DNA Analysis
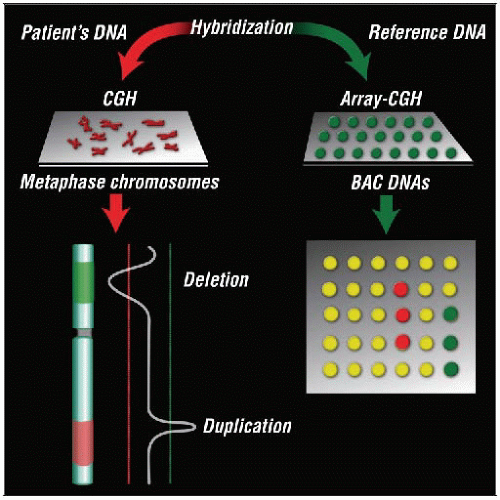 Labeled DNA from a tumor is hybridized to either metaphase chromosomes (CGH) or a DNA microarray (array-CGH). Deletions or duplications of DNA are detected by comparison with reference normal DNA. |
TERMINOLOGY
Abbreviations
Conventional cytogenetics (CC)
Comparative genomic hybridization (CGH)
Loss of heterozygosity (LOH)
Copy number alterations (CNA)
Definitions
Completion of human genome project will expand knowledge of human DNA sequences
Development of new genomic technologies provide new tools to study human disease
CC: Visualizes all chromosomes; detects large translocations and chromosomal duplication or loss
CGH: Detects gains or losses of short DNA segments throughout genome
LOH: Detects loss of 1 of 2 alleles of specific genes
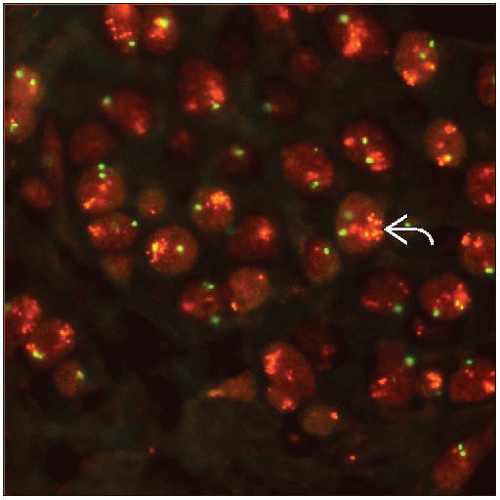
FISH: Uses specific probes to visualize increased copies of genes or to identify gene translocations
These genomic technologies can be applied to clinical breast cancer samples
Possible to study complex genomic aberrations and recurrent genomic imbalances characterizing tumors
CGH allows genome-wide investigation of CNA
Has resulted in major advances in understanding of breast carcinogenesis and tumor progression
ETIOLOGY/PATHOGENESIS
Histogenesis
Breast cancers are characterized by complex genomic changes and CNA
Common and specific recurrent genomic changes described in breast tumors
Most common recurrent chromosomal genomic gains: 1q, 8q, 17q, 20q, and 11q
Most common recurrent chromosomal genomic losses: 8p, 11q, 13q, 16q, and 18q
Many of these changes occur in regions that harbor known proto-oncogenes and tumor suppressor genes
May cause gene overexpression or loss of function
CLINICAL IMPLICATIONS
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree