportunities in a fashion of subgroup-specific targeted therapies. A large number of therapeutic drugs inhibiting these molecular pathways (e.g. MAPK-, EGFR-, PDGF-, VEGF- and integrin-inhibitors) are already approved for other cancers and/or in various stages of clinical development, including those for pediatric patients. Carefully designed clinical studies will be needed to assess the potential of these agents to complement current standard therapies (surgery, radiotherapy) and/or other investigational therapies, such as chemotherapy. Of note, an integrated in vivo high-throughput drug screen using the preclinical supratentorial subgroup D-specific model [19] recently showed that the most active compounds against ependymoma which showed the least toxicity on NSCs were 5-FU and bortezomib, a proteasome inhibitor [19].
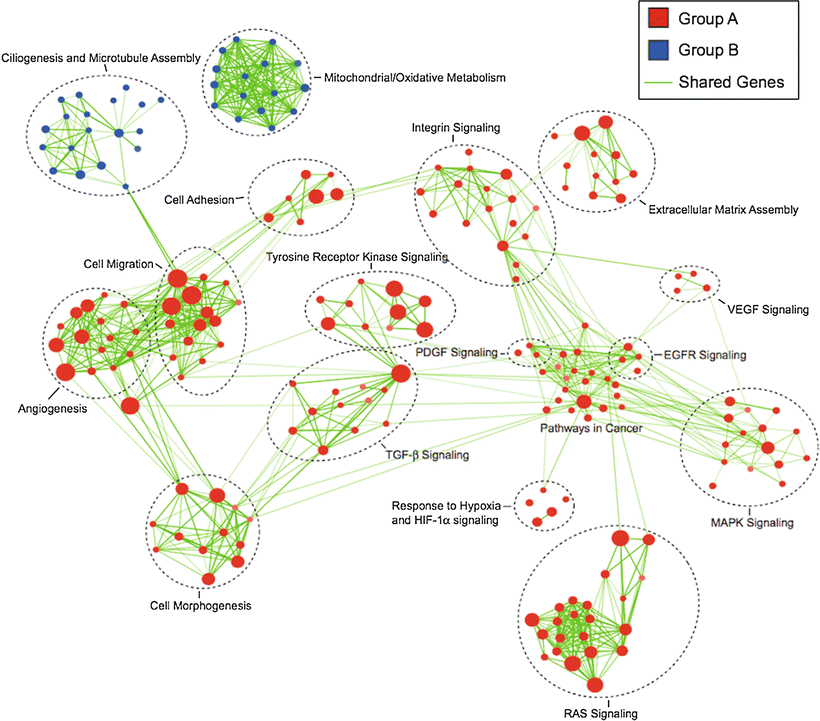

Fig. 5.2
Group A (red) and Group B (blue) posterior fossa ependymomas show distinct activation of signaling pathways and biological functions. The map was created by geneset enrichment analysis of transcriptomes of the two molecular subgroups of posterior fossa ependymomas, using Cytoscape and Enrichment Map (Adapted from Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143-57, with permission).
Other signaling pathways implicated in ependymoma biology are the Notch pathway, p53 and ERBB (EGFR and ERBB2/3/4), among others. Notch activation is associated with ependymoma progression [11], and inhibition of the Notch pathway using gamma-secretase inhibitors reduces neurosphere formation in vitro [11]. Aberrant expression of p53 was identified as an unfavorable prognostic marker in ependymoma, whereas its regulator MDM2 did not show an association with prognosis [12, 32]. The molecular mechanism of p53 pathway dysregulation in ependymoma is not yet understood. Despite frequent p53 overexpression in ependymoma, known mechanisms such as TP53 mutation or promoter hypermethylation, MDM2 overexpression, P14ARF promoter hypermethylation or increased PAX5 expression are not observed in ependymoma. Furthermore, some studies implied the RTK1 family of proteins including EGFR, ERBB2, ERBB3, and ERBB4, in ependymoma biology, promoting growth, motility, and survival of ependymoma cells, whereas ERBB2 overexpression may potentiate radial glia proliferation [14]. In two studies, EGFR expression was found to be associated with an increased risk of disease recurrence [7, 12].
Molecular Targeted Therapies
Radical surgical resection, whenever feasible, remains the mainstay of ependymoma treatment and may be sufficient in a subset of patients with supratentorial ependymomas [33]. Because of high rates of local recurrence without additional therapy, adjuvant involved-field radiation therapy is generally employed in current standard treatment protocols. Nevertheless, local or distant disease recurrence is common, including for approximately half of all patients with posterior fossa ependymoma. In case of metastatic dissemination at diagnosis, craniospinal radiation therapy is typically used. The role of chemotherapy in the treatment of ependymoma is not well established, and response rates to single agent or combination chemotherapy in recurrent ependymoma are disappointing [34]. It has been shown, however, that chemotherapy can be effectively used to delay the beginning of radiotherapy in very young children, without compromising their prognosis [35]. Recent and ongoing clinical trials are examining the role of neoadjuvant chemotherapy in unresectable or disseminated disease, as well as in the adjuvant setting post radiation therapy for high-risk patients [36]. Due to the generally limited efficacy of classical chemotherapy in disseminated and recurrent ependymoma, however, novel therapies are urgently needed.
Based on the discovery of molecular signaling pathways relevant to ependymoma biology, several targeted therapy approaches are currently in development, including chromatin-modifying drugs. Early phase clinical trials are currently investigating compounds targeting Notch, EGFR, HDACs, and ERBB among others, as single agents or in combination with chemotherapy, in children with ependymoma.
Targeting the Notch pathway in a phase I clinical trial by using the gamma-secretase inhibitor MK-0752 showed that the MK-0752 is well tolerated in children, and response was seen in one ependymoma and one glioblastoma patient [37]. Promising preclinical data on EGFR inhibitors in ependymoma show success alone or in combination with phosphoinositide 3-kinase inhibitors [38, 39]. In vitro, ependymoma cells are sensitive to HDAC inhibitor (HDACi) treatment, which induced differentiation in subgroup C ependymoma cells [40] and increased apoptosis in others [41]. As a result, HDACis, such as vorinostat (suberoylanilide hydroxamic acid, SAHA), are being investigated in recent and ongoing clinical studies including patients with ependymoma [42, 43].
For both traditional chemotherapy and targeted agents, however, sufficient drug penetration into CNS tumor tissue and efficient target inhibition in vivo represent formidable challenges for successful translation of preclinical discoveries into effective clinical therapy. For example, a recent clinical and molecular biology study using the EGFR/ERBB2 inhibitor lapatinib in children with refractory brain tumors revealed that the drug failed to achieve meaningful concentration in the tumor tissues and, as a result, failed to inhibit the molecular targets [44].
Recently, two studies highlighted genetic and epigenetic alterations as therapeutic targets of different subtypes of ependymoma.
Posterior fossa ependymoma harbor nonrecurrent somatic mutations in a cohort of 47 tumors using whole-exome and whole-genome sequencing technologies [45]. Notably, a very low mutation rate was found in these tumors regardless of subgroups, with an average of only five somatic mutations per tumor (4.6 and 5.6 somatic mutations in Group A and Group B ependymomas, respectively). In contrast, DNA methylation patterns were highly dissimilar between both subtypes. When comparing only PF ependymoma subtypes, Group A ependymomas display a much higher proportion of methylated CpG-islands within the promoter regions as compared to Group B ependymomas. Based on this distinct pattern of epigenetic alteration, Group A tumors show a CpG-island methylator phenotype (CIMP). Additionally, Group A/CIMP-positive tumors show a greater extent of epigenetic silencing of targets of the polycomb repressive complex 2, including downregulation of differentiation genes through histone H3-lysine 27 (H3K27) trimethylation. To investigate if epigenetic agents can be used as potential novel treatment option for Group A tumors, in vitro and in vivo tests were performed. The preclinical treatment approaches using either 5-aza-2′-deoxycytidine, 3-deazaneplanocin A, or GSK343 (a selective inhibitor of the H3K27 methyltransferase EZH2) have shown very good response of cells and mice bearing Group A tumors. These results are promising treatment strategies targeting DNA CpG methylation, PRC2/EZH2, and/or histone deacetylases of this chemotherapy-resistant disease.
Another study, using whole-genome sequencing and/or RNA sequencing of 77 ependymomas, identified a novel gene fusion affecting RELA and C11orf95 [46]. In line with findings of the study by Mack and colleagues, no recurrent somatic mutations were detected in posterior fossa ependymomas, including Group A and Group B. Notably, among supratentorial ependymomas Parker and colleagues discovered a frequent translocation within a region of chromosome 11q, which is possibly caused by chromotripsis (a recently discovered phenomenon of genomic rearrangement arising during a single genome-shattering event) and resulted in a C11orf95–RELA gene fusion in about 70 % of cases. RELA is a downstream target of the NF-κB signaling pathway, acting as a transcription factor and regulating several biological actions of cell maintenance. Importantly, a genetically engineered mouse model was successfully developed based of the C11orf95–RELA gene fusion. NSC from a Ink4a/Arf-null background were transduced with the retroviruses carrying the C11orf95–RELA fusion. These transgenic NSCs were then implanted into the cerebrum and developed supratentorial ependymomas within a few days. Hence, this model delivers excellent opportunities for preclinical drug testing in vivo of a supratentorial subtype of ependymomas.
Summary
As has been shown, genomic and gene expression profiling in ependymomas not only identifies biologically distinct subgroups but also allows for the stratification of patients into clinically meaningful prognostic subgroups. As demonstrated by the delineation of Group A and B posterior fossa ependymomas, the tight association between molecular profile and clinical behavior is of high practical relevance for the individual patient. One simple consequence of the identification of a Group A vs. Group B tumor for the patient is the new possibility of truly risk-adapted adjuvant treatment of previously equally treated tumors. Thus, the identification of the molecular profile adds considerable additional information to the classical histopathological analysis, enabling better informed clinical decisions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


