Bunyaviridae
Richard M. Elliott
Connie S. Schmaljohn
History
The family Bunyaviridae takes its name from Bunyamwera virus (BUNV), which was isolated originally from Aedes mosquitoes in the Semliki Forest, Uganda, during a yellow fever study in 1943.499 Following the isolation of BUNV, several additional arboviruses were discovered that clearly did not fit into the classic arbovirus group A and B antigenic groups (which now are included in the Flaviviridae family or the Alphavirus genus of the Togaviridae family) and were assigned to what became known as group C arboviruses.83 During the next decade, further virus discovery and detailed serologic studies, together with biochemical and morphologic analyses,363 led to the concept of a Bunyamwera supergroup consisting of groups of viruses that could be linked by repeatable serologic cross-reactions. Serogroups (viruses related by their reactivity in any serologic test) and complexes (closely related members of a serogroup) were further defined for these viruses.73
The family Bunyaviridae was formally established in 1975.423 The International Committee on Taxonomy of Viruses (ICTV) approved the creation of the Bunyavirus genus in 1980, along with viruses morphologically and biochemically similar but antigenically distinct, that formed the Uukuvirus, Phlebovirus, and Nairovirus genera.46 The discovery of a novel group of rodent-borne viruses and another group of plant infecting viruses with conforming molecular properties respectively led to the additions of the Hantavirus genus in 1985, and the Tospovirus genus in 1991. Further studies demonstrated a close biochemical similarity between uukuviruses and phleboviruses, resulting in their combination into the Phlebovirus genus in 1991.71 To prevent confusion when discussing viruses in the Bunyavirus genus as opposed to the family as a whole, the former was renamed Orthobunyavirus in 1995.142 Viruses within each genus are thus referred to as hantaviruses, nairoviruses, orthobunyaviruses, and so on, whereas the term “bunyavirus” is used to refer to any member of the family. Currently more than 350 named viruses are included in the family, making it one of the largest families of RNA viruses.
Except for the hantaviruses, which are transmitted in aerosolized rodent excreta, bunyaviruses are carried and
transmitted by arthropods, primarily mosquitoes, ticks, sand flies, or thrips.
transmitted by arthropods, primarily mosquitoes, ticks, sand flies, or thrips.
BUNV is the prototype of the family, and remains an important research model. It was the first bunyavirus whose genome was completely sequenced,141,306 and significantly was the first segmented genome negative-sense RNA virus that was recovered (rescued) entirely from cloned complementary DNA (cDNA).62,450
Table 42.1 Pattern of Sizes of Viral RNA Segments in Bunyaviridae Genera | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||
Classification
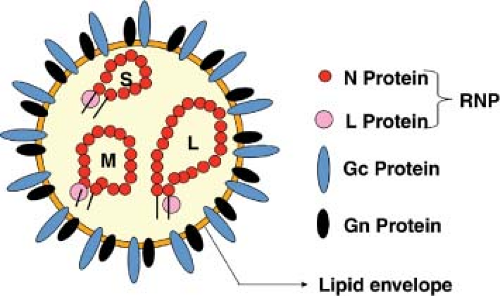
The essential criteria for inclusion in the family are enveloped virions of 80 to 120 nm diameter (Fig. 42.1), containing a tripartite, single-stranded RNA genome of negative- or ambi-sense polarity, which replicate in the cytoplasm, and usually assemble at the Golgi complex. Assignment to one of the five genera is based on lack of serologic cross-reactivity with members of other genera, the patterns of sizes of virion proteins (Table 42.2) and genome segments (Table 42.1), gene expression strategy (Fig. 42.2), and conserved terminal nucleotide sequences of the genomic RNAs (Table 42.3). The ICTV, through the Bunyaviridae Study Group, describes criteria for the designation of individual species within each genus.372 These designations, which are based on rather limited molecular data, should be treated with caution and as being fluid. The field requires large-scale, organized sequencing projects to more accurately understand the relationships between these viruses, and in particular to investigate the extent of genome segment reassortment among isolated viruses.65,378,390,391 A potential prototype of a sixth genus, Gouleako virus, has recently been described.329 This virus is most similar to phleboviruses but appears restricted to mosquitoes and could not infect vertebrate cells in culture. Table 42.4 lists the most notable members in the currently defined genera, and a brief introduction to each genus follows.
Orthobunyavirus Genus
The largest genus of bunyaviruses is the Orthobunyavirus genus, which contains more than 170 named viruses that are found throughout the world. Almost all of these viruses are transmitted by mosquitoes and have amplification cycles in a variety of vertebrate hosts.73,372 Among important orthobunyavirus pathogens in humans are La Crosse virus (LACV) that causes pediatric encephalitis, Oropouche virus (OROV) that causes a debilitating febrile illness, and Ngari virus that causes hemorrhagic fever, whereas Aino, Akabane, and Cache Valley viruses are examples of viruses causing disease in domestic animals (Table 42.4).
Classification of orthobunyaviruses has proven to be a complex issue. The majority of viruses have been placed in one of 18 serogroups based on serologic relatedness of complement fixing antibodies (mediated by the N protein) and hemagglutinating and neutralizing antibodies (mediated by the glycoproteins), although a number of viruses classified into the Orthobunyavirus genus are currently not assigned to any of these serogroups.73,372 The 18 serogroups are Anopheles A, Anopheles B, Bakau, Bunyamwera, Bwamba, Group C, Capim, California, Gamboa, Guama, Koongol, Minatitlan, Nyando, Olifanstlei, Patois, Simbu, Tete, and Turlock. Serologic relatedness varies within a serogroup and is further complicated by the occurrence of natural reassortant viruses (e-Fig. 42.1), such that viruses may be more related to members of one group or another depending on the assay used.72
The latest report of the ICTV delineates the orthobunyaviruses into 44 species,372 although as described above such classification should be regarded as fluid because of the paucity of molecular data. Comprehensive molecular genetic studies have involved viruses in only 4 serogroups, namely Bunyamwera, Group C, California, and Simbu,57,126,378,457 but S segment nucleotide sequences have been obtained for one or two representatives of the Anopheles A, Anopheles B, Bakau, Bwamba, Nyando, Tete, and Turlock serogroups.292,351,588 In most cases the S genome segment encodes two proteins, N and, in an overlapping reading frame, a small nonstructural protein termed NSs. N and NSs proteins are translated from the same
viral messenger RNA (mRNA) species as the result of alternate AUG-initiation codon selection.48,144 However, viruses in the Anopheles A, Anopheles B, and Tete serogroups do not show evidence of having an NSs ORF. In addition, these viruses have rather longer N proteins than those in the other serogroups.351
viral messenger RNA (mRNA) species as the result of alternate AUG-initiation codon selection.48,144 However, viruses in the Anopheles A, Anopheles B, and Tete serogroups do not show evidence of having an NSs ORF. In addition, these viruses have rather longer N proteins than those in the other serogroups.351
Table 42.2 Pattern of Sizes of Viral Structural Proteins in Bunyaviridae Genera | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||
Phlebovirus Genus
The Phlebovirus genus contains more than 80 viruses, with about half classified into nine antigenic complexes that are regarded as species, whereas 33 are considered as tentative species in the genus. The viruses of the genus Phlebovirus are present throughout the world, with the exception of Australia, and are more diverse in terms of arthropod vector than those of the other arthropod-borne genera. Most virus members are associated with phlebotomine sandflies, hence the genus name Phlebovirus (see Table 42.4). However, there are prominent exceptions, such as Rift Valley fever virus (RVFV), a medically and agriculturally important virus in Africa, which is primarily associated with Aedes species mosquitoes. In addition, Uukuniemi virus (UUKV), which has been used in a number of laboratory studies, is associated with the tick Ixodes ricinus. Recently a newly emerged phlebovirus was described in China (Huaiyangshan virus or severe fever with thrombocytopenia syndrome virus) that is associated with significant human mortality, and is transmitted by Haemaphysalis ticks.595
RVFV was first isolated in 1930 by Daubney et al. from an infected newborn lamb as part of an investigