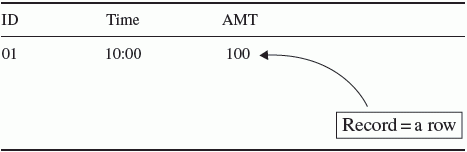
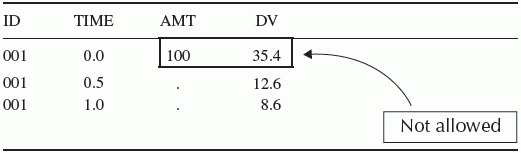
Chapter 4 Analysis of pharmacometric datasets intended for population analysis requires a potentially complex arrangement of information in a format upon which the analysis program can act. Pharmacometric datasets are generally time-ordered arrangements of records representing the time course of events related to drug administration, resulting plasma concentrations, and/or pharmacodynamic (PD) responses occurring within the individuals enrolled in a clinical trial or in a particular patient. The complexity of the dataset is dependent upon the complexity of the features of the system being modeled. Discussions of dataset construction are sometimes overlooked when the modeling process is considered. Dataset construction can be tedious work that must be done before the interesting work of modeling can begin. Datasets are a critical component of modeling; therefore, the importance of skills in constructing datasets should not be underestimated. The task of dataset construction is not trivial, especially when large datasets are assembled from multiple studies or messy clinical trial data, such as when covariates are recorded in mixed units (e.g., weight in pounds and kilograms for different individuals) or the time and date information for concomitant medication usage needs to be sequenced with study drug administration and blood sample collection in the analysis dataset. The amount of time and effort required to construct a complex dataset can sometimes rival the amounts required to perform the modeling. Quality control is critical in constructing datasets that reflect the events that gave rise to the data. If errors in stating the times of doses or concentrations are made, the analysis becomes flawed by the errors in the data. Documentation of that quality control process is especially important when models are to be used for regulatory and/or decision-making purposes. The activities of good quality control add to the effort of constructing datasets. Modeling pharmacokinetic (PK) data generally requires specification of the occurrence of doses (dosing events) in a study, the values of independent variables (e.g., time, body weight, age), and the values of the dependent variable (DV) (i.e., concentrations of drug). The model is then fit to, or is used to simulate, the time course of drug concentrations resulting from those doses. Datasets for PD models may or may not include dosing events in the dataset, depending upon the structure of the system being modeled, whether the data represent the biological process of disease progression or placebo effects, and whether drug concentrations are included in the model. Additionally, some PD models may not require that the data be structured according to time-ordered events, as in the simple linear or nonlinear regression of QTc versus peak drug concentration. Models that do not use time-ordered data do not use PREDPP and are expressed in a user-supplied PRED routine. Datasets for these analyses associate independent variable(s) and DV on a record within an individual, but are not necessarily time ordered and do not contain separate dose records. Models implemented using PREDPP require that the data be time oriented. When modeling the time course of changes in the concentration of endogenous compounds, or whenever the initial conditions of the system are not zero, the initial circulating amount or concentration of the compound must be input into the system. Similarly, if the baseline values of a PD response variable are nonzero, then an initial nonzero value must be specified for modeling the response variable. Baseline values may be entered in the dataset or assigned through certain options in the control file. In general, the pharmacometric dataset will have elements of input mass, time, covariates, and the observations of events which the model is intended to describe. In addition, there are NONMEM-required variables that allow NONMEM to appropriately interpret and process the data in the manner intended by the analyst. The data file can be generated using many different data management packages, spreadsheets, programming languages, or text editors. The use of programming languages such as R (R Development Core Team 2013) or SAS Institute Inc. (2002–2010) has significant advantages in processing data in a consistent, repeatable, and verifiable manner. However, the use of such systems requires skill with the language. Using a spreadsheet for data assembly and formatting may be easier when working with small datasets or for a novice but is prone to errors that are not easily discovered, such as a key being unintentionally pressed that leads to unintended changes. Such errors are not easily found since spreadsheets do not keep a record of changes made in a file. Federal regulation 21 CFR Part 11 requires accountability of data and a record of any changes made, which cannot easily be done in a spreadsheet that doesn’t keep a log of all changes. When built using a spreadsheet, validation of the analysis dataset would require 100% verification against the source data. The use of a programming language to construct datasets allows one to track every change made to the dataset. Spreadsheets might be considered for the analysis of simple data from a small number of individuals or from a single Phase 1 study; however, when the analysis includes more complex dosing histories, merging data from multiple Phase 1 and 2 studies, or data from a Phase 3 study, use of a programming language becomes essential. Dataset assembly according to regulatory standards is best accomplished by the use of a programming language in order to comply with quality management requirements. For most