Caliciviridae: The Noroviruses
Kim Y. Green
The family Caliciviridae is composed of small (27 to 40 nm), nonenveloped, icosahedral viruses that possess a linear, positive-sense, single-stranded RNA (ssRNA) genome. The five genera of the family are Norovirus, Sapovirus, Nebovirus, Lagovirus, and Vesivirus. The major human pathogens in the family are the noroviruses and sapoviruses, which cause acute gastroenteritis. Important veterinary pathogens include vesiviruses such as feline calicivirus (FCV), which causes a respiratory disease in cats, and lagoviruses such as rabbit hemorrhagic disease virus (RHDV), which causes an often fatal hemorrhagic disease in rabbits. This chapter provides a description of the family Caliciviridae, with major emphasis on the noroviruses because of their prominent role in sporadic and epidemic gastroenteritis.147
History
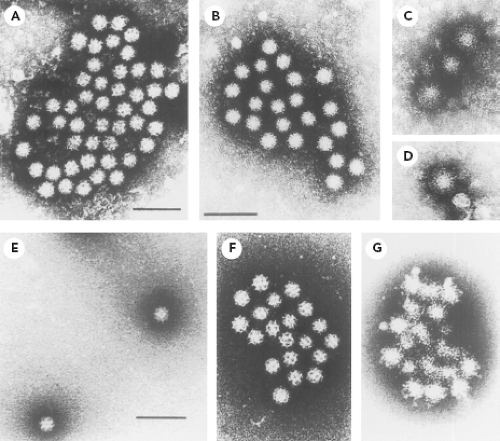
The establishment of a viral etiology for gastroenteritis in humans was a decades-long process that was hampered by the fastidious nature of many of these viruses for growth in cell culture.213 Volunteer studies carried out in the 1940s and 1950s in the United States and Japan played a major role in establishing that filterable, nonbacterial infectious agents can cause enteric disease.213 An important advance occurred in 1972 with the discovery of Norwalk virus (NV) by Kapikian et al.214 Stool material from a rectal swab obtained from an ill individual involved in a gastroenteritis outbreak that had occurred at an elementary school in Norwalk, Ohio, in October 1968 was administered to adult volunteers as a bacteria-free filtrate and serially passaged to other volunteers, inducing acute gastroenteritis in certain individuals.112,113 Stool material from these volunteers was then examined for the presence of viruses by the technique of immune electron microscopy (IEM), which involves the direct observation of antigen–antibody complexes by EM.11,13,18,214 The fecal filtrate was incubated with prechallenge or convalescent-phase serum from a volunteer who had become ill following ingestion of the filtrate. Figure 20.1 shows the striking difference between the appearance of the small, round virus particles (described as 27 to 30 nm in diameter with a hint of surface structure) after incubation with the prechallenge serum (part A and B) and after incubation with convalescent-phase serum (parts C and D). The increase in visible virus-specific antibodies in the convalescent serum from individuals involved in the original Norwalk outbreak, as well as in volunteers challenged with the virus derived from the outbreak, led to the conclusion that NV was the etiologic agent.214 Norwalk virus would become the prototype strain for a large group of related Norwalk-like viruses or small round structured viruses, known now as the noroviruses.
The techniques used in the discovery of the NV proved instrumental in the subsequent characterization of other fastidious enteric viruses (including human rotaviruses, astroviruses, parvoviruses, enteroviruses, and hepatitis A virus) by an approach that became known as particle or direct virology.213 Norovirus reference strains such as Hawaii virus (from a family outbreak of gastroenteritis that occurred in Honolulu in 1971) and Snow Mountain virus (from an outbreak in a Colorado resort camp in 1976) were discovered in 1977 and 1982, respectively.116,468 In 1976, Madeley and Cosgrove296 reported the presence of “typical” caliciviruses in the stools of infants and young children (2 to 18 months of age), and these viruses showed a striking morphologic similarity to previously characterized animal caliciviruses that were known to exhibit “classical” distinct cup-like depressions on the surface of the virion. That same year, Flewett and Davies130 observed similar calicivirus particles in a sample from the intestinal lumen of a child at autopsy and in the feces of a child with gastroenteritis. Chiba et al90 described a “classical” calicivirus, associated with gastroenteritis, in infants and young children living in an infant home in Sapporo, Japan, in 1977. Another virus (later designated Sapporo/82/Japan) that exhibited classical calicivirus morphology was subsequently detected from this same infant home (Fig. 20.1E).340 Hyperimmune serum was raised in guinea pigs against Sapporo/82/Japan virus particles purified from stool material, and IEM was used to establish its antigenic characteristics (Fig. 20.1F and 20.1G, respectively). The Sapporo virus would become the prototype strain for the Sapporo-like viruses or classical caliciviruses, known now as the sapoviruses.
In the late 1980s, the stool filtrate derived from the Norwalk, Ohio, outbreak was again fed to adults in volunteer studies to obtain adequate quantities of virus particles to
characterize the viral genome, a major advance by Jiang et al199 that established the classification of Norwalk virus as a member of the Caliciviridae. The complete RNA genome sequences of Norwalk virus and the closely related Southampton virus were determined and found to be organized into three major open reading frames (ORF1, −2, and −3) with a polyadenylated 3′ end176,202,253,254 (see Genome Structure and Organization). The ORF1 was shown to encode a large polyprotein that was proteolytically processed into the mature nonstructural proteins.273,276 The ORF2 encoded the major capsid protein, VP1, and ORF3 encoded a minor structural protein, VP2. The human noroviruses initially segregated into two major phylogenetic groups within what would become the genus Norovirus of the Caliciviridae that were designated as genogroups I (GI) and II (GII), with NV belonging to GI and the Hawaii and Snow Mountain viruses belonging to GII.266,487 In addition, sequence analysis of the “classical” caliciviruses confirmed that they were distinct from the noroviruses, ultimately forming a separate genus, Sapovirus, within the Caliciviridae.252,274,309
characterize the viral genome, a major advance by Jiang et al199 that established the classification of Norwalk virus as a member of the Caliciviridae. The complete RNA genome sequences of Norwalk virus and the closely related Southampton virus were determined and found to be organized into three major open reading frames (ORF1, −2, and −3) with a polyadenylated 3′ end176,202,253,254 (see Genome Structure and Organization). The ORF1 was shown to encode a large polyprotein that was proteolytically processed into the mature nonstructural proteins.273,276 The ORF2 encoded the major capsid protein, VP1, and ORF3 encoded a minor structural protein, VP2. The human noroviruses initially segregated into two major phylogenetic groups within what would become the genus Norovirus of the Caliciviridae that were designated as genogroups I (GI) and II (GII), with NV belonging to GI and the Hawaii and Snow Mountain viruses belonging to GII.266,487 In addition, sequence analysis of the “classical” caliciviruses confirmed that they were distinct from the noroviruses, ultimately forming a separate genus, Sapovirus, within the Caliciviridae.252,274,309
Table 20.1 Taxonomic Structure of the Caliciviridae | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||
Classification
Members of the virus family Caliciviridae have a virion protein, genome (VPg)-linked, positive-sense RNA genome that is polyadenylated and surrounded by a nonenveloped, icosahedral capsid of 27 to 40 nm in diameter. The capsid is constructed predominantly from a major structural protein, VP1, of approximately 60,000 D. The five genera of the family Caliciviridae—Norovirus, Sapovirus, Lagovirus, Nebovirus, and Vesivirus—each represent a distinct phylogenetic clade in the family92,152 (e-Fig. 20.1). Within each genus, one or more species has been defined based primarily on genetic relatedness, and the current taxonomic structure of the Caliciviridae is shown in Table 20.1. It is possible that additional genera will be established, following characterization of the unique genomes of Tulane virus (simian),128 St. Valérian virus (porcine),251 and Bayern virus (avian)509 (e-Fig. 20.1). Below the level of species in certain genera (Norovirus and Sapovirus), provisional genetic typing systems (consisting of genogroups subdivided into genetic clusters or genotypes) have proven useful in epidemiologic studies (see Molecular Epidemiology).
Virion Structure
Calicivirus virions exhibit T = 3 icosahedral symmetry. The capsid contains 90 dimers of the VP1 capsid protein that form a shell from which 90 arch-like capsomeres protrude at the local and strict twofold axes.86,381,382,384 These arches are arranged in such a way that 32 large hollows are seen at the icosahedral five- and threefold positions, and these hollows are seen as cup-like structures on the surface of caliciviruses (calici is derived from the Latin word calyx, or “cup”). Electron cryomicroscopy and computer image processing studies of representative caliciviruses show subtle variation in the capsid structures that are consistent with their differences in appearance by negative-stain EM, which can range from a feathery appearance (many noroviruses, such as Norwalk virus in Fig. 20.1A) to the presence of sharply defined “cups” (many sapoviruses, such as Sapporo virus in Fig. 20.1E and the vesiviruses).86,383
Self-assembly of the norovirus VP1 into virus-like particles (VLPs) is an efficient process and does not require RNA201 or the minor capsid protein, VP2.256,263 This feature has been especially useful in the study of the fastidious caliciviruses, because recombinant (r) VLPs expressed in the baculovirus system have served as a surrogate for native virions.154,201,203 The NV rVP1 (180 copies) characteristically self-assembles into 38-nm particles with T = 3 symmetry, but it can form smaller VLPs (23 nm) with T = 1 symmetry composed of 60 copies of VP1.498
The atomic structure of the Norwalk rVLP has been determined by x-ray crystallography.382 These structural studies
have defined two major domains in the VP1—the shell (S) and the protruding (P) arm (Fig. 20.2). The S domain forms the inner part of the capsid that surrounds the RNA genome and maintains the icosahedral contacts of the T = 3 structure, and the P domain forms the arch-like protrusions that emanate from the shell and contain the dimeric contacts.382 The amino acid residues that correspond to the S and P subdomains in the NV VP1 (530 amino acids in length) are diagrammed in Figure 20.2A. A ribbon model of the VP1 dimeric subunit derived from the crystallographic structure of the capsid shows a more detailed view of these domains and their interactions (Fig. 20.2B, C). The NH2-terminal (N) arm, located within the S domain, consists of residues 1 to 49 and faces the interior of the capsid. The part of the S domain that forms a classic eight-stranded antiparallel β-sandwich fold consists of amino acids (aa) 50 to 225 (which includes a flexible hinge). The entire S domain (aa 1 to 225) corresponds to the N-terminal region of the capsid protein that is relatively conserved among noroviruses in sequence comparisons. The P domain, which is linked to the S domain through a flexible hinge (aa 219 to 225), corresponds to the C-terminal half of the VP1, which is more variable in amino acid sequence. The P domain is divided into the P1 subdomain, encompassing aa 226 to 278 and 406 to 520, and the P2 subdomain, encompassing aa 279 to 405. The P1 subdomains form the sides of the arch of the capsomeres and position the highly variable P2 subdomain at the top of the arch. In the dimeric form of the capsid protein, two P2 subdomains form what appears to be a bilobed structure at the surface of the virion. The exposure of the variable P2 region on the surface is consistent with its role in the formation of a major antigenic site and in receptor binding.10,47,85,86,107,110,172,178,185,221,271,281,348,382,398,459,497 It has been proposed that the highly conserved S domain may function as an icosahedral scaffold with the N-terminal arm providing a switch to facilitate the appropriate curvature, and that the P domain may be a replaceable module for conferring strain differences and antigenic specificity.85,86,382 Structural studies of other caliciviruses have shown a similar organization, with an internal shell domain serving as the scaffold for the more variable protruding domain.85,220
have defined two major domains in the VP1—the shell (S) and the protruding (P) arm (Fig. 20.2). The S domain forms the inner part of the capsid that surrounds the RNA genome and maintains the icosahedral contacts of the T = 3 structure, and the P domain forms the arch-like protrusions that emanate from the shell and contain the dimeric contacts.382 The amino acid residues that correspond to the S and P subdomains in the NV VP1 (530 amino acids in length) are diagrammed in Figure 20.2A. A ribbon model of the VP1 dimeric subunit derived from the crystallographic structure of the capsid shows a more detailed view of these domains and their interactions (Fig. 20.2B, C). The NH2-terminal (N) arm, located within the S domain, consists of residues 1 to 49 and faces the interior of the capsid. The part of the S domain that forms a classic eight-stranded antiparallel β-sandwich fold consists of amino acids (aa) 50 to 225 (which includes a flexible hinge). The entire S domain (aa 1 to 225) corresponds to the N-terminal region of the capsid protein that is relatively conserved among noroviruses in sequence comparisons. The P domain, which is linked to the S domain through a flexible hinge (aa 219 to 225), corresponds to the C-terminal half of the VP1, which is more variable in amino acid sequence. The P domain is divided into the P1 subdomain, encompassing aa 226 to 278 and 406 to 520, and the P2 subdomain, encompassing aa 279 to 405. The P1 subdomains form the sides of the arch of the capsomeres and position the highly variable P2 subdomain at the top of the arch. In the dimeric form of the capsid protein, two P2 subdomains form what appears to be a bilobed structure at the surface of the virion. The exposure of the variable P2 region on the surface is consistent with its role in the formation of a major antigenic site and in receptor binding.10,47,85,86,107,110,172,178,185,221,271,281,348,382,398,459,497 It has been proposed that the highly conserved S domain may function as an icosahedral scaffold with the N-terminal arm providing a switch to facilitate the appropriate curvature, and that the P domain may be a replaceable module for conferring strain differences and antigenic specificity.85,86,382 Structural studies of other caliciviruses have shown a similar organization, with an internal shell domain serving as the scaffold for the more variable protruding domain.85,220
 Figure 20.2. Organization of the norovirus major capsid protein, VP1. A: The Norwalk virus major capsid protein VP1 is 539 amino acids (aa) in length and is organized into two major parts, shell (S) and protruding (P), connected by a hinge (H) region. The aa borders of the defined domains are N-terminal (N) arm, 1 to 49; S, 50 to 218; hinge, 219 to 225; P1, 226 to 278 and 406 to 530; and P2, 279 to 405. (Adapted from Prasad BV, Hardy ME, Dokland T, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999;286:287–290.) B: Model of the T = 3 Norwalk virus whole capsid, determined at 3.4-Å resolution.382 The capsid is composed of 90 dimers of VP1, with the S, P1, and P2 domains shown in blue, red, and yellow, respectively. The box highlights the arrangement of a VP1 dimer as it is displayed on the surface of the virion. C: Three-dimensional ribbon representation of a VP1 dimer derived from x-ray crystallography studies of Norwalk virus recombinant virus-like particles (rVLPs) at 3.4 Å, showing the presentation of the P2 domain at the top of an arch supported by two P1 domain “arms”.382 The S domain forms the internal scaffold of the virion that surrounds the RNA genome and positions the arch to the virion surface. The N-terminus (N) and C-terminus (C) of the VP1 are indicated. D: Three-dimensional ribbon representation of a P domain–only dimer, determined at 1.4-Å resolution.91 The box highlights the location of a histo-blood group antigen (HBGA) carbohydrate binding site mapped within the P2 domain (see Fig. 20.7). (Images provided by B. V. V. Prasad.) |
Knowledge of the capsid structure has informed study of capsid assembly and facilitated the expression of subviral forms of the capsid that have proven useful in several areas of research (e-Fig. 20.2).460 The extreme N-terminus of the NV VP1 protein (first 20 aa) was not required for the assembly of native-sized 38-nm VLPs with T = 3 symmetry.36 Expression of the NV VP1 S region alone (beginning with the extreme N-terminus of the VP1 and including amino acid residues 1 to 227) resulted in the formation of smooth particles (designated “S” particles in e-Fig. 20.2) of approximately 30 nm in diameter.36
Although initial expression of the NV P region alone did not yield VLPs in baculovirus or bacterial systems,36,458 the native P domain from NV and other noroviruses formed P dimers that were recognized by antibodies and carbohydrates similarly to intact VLPs.458 The ability to produce rapidly P dimers (and mutagenized forms) accelerated structure and function studies of the norovirus capsid protein.61,463,464,465 Further genetic engineering to include a four to seven arginine-rich sequence at the C-terminus of the P domain resulted in the generation of small, subviral particles termed P particles that were assembled from 12 dimers of the P domain and that showed T = 1 symmetry460,463 (e-Fig. 20.2). Recently, expression of the P domain with a further modified terminus allowed the production of small P particles.457
Although initial expression of the NV P region alone did not yield VLPs in baculovirus or bacterial systems,36,458 the native P domain from NV and other noroviruses formed P dimers that were recognized by antibodies and carbohydrates similarly to intact VLPs.458 The ability to produce rapidly P dimers (and mutagenized forms) accelerated structure and function studies of the norovirus capsid protein.61,463,464,465 Further genetic engineering to include a four to seven arginine-rich sequence at the C-terminus of the P domain resulted in the generation of small, subviral particles termed P particles that were assembled from 12 dimers of the P domain and that showed T = 1 symmetry460,463 (e-Fig. 20.2). Recently, expression of the P domain with a further modified terminus allowed the production of small P particles.457
Genome Structure and Organization
Caliciviruses have a linear, single-stranded, positive-sense RNA genome (ranging from approximately 7.3 to 8.5 kb [kilobases] in length) (Fig. 20.3A). Genomes characteristically begin with a 5′ end terminal pGpU sequence that is covalently linked to a small protein, VPg. A short conserved region (CR) at the 5′ end is repeated internally in the genome near the beginning of a subgenomic-sized RNA transcript that is co-terminal with the 3′ end of the genome.179,254,324,331,344 The nonstructural proteins are encoded beginning near the 5′ end of the genome, and the structural proteins (VP1 and VP2) are encoded toward the 3′ end of the genome in the region corresponding to a subgenomic RNA. Calicivirus genomes are organized into two or more major ORFs, depending on the genus. The noroviruses and vesiviruses encode the VP1 structural protein in a separate ORF (ORF2), whereas sapoviruses, lagoviruses, and neboviruses encode a VP1 that is contiguous with the large nonstructural polyprotein in ORF1 (Fig. 20.3B). All caliciviruses have a relatively small ORF near the 3′ end that encodes the minor structural protein, VP2. The VP2 is variable in size (12,000 to 29,000 D) and sequence identity among the caliciviruses.65,418 Murine norovirus genomes analyzed thus far contain a unique conserved ORF (ORF4).467 with an encoded protein of 23,800 D
(designated VF-1) that has been implicated in modulation of the host innate immune response in vivo.318
(designated VF-1) that has been implicated in modulation of the host innate immune response in vivo.318
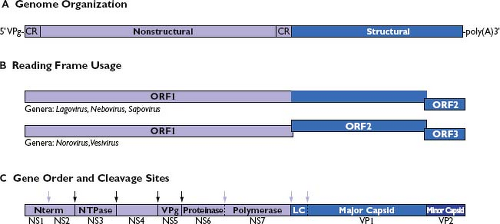 Figure 20.3. Comparative features of calicivirus genomes. A: The positive-sense RNA genome is covalently linked to a virion protein genome (VPg) at the 5′ end and polyadenylated at the 3′ end. Nonstructural proteins are encoded beginning from the 5′ end of the genome and the structural proteins are encoded toward the 3′ end. A conserved region (CR) of nucleotide sequence is shared between the 5′ end of the viral genome and the 5′ end of an abundant subgenomic RNA species produced during replication that serves as a template for translation of the viral structural proteins. B: Calicivirus genome organization. Calicivirus genomes are organized so that the major capsid protein coding sequence is either in frame or not with the upstream nonstructural protein sequence. The genomic organization and reading frame usage are shown for representative strains in the genera Lagovirus (Ra/LaV/RHDV/GH/1988/DE, GenBank Accession Number M67473), Nebovirus (Bo/NeV/NBV/Newbury-1/1976/UK, GenBank Accession Number DQ013304), Sapovirus (Hu/SaV/Manchester/1993/UK, GenBank Accession Number X86560), Norovirus (Hu/NoV/Norwalk/1968/US, GenBank Accession Number M87661), and Vesivirus (Fe/VeV/FCV/F9/1958/US, GenBank Accession Number M86379). C: A large polyprotein (encoded in open reading frame 1 [ORF1]) is translated from the viral RNA genome, and it is processed into precursors and final products by a virus-encoded protease. The proteolytic processing strategy varies among the caliciviruses, but all viruses encode domains for at least seven protein functions indicated here as nonstructural (NS) proteins NS1 through NS7.434 An extra cleavage site is present in the ORF1 of caliciviruses in which the capsid protein sequence is in frame with the nonstructural polyprotein encoded in ORF1. Cleavage at this site is thought to release the VP1 from the polyprotein so that the RNA-dependent RNA polymerase (NS7) can adopt an active conformation early in the replicative cycle. A cleavage site unique to the vesiviruses is present to release the leader of the capsid protein (LC) from a capsid precursor encoded in ORF2.438 Mapped cleavage sites conserved and utilized among all calicivirus genera are indicated with a dark arrow; cleavage sites that vary in utilization among the family are indicated with a light arrow.238,273,276,358,359,434,435,438,506 |
Viral Proteins
Structural Proteins
Three proteins are found in mature calicivirus virions: VP1, VP2, and VPg.436 The VP1 (∼60,000 D), which is the major structural protein of the virus, is present in 180 copies (90 dimers) per virion.383 The predominance of VP1 in the formation of the viral capsid structure (see Virion Structure) is consistent with its critical role in determining the antigenic phenotype of the virus and its interactions with host cells.
The VP2 (12,000 to 29,000 D) is considered a minor structural protein because it is present in only one to two copies per virion, and its function is unknown.145,146,436 The ablation of VP2 expression in an infectious FCV complementary DNA (cDNA) clone by the introduction of a stop codon in its reading frame did not abolish RNA replication; however, infectious virions could not be recovered without an intact VP2.433 Although VP1 can self-assemble into rVLPs independently of the presence of VP2,256,263 the presence of VP2 may increase the efficiency of VP1 expression and enhance the stability of rVLPs generated in the baculovirus expression system.35 Evidence for a direct interaction between the VP1 and VP2 capsid proteins has been reported for both NV74,145,146 and FCV,108,211 further suggesting a role in particle maturation or stability.
The VPg is covalently linked to the genomic and subgenomic RNA in infected cells,120,180 and is a minor component in virions at an estimated one or two copies per particle.410,436 Although the VPg is present in virions, it likely functions primarily as a nonstructural protein during replication (see Nonstructural Proteins).
Nonstructural Proteins
Caliciviruses derive their mature nonstructural proteins (designated here as NS1 through NS7) by proteolytic cleavage of a large polyprotein encoded in ORF1 (Fig. 20.3C). The length of the noncleaved polyprotein precursor is approximately 200,000 D (excluding the in-frame VP1 capsid protein sequences of the lagoviruses, neboviruses, and sapoviruses). This large precursor has never been observed, most likely because proteolytic processing is rapid and co-translational.273,276,439 The proteolytic cleavages are mediated by a virus-encoded cysteine proteinase (NS6pro).50 The location of the cleavage sites in the ORF1 polyprotein that define the borders of the final nonstructural protein cleavage products has been determined for calicivirus strains representing the genera Norovirus,34,273,276,434 Vesivirus,435 Lagovirus,238,506 and Sapovirus.359 Some variation is seen among the proteolytic processing strategies, but the overall gene order of the calicivirus nonstructural proteins is conserved. The lagoviruses, represented by RHDV, have the highest number (six) of mapped cleavage sites. The polyprotein of RHDV is cleaved at these six sites to release seven final products (designated as NS1 through NS7 in Fig. 20.3C and Table 20.2). Five cleavage sites for the norovirus ORF1 polyprotein (represented by Southampton virus) have been mapped that would release six mature products.273,276 The noroviruses differ from the lagoviruses in that a protease cleavage site has not been directly mapped in the extreme N-terminal protein, although evidence suggests that additional processing (or modification) of the protein can occur in cells.419,434 The vesivirus cleavage map (represented by FCV) contains five mapped cleavage sites to release six mature nonstructural proteins, and each cleavage event is essential in the virus replication cycle.435 The vesiviruses show yet another variation in processing, in that no evidence exists for efficient viral protease-mediated cleavage between the Pro and polymerase (Pol) proteins, even in virus-infected cells.306,357,435,439 In addition, vesiviruses bear a unique cleavage site in a capsid protein precursor protein that is processed by NS6pro to release the leader of the capsid (LC) and the major capsid protein, VP1438,470 (Fig. 20.3C). The predicted nonstructural protein cleavage map of human sapovirus strain, Mc10, shows an overall similarity with that of the vesiviruses.359 Sapoviruses (and other caliciviruses in which the capsid region is in frame with the nonstructural polyprotein) bear an additional protease cleavage site in the ORF1 polyprotein between the polymerase and capsid coding regions. A number of stable precursor proteins have been described also for the caliciviruses, and it is likely that these proteins have defined functions in replication.34,39,93,238,325,326,434,435
The calicivirus dipeptide cleavage recognition sites are consistent with those described for the picornavirus 3C cysteine proteinase.174 The calicivirus cleavage sites identified thus far have either a negatively charged glutamic acid (E) or polar glutamine (Q) in the first position (designated P1). More variation exists within the second position of the dipeptide cleavage site (designated P1′). Studies of the calicivirus proteinase substrate specificity have shown some tolerance in the P1′ position at certain cleavage sites.174,438,505 The conformation of the protein surrounding the dipeptide recognition site is also important for efficient cleavage by the proteinase.174,439
The availability of proteolytic cleavage maps for the calicivirus nonstructural polyproteins has enabled studies that elucidate the functions and structures of individual proteins.175 Biochemical studies first confirmed enzymatic activities in calicivirus proteins corresponding to an NTPase (NTPase or NS3NTPase),300 a chymotrypsin-like cysteine proteinase (Pro or NS6pro),50 and an RNA-dependent RNA polymerase (Pol or NS7pol).479 The three-dimensional structures for the latter two enzymes have been reported. The norovirus Pro shares structural similarities with classical chymotrypsin-like serine proteases188,189,339,518 (Fig. 20.4A). A cleft containing the active site catalytic residues (His 30, Glu 54, and Cys 139 for Norwalk virus) involved in substrate cleavage is located between two domains. The N-terminal domain (blue) starts with an α-helix followed by a five-stranded twisted antiparallel β-sheet. The structure of this domain contains features of both the simpler four-stranded β-sheet found in the N-terminal domain of picornaviral 2A proteinases and the more commonly found complete β-barrels observed in the N-terminal domains of most chymotrypsin-like proteinases. The C-terminal domain (red) adopts the structure of a classical six-stranded β-barrel found in a wide range of viral and nonviral chymotrypsin-like proteinases. Co-crystallization of the protease with an active site-directed peptide inhibitor (acetyl-Glu-Phe-Gln-Leu-Gln-propenyl ethyl ester) has given detailed insight into the interaction of substrate with the catalytic residues such as Cys 139 (the active site nucleophile) within the cleft189 (Fig. 20.4B). The norovirus RNA-dependent RNA polymerase has a classical “right hand” (finger, thumb, and palm) organization,346,347,517 with the C-terminus of the protein positioned in the active site cleft (Fig. 20.5A). Modeling of the
interaction of Pol with an RNA template in the presence of manganese and cytidine triphosphate (CTP) shows that the initiation of RNA synthesis occurs within the active site cleft (Fig. 20.5B–D). Binding of the primer/template RNA duplex displaces the C-terminal tail away from the active site, allowing the central helix of the thumb domain to position itself for interaction with the primer strand and minor groove of the primer–template duplex.517 Two divalent metal ions (likely Mg2+ in cells) help mediate catalysis by forming coordination bonds with three highly conserved aspartic acid residues and the nucleoside triphosphate (NTP). After nucleotidyl transfer has occurred, the pyrophosphate is released from the enzyme. The primer–template duplex is predicted to translocate in a manner that places the newly incorporated nucleotide into the same position as that of the 3′ end of the primer strand immediately prior to nucleotidyl transfer. This translocation process
provides the space needed to form the binding site for the next incoming nucleoside triphosphate.
interaction of Pol with an RNA template in the presence of manganese and cytidine triphosphate (CTP) shows that the initiation of RNA synthesis occurs within the active site cleft (Fig. 20.5B–D). Binding of the primer/template RNA duplex displaces the C-terminal tail away from the active site, allowing the central helix of the thumb domain to position itself for interaction with the primer strand and minor groove of the primer–template duplex.517 Two divalent metal ions (likely Mg2+ in cells) help mediate catalysis by forming coordination bonds with three highly conserved aspartic acid residues and the nucleoside triphosphate (NTP). After nucleotidyl transfer has occurred, the pyrophosphate is released from the enzyme. The primer–template duplex is predicted to translocate in a manner that places the newly incorporated nucleotide into the same position as that of the 3′ end of the primer strand immediately prior to nucleotidyl transfer. This translocation process
provides the space needed to form the binding site for the next incoming nucleoside triphosphate.
Table 20.2 Calicivirus Nonstructural Proteins | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
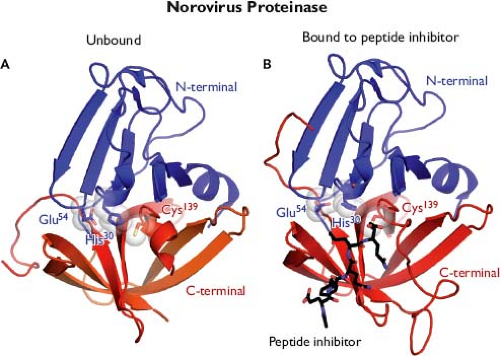 Figure 20.4. Structure of the norovirus proteinase. A: Three-dimensional ribbon representation of the Norwalk virus protease structure resolved at 1.5 Å shows that it adopts the conformation of a classical serine protease, in which a cleft containing the enzymatic active site is located between two β-barrel domains. The active site is positioned at the opening of the cleft and composed of residues His 30, Glu 54, and Cys 139 (the catalytic triad). The conformation of the loops and β-strands are thought to play a role in positioning the active site residues for proteolysis. B: Modeled interaction of Norwalk virus protease with an inhibitor that blocks proteolysis.188,189,339,518 (Images provided by K. Ng.) |
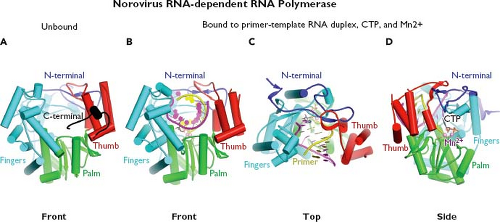 Figure 20.5. Structure of the norovirus RNA-dependent RNA polymerase. A: The norovirus RNA-dependent RNA polymerase adopts the classic “right hand” structure characteristic of polynucleotide polymerases as shown in this stick model. The fingers (blue) and palm (green) domains form a rigid unit, while the thumb (red) domain is flexible and can assume either a “closed” or “open” conformation. An N-terminal domain bridges the fingers and thumb domains. When the polymerase is unbound to template, the C-terminal end of the protein appears to lie within the active site cleft.347 B–D: The front, top, and side views, respectively, of the Norwalk RNA polymerase bound to a primer–RNA duplex, cytidine triphosphate (CTP), and metal divalent cation, Mn2+.346,347,517 (Images provided by K. Ng.) |
Although the gene order of the nonstructural proteins and strong structural and functional homology in the Pro and Pol enzymes suggest a common ancestor for the caliciviruses and picornaviruses,342 it is striking that several proteins encoded in the calicivirus ORF1 share little or no detectable sequence relatedness with the picornaviruses. These include the extreme N-terminal proteins NS1 and NS2 (corresponding in gene order to the picornavirus 2A and 2B proteins), the NS4 (corresponding to the picornavirus 3A protein), and the NS5 (corresponding to the VPg). Although the roles of these proteins in replication require further investigation, evidence for some functional homology with the picornaviruses has been
proposed.25,129,422 A summary of the calicivirus nonstructural proteins and their known properties is shown in Table 20.2.
proposed.25,129,422 A summary of the calicivirus nonstructural proteins and their known properties is shown in Table 20.2.
Stages of Replication
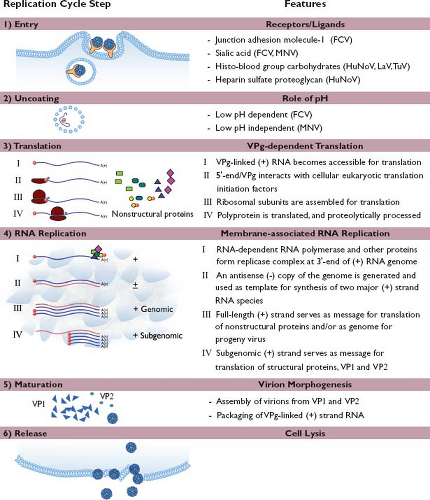
Replication Strategy
The replication strategy elucidated thus far for the caliciviruses shares many features with those of other positive-strand RNA viruses (Fig. 20.6). Caliciviruses attach and enter the cell, the RNA genome is released, and translation of the genome occurs via the host cell machinery. Certain newly translated viral proteins interact with the host cell to establish defined sites of virus replication (characteristically involving reorganized intracellular membranes), while other proteins function as replicative enzymes. Newly synthesized positive-strand RNA genomes are covalently linked to VPg and packaged into virions that are released from lysed cells. The replication cycle of a calicivirus is rapid: new viral progeny can be detected within hours after infection.
Reverse genetics systems have been developed for several caliciviruses, including FCV,432 porcine enteric calicivirus,77 Tulane virus,492 RHDV,277 and murine norovirus,83,489,515 based on the construction of infectious full-length cDNA clones of the viral genome. Murine norovirus is presently the only norovirus that grows efficiently in cell culture,507 making this virus an important model.508 Human norovirus infectious cDNA clones have remained elusive in the absence of a fully permissive cell culture system to verify recovery of virus, but replication in cells can be studied following expression of proteins from full-length cDNA clones21,219 or in stable RNA replicon-bearing cell lines.79
Mechanism of Attachment
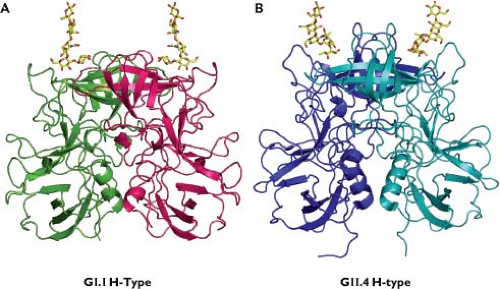
Calicivirus virions must first interact with the host cell. Carbohydrates, including those present on various histo-blood group antigens (HBGAs), have been implicated in the binding of a number of calicivirus strains to cells245,301,403,443,455,462,516 (Fig. 20.6). Structural studies have verified that the norovirus VP1 interacts with HBGA carbohydrates61,91 (Fig. 20.7). Receptor recognition is essential for the caliciviruses, as transfection of infectious calicivirus RNA into nonpermissive cells (i.e., those that cannot be infected with virions) allows replication and recovery of infectious progeny virus.162,245,298 The junction adhesion molecule-1 (JAM-1), an immunoglobulin-like cellular membrane protein, has been identified as a functional receptor for FCV,299 making this the first experimentally verified cellular receptor in the family. Structural and modeling studies have shown that FCV interacts with feline (f) JAM-1 through binding of the P2 domain of the capsid to the distal membrane domain (D1) of f JAM-137,38,363 (e-Fig. 20.3).
Nearly all norovirus VLPs bind to one or more HBGA carbohydrates, and as noted earlier, structural studies have defined interactions with certain HBGA saccharides at the amino acid level61 (Fig. 20.7). A correlation was found between susceptibility to Norwalk virus infection in adult volunteers and HBGA secretor status, which is linked to the FUT2 gene269 (see Cell and Tissue Tropism). A similar pattern of genetic susceptibility to RHDV (lagovirus) infection in rabbits was associated with genes involved in the modification of HBGAs, indicating a role for blood group carbohydrates in host cell recognition by lagoviruses.161 Experiments to verify that HBGAs can serve as a functional receptor for the noroviruses have been unsuccessful, hampered in part by the unavailability of a permissive cell culture system. Expression of the human FUT2 gene product (fucosyltransferase-2) in nonpermissive cells enhanced binding of Norwalk virus VLPs to cells in vitro but did not render them permissive for the virus.162
Mechanism of Entry and Intracellular Trafficking
Variation has been noted among caliciviruses in their requirements for entry into cells and subsequent infection. Feline calicivirus replication was inhibited by chloroquine, a reagent that raises lysosomal pH, indicating dependence on a low pH step during entry.244,444 In contrast, murine norovirus (strain MNV-1) was not dependent on the acidification of endosomes for infectivity.143,373 Feline caliciviruses use clathrin-mediated endocytosis for entry into mammalian cells,444 whereas MNV-1 apparently does not enter via clathrin- or caveolin-mediated pathways.143,373 An endocytic pathway was proposed for MNV-1 entry that likely involves cholesterol-sensitive lipid rafts and dynamin II (at least in RAW264.7 cells).143,373,374
Uncoating
The uncoating events that allow the calicivirus positive-sense RNA genome to become accessible to the cellular translational machinery have not been defined. Uncoating is a rapid process: MNV genome was released within 1 hour after infection.374 Binding to the cellular receptor induces a conformational change in the capsid of FCV, suggesting that uncoating may, in part, be receptor binding mediated.37 FCV infection is associated with an increase in permeability of the host cell membrane soon after virus entry via clathrin-coated endosomes and exposure to low pH.444 It was proposed that this acidification process might change the conformation of the FCV virion structure to facilitate release of the viral RNA.444 Analysis of norovirus (Norwalk) VLPs by mass spectrometry has shown that they are stable under acidic conditions; a disassembly model has been proposed in which T = 3 particles (containing 180 copies of VP1) disassociate into predominantly dimers in the presence of alkaline conditions.426
Translation
Calicivirus genomic RNA in virions requires the presence of the covalently linked VPg protein in order to establish an infection after release (or transfection) into cells.58 The initiation of translation of the incoming positive-strand genome is likely mediated through interactions of the VPg protein with the cellular translation machinery.82,103,148,165 Calicivirus replication is associated with an inhibition of host cell translation, and the viral proteinase (NS6pro) has been shown to cleave certain cellular proteins involved in translation, which may, in part, give the viral RNA a competitive advantage.250 The ORF1 of the virus is translated first to produce a large polyprotein, which is processed rapidly into precursors and products by NS6pro at several essential cleavage sites.435 Some of these nonstructural proteins and their precursors function to set up replication sites within the host cell,25,194 while others (such as the RNA-dependent RNA polymerase, NS7pol) play a role in replication of the viral RNA (Table 20.2).
