5
Classification of Medically Important Bacteria
CHAPTER CONTENTS
PRINCIPLES OF CLASSIFICATION
The current classification of bacteria is based primarily on morphologic and biochemical characteristics. A scheme that divides the medically important organisms by genus is shown in Table 5–1. For pedagogic purposes, this classification scheme deviates from those derived from strict taxonomic principles in two ways:
TABLE 5–1 Classification of Medically Important Bacteria
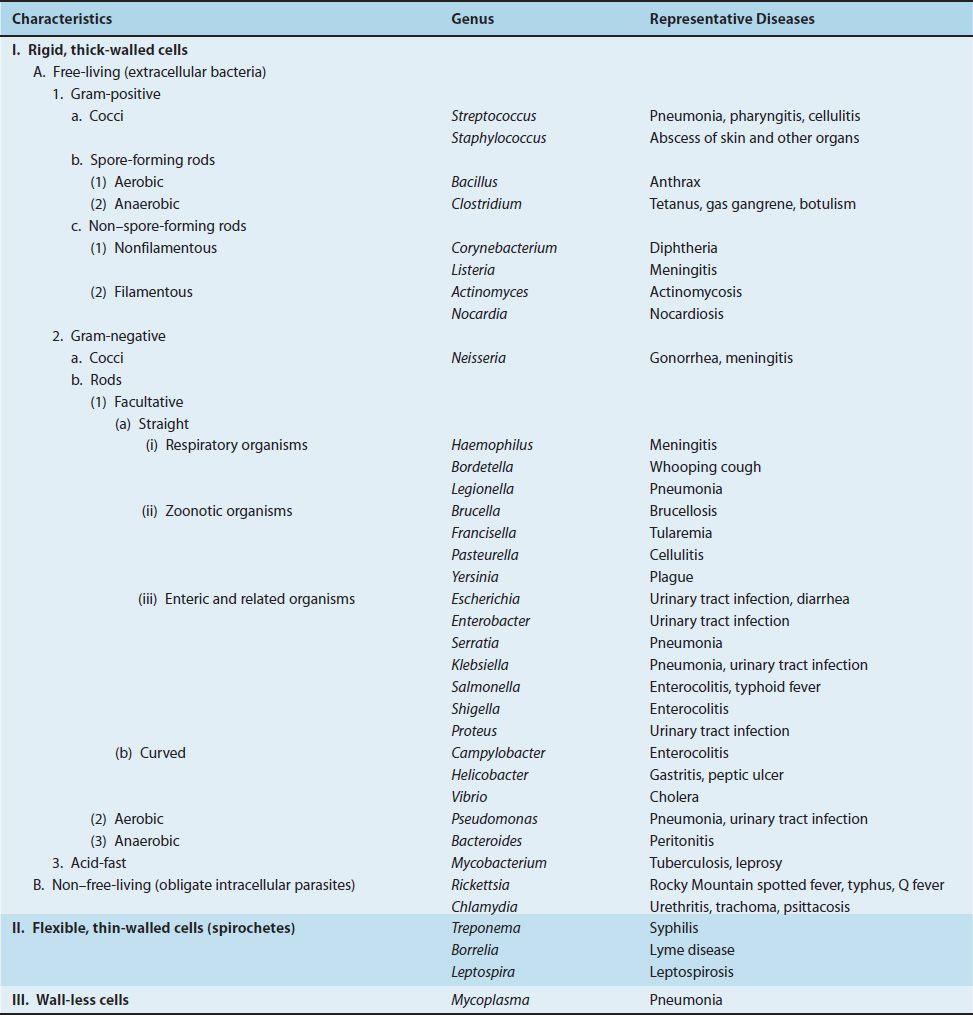
(1) Only organisms that are described in this book in the section on medically important bacteria are included.
(2) Because there are so many gram-negative rods, they are divided into three categories: respiratory organisms, zoonotic organisms, and enteric and related organisms.
The initial criterion used in the classification is the nature of the cell wall (i.e., is it rigid, flexible, or absent?). Bacteria with rigid, thick walls can be subdivided into free-living bacteria, which are capable of growing on laboratory medium in the absence of human or other animal cells, and non–free-living bacteria, which are obligate intracellular parasites and therefore can grow only within human or other animal cells. The free-living organisms are further subdivided according to shape and staining reaction into a variety of gram-positive and gram-negative cocci and rods with different oxygen requirements and spore-forming abilities. Bacteria with flexible, thin walls (the spirochetes) and those without cell walls (the mycoplasmas) form separate units.
Using these criteria, along with various biochemical reactions, many bacteria can be readily classified into separate genus and species. However, there have been several examples of these criteria placing bacteria into the same genus when DNA sequencing of their genome reveals they are significantly different and should be classified in a new or different genus. For example, an organism formerly known as Pseudomonas cepacia has been reclassified as Burkholderia cepacia because the base sequence of its DNA was found to be significantly different from the DNA of the members of the genus Pseudomonas.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