platform configurations, the data file could not contain embedded tabs when using older versions of NONMEM. In current versions, column delimiters such as spaces, commas, or tabs are acceptable. The NONMEM (NM-TRAN) system has certain structural requirements of analysis datasets. Data must be present in a text (i.e., ASCII) format flat file with appropriate delimiters. The data file has a two-dimensional arrangement of data in rows and columns. All NONMEM data should be numerical except for certain formats of DATE and TIME, which may include alphabetic characters. A data record refers to a line, or row, of data, as shown in Table 4.1. Table 4.1 Example of a data record The record contains a specific collection of associated data elements. When modeling a time-dependent system, the record is the collection of concurrent values of data and variables at a specific point in time. The terms variable, field, or data item are used to refer to the information contained in a column of a space-delimited file or a specific position within a record of a comma-delimited file. A space-delimited file uses one or more spaces to separate data items in each row of the dataset. Space-delimited files with data arranged in columns have the advantage of readability to the analyst. If problems arise in fitting the data of a particular individual, or an error is reported in processing the data on a particular record, data arranged in space-delimited columns are easier for the analyst to review than data in comma-delimited files. Each dataset record must contain the same number of variables. The order of variables is arbitrary but must be consistent across all records (within a subject and across subjects). Missing values should have a placeholder such as “.” or “0”; blanks are not acceptable for missing data in space-delimited files. While it is not always required, it may be advisable to keep the format of a variable consistent across all records. NONMEM will allow some mixed format entry of some variables, but for clarity to the analyst and prevention of unanticipated errors, consistency might be of value. Consider the example in Table 4.2. TIME could be expressed as hh:mm for one record and as a real number (elapsed time) for another record. If the analyst intended 8:00 to be time zero and 0.5 hours to be the time of an event occurring one-half hour later, the records in Table 4.2 would result in a data error in the NONMEM report file stating that ELAPSED TIME MAY NOT BE NEGATIVE. The values that NONMEM assigns to TIME for each record can be reviewed in the FDATA file. In this case, the event at 0.5 hours would evaluate to –7.5 hours. The first record time of 8:00 is assigned the TIME = 0 in FDATA, but the 0.5 record is converted to –7.5 hours. Table 4.2 Example of data records in unlike units that lead to an error in analysis If other variables such as covariates were included in the dataset using different units, one would need to capture the differences and convert them to like units within the control stream file. For example, if estimated creatinine clearance (CrCl) was expressed in common clinical units of mL/min for data from one study and SI units of mL/s from another study, the control stream would need to adjust the units of one or the other form before CrCl is used in a covariate model equation. In the authors’ experience, constructing the analysis dataset to have like units is best achieved through programmatic adjustments during the construction of the analysis dataset. In general, good documentation and consistency can lead to reduced errors during analysis. The maximum number of variables (data items) that can be read into and used for any one model run in NONMEM 7 is 50. A greater number of items may be contained in the dataset, but some of them must be ignored on each execution using the DROP option on the $INPUT statement. This allows NM-TRAN to process a dataset with more than the maximum number of variables by simply ignoring or dropping some of the variables not currently being used. Earlier versions of NONMEM allowed a maximum of only 20 data items and were thus quite limited in this regard. The control file includes a $INPUT statement to specify to NONMEM the order of variables in the dataset record. Variable names (data labels) in this statement may be up to 20 characters long in NONMEM 7 but in prior versions must have been between 1 and 4 characters in length. This variable name will be used in the control file to identify the data read from the dataset. Certain variable names are reserved in NONMEM (and PREDPP) and must be used in accordance with the reserved definitions. The following are examples of reserved variable names: ID, DATE, DAT1, DAT2, DAT3, TIME, DV, AMT, RATE, steady state (SS), interdose interval (II), ADDL, event identification (EVID), missing dependent variable (MDV), CMT, prediction compartment (PCMT), CALL, CONT, L1, and L2. In addition, PK parameter names have reserved definitions in accordance with the ADVAN subroutine being used. For example, KA, CL, V, F1, F2, ALAG1, ALAG2, S1, and S2 are reserved for ADVAN2. Unlike some other analysis programs, the PREDPP routines of NONMEM require the use of an event-type structure to the records of the dataset. Each record contains information about a particular event. In the case of some dosing options, the dose record may imply the occurrence of other past or future events as well. Events may be of the following types: (i) dosing event, (ii) observation event, or (iii) other-type event. Dosing events introduce an amount (recorded in the variable AMT) of the DV (e.g., drug mass or response variable in the case of PD models) into the system at a particular time. Observation events (recorded in the variable DV) record the concentration or effect measure at a particular time. Other-type events record a change in some other system parameter such as a physiological condition like a change in body weight or the administration of a concomitant medication. Other-type events may also be included as part of a dose or observation record. However, administered doses cannot be present on the same record as an observed concentration or effect measure. Similarly, observation data cannot appear on the same record with an administered dose. Either AMT or DV, or both, must be missing on each record. Values other than 0 or “.” for these variables cannot appear together on the same record, as shown in Table 4.3. Table 4.3 Example of incorrect expression of simultaneous events in the analysis dataset Sometimes two events, such as a dose and an observation, need to be recorded at the same time in an individual. When this is needed, as illustrated in Table 4.4, the two events should be reported in separate, consecutive records with the same value for TIME. Table 4.4 Example of correct expression of simultaneous events in the analysis dataset Data items that report descriptive information (covariates) about a patient, for example, age or body weight, should be nonmissing on all records (dose, observation, and other events). If these items are used in the model, they must be present on every record as it is processed. If the value of a covariate is missing, the model will be evaluated with the missing value (interpreted by NM-TRAN as 0), and numerical errors or unanticipated erroneous results may occur. Datasets for models not using PREDPP do not use all the elements of the event-type structure. Some such datasets are used for a variety of PD models, and each record is an observation event. In these datasets, there is generally no concept of a dose record. These datasets are set up for a specific nonlinear regression approach and must be constructed in a fashion consistent with the model to be applied. Some of the efficiency and flexibility of NONMEM comes through the dataset construction. A variety of data items are available to communicate the events to be modeled with NONMEM and PREDPP. Some data items are required by NONMEM for all population models (e.g., ID and DV), while others are required only in certain circumstances and may be optional in other situations. When using PREDPP, TIME and AMT are required for all datasets. Some data items that have specific requirements for use are outlined in the following sections. In a time-oriented dataset (intended for use with PREDPP), the value of TIME must be nonmissing and non-negative on all records. All records within an individual must be arranged in increasing (or equal) values of TIME, unless there is a record indicating the occurrence of a reset event that allows the elapsed time interval to restart. The value of TIME may be formatted as clock time (e.g., 13:30) or decimal-valued time (13.5) but should be of the same type on all records. If a clock time format for TIME is selected, however, it is critical that times be recorded using military time to prevent a syntax error from NM-TRAN regarding a potential nonconsecutive time sequence for values recorded past noon. Decimal-valued time represents the elapsed time since some initial event; however, there is no requirement that the first record for each subject has a TIME value of zero. The DATE data item is not required, but may be useful in constructing the sequence of events in time. It is used in conjunction with TIME to construct the time sequence of event records. The DATE data item may be expressed as an integer (e.g., representing the study day) or as a calendar date. Several variables are available for expressing date values as calendar dates using different formats, as shown in Table 4.5. Table 4.5 Date formats for input of calendar dates Using these formats, the year can be expressed using from 1 to 4 digits (e.g., 9, 09, or 2009). If the year is specified using 1–2 digits, then the LAST20 = n option should be specified on the $DATA line in the control file, where n is an integer from 0 to 99. The value of n determines which century is implied for the year. The default value of n is 50. When the value of year (YY) is greater than n, the year is assumed to be 19YY, and when YY ≤ n, it is assumed to be 20YY. For example, if n = 50 and YY = 9 or YY = 09, the year is interpreted as 2009. Whenever DATE is formatted as a calendar date (i.e., using “/” or “-” as a delimiter), $INPUT must specify DATE = DROP. In this case, the data preprocessor will use the DATE item to compute the relative time of events but will drop the item from the dataset. Since DATE is dropped, its value cannot be output to the $TABLE file. Whenever the DATE variable is used in the dataset, values of TIME are restricted to a range from 0 to 24 hours. To specify a greater TIME interval than 24 hours, the DATE must be incremented by the appropriate amount so that the value of TIME will fall within 0–24 hours, for example, if a dose is given at 0:00 on 1/1/2013 and a sample is collected 36 hours later. The sample record could be coded as shown in Table 4.6 but not be recorded as shown in Table 4.7. The data preprocessor will use DATE and TIME together to compute the relative or elapsed time of events since the first record within the individual. It is important to note that if TIME is output to the $TABLE file, the value output will be the relative time, not the TIME value specified in the original data file. If a DATE data item is not used, then TIME must be defined as the elapsed time since the first event. Events spanning more than 1 day will have time values greater than 24 hours. Even when the time sequence of events is defined using TIME and DATE, we have often found it useful to include a time variable that describes the elapsed time of all events within a subject. We frequently refer to this as the time since first dose (TSFD). A second time value that is frequently useful is the time since the most recent dose was administered, or time after dose (TAD). These values of time are often useful for graphing the data, producing goodness-of-fit plots, and as a merge key for certain data handling operations. For population data, the dataset must have an ID variable, which is the subject identifier. All records for a single subject must be contiguous and, for PREDPP, sorted in order of increasing time. If records from a particular subject are separated in the dataset by those of another ID number, the separated records will be treated by NONMEM as separate individuals (even though they have the same ID value). As such, an ID number could be recycled and seen as different subjects, but this is generally not a good practice and could lead to unexpected errors when plotting, sorting, counting individuals, or otherwise handling the dataset. One practice we have found helpful is to create a unique patient identifier variable for the analysis dataset. This value can be particularly useful when data are to be combined from multiple studies. In this case, the unique ID number value may be generated by concatenating unique information from the clinical study number and the subject number in the study. For instance, subject 126 from Study 10 might have a new ID value of 10126. In this way, subject 126 in Study 10 would not be confused with subject 126 in Study 7 (whose ID would be 7126). This reassignment of ID occurs during the process of building the analysis dataset, and of course, accurate documentation of the assignment process is required so that identification of every subject in the dataset can be maintained with the source data. With NONMEM 7, ID values can be up to 14 digits in length. However, using a value with greater than 5 digits in length may create an unanticipated problem in the table files. NONMEM will read the analysis dataset correctly with up to 14 digits in the ID field. However, with the default settings, the table file will report the value of ID with only 5 unique digits. The default table format for all fields is s1PE11.4. This format allows a total width of 11 characters, including sign and decimal places. Four decimal places are used, with one digit to the left of the decimal place. An example default value is as follows: –2.4567E+01. Therefore, 5 unique digits are the most that will show by the default settings. Two consecutive subjects in a dataset with the ID values of 12345678901234 and 12345678901235 would be seen by NONMEM as different individuals, and model estimation would be performed as anticipated. However, the ID value output in the table would be 1.2346E+13 for both individuals. Thus, when postprocessing for summary values, plots, or tables, the records from the two individuals would be seen as arising from a single subject. Longer ID records, or any other data item, can be accommodated by changing the output format used in the table file. The LFORMAT and RFORMAT options can be used to accomplish this, using FORTRAN syntax. For example, the following code could be used to generate table file in which the ID numbers given earlier could be distinguished: This example gives a real number output of the 14-character ID, plus one space for a decimal, and uses the default format for the remaining 7-item output in the table file (3 named items, plus 4 items automatically appended to the table file). The reader is referred to the user’s manuals and help files for more details on the use of LFORMAT and RFORMAT. DV is the dependent variable data item. The values in this field represent the observations to be modeled. These observations could be drug concentrations for a PK model and drug effects for a PD model, or the DV field might contain observations of both PK and PD types in a PK/PD model. The DV item is used in conjunction with the compartment (CMT) data item, which specifies the model compartment in which the observation was made to the program. The value of DV must be missing on all dose records. It would also be missing on other-type event records. However, if an other event and DV appear on the same record, the record would be understood by NONMEM to be an observation record with DV equal to a nonmissing value that also includes a change in another variable (e.g., weight). The MDV data item allows the user to inform NONMEM whether or not the value in the DV field is missing. MDV is a logical variable, so MDV should have a value of 1 (i.e., true) on records for which DV is missing, such as on dosing records. MDV should have a value of 0 (i.e., false) on observation records where DV is not missing. If DV is missing on an observation record and MDV is not specified or MDV = 0, then an error will be reported. MDV is not a required data item. NM-TRAN will assign a value from the context of the dataset if it is not included. We find it a good practice, however, to assign the value of MDV in the construction of the dataset, in order to be certain that the behavior is what we expect.
Datasets
4.1 Introduction
4.2 Arrangement of the Dataset
ID
TIME
001
8:00
001
0.5
ID
TIME
AMT
DV
001
0.0
100
.
001
0.0
.
35.4
001
0.5
.
12.6
001
1.0
.
8.6
4.3 Variables of the Dataset
4.3.1 TIME
4.3.2 DATE
Variable name
Format
DATE
MM/DD/YY or MM-DD-YY
DAT1
DD/MM/YY or DD-MM-YY
DAT2
YY/MM/DD or YY-MM-YY
DAT3
YY/DD/MM or YY-DD-MM
4.3.3 ID
$TABLE ID DOSE WT TIME NOPRINT
FILE=./filename.tbl
RFORMAT=”(F15.0,”
RFORMAT=”7(s1PE11.4))”
4.3.4 DV
4.3.5 MDV
4.3.6 CMT
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