An abundant VPg-linked subgenomic positive-strand RNA serves as a bicistronic message for translation of the structural proteins VP1 and VP2,179,344 and the regulation of translation of VP2 from the subgenomic RNA (translated at approximately 20% of the levels of VP1) was mapped to an upstream RNA sequence element in the VP1 coding region of approximately 70 nucleotides designated as the termination upstream ribosomal binding site (TURBS).322,323 The TURBS site contains two motifs, one of which mediates base pairing between the viral subgenomic messenger RNA and the cellular 18S ribosomal subunit.322 This interaction may function as a “tether,” positioning the ribosome for immediate reinitiation of translation following termination of translation of the VP1 gene.291,292
Replication of Genomic Nucleic Acid
As with all other positive-strand RNA viruses, the replication of calicivirus RNA is associated with host cell membranes.155 A marked rearrangement of intracellular membranes occurs, and evidence exists for the initiation of RNA replication in a perinuclear site that contains membranes associated with endoplasmic reticulum, trans-Golgi network, and endosomal membrane markers.25,194 The initiation of synthesis of an antisense (negative)-strand RNA from the genomic RNA template occurs beginning at the 3′ end of the genomic positive-strand RNA and likely involves interactions with cellular proteins.166 The negative-strand RNA, in turn, serves as a template for transcription of two major positive-strand RNA species corresponding to the full-length genome (genomic RNA) and the approximately terminal one-third of the genome toward the 3′ end (subgenomic RNA).344
The calicivirus RNA genome bears conserved regions of secondary structure,292,378,429 and functional RNA regulatory elements have been mapped both internally and near the ends of the genome.26 Transcription from the start site for the subgenomic RNA species on the negative-strand template (nt 5,296 of RHDV) was found to require an upstream sequence of 50 nt for full polymerase activity in in vitro studies,331 consistent with its role in the formation of a subgenomic promoter. The corresponding region of MNV was mapped also as bearing a promoter for subgenomic RNA synthesis from the negative-strand template.429
Assembly
A two-stage process has been proposed for the maturation of calicivirus (FCV) particles.237 The first stage involves the rapid aggregation and assembly of capsid precursor proteins into 5S subunits, which then pass through several intermediate forms (with varying stability) to form stable 15S subunits. The second stage involves the association of 15S subunits with newly synthesized RNA genomes to form infectious particles (that sediment at 170S). Protein–protein interactions have been detected between the FCV VPg (covalently linked to the RNA genome) and the capsid precursor as well as between the RNA-dependent RNA polymerase NS7pol and the capsid precursor, suggesting that these interactions may be related to packaging of the newly synthesized RNA into the viral capsid.211 In addition, successful assembly of the infectious FCV virions was linked to an efficient expression of the virus minor capsid protein, VP2.433
The VPg-linked genomic and subgenomic positive-strand RNA species are found in FCV and RHDV particles of distinct densities, indicating that they are not packaged together in the same virion.324,343 The incorporation of subgenomic RNA into the lower-density (LD) particles suggests that a packaging signal is located within the 3′ terminal 2,400 nucleotides of the genome. It has been suggested that LD particles (which would not be infectious) may be associated with FCV strains of higher virulence, but their function, if any, in the virus life cycle is unknown.343
Release
Calicivirus-infected cells undergo lysis, and it is presumed that the majority of progeny viruses are released during this process. The triggering of apoptosis has been associated with calicivirus infection both in vitro7,49,135,341,437 and in vivo,12,84,208,223,334,337,386,472 and apoptotic changes in cellular membranes may be one of the mechanisms by which cells lyse and release viral particles.
Pathogenesis and Pathology
Caliciviruses cause a broad range of diseases in many different animal hosts (Table 20.3). This section will focus on the noroviruses, but there are important common themes in pathogenesis shared by all caliciviruses. Illnesses range from
mild to life-threatening, and evidence exists for the emergence of calicivirus strains with increased virulence. Asymptomatic infection occurs in susceptible populations,106,377 and shedding of virus can extend days to weeks after resolution of acute symptoms.24,140
mild to life-threatening, and evidence exists for the emergence of calicivirus strains with increased virulence. Asymptomatic infection occurs in susceptible populations,106,377 and shedding of virus can extend days to weeks after resolution of acute symptoms.24,140
Table 20.3 Pathogenesis and Disease Manifestations of Representative Calicivirus Strains | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree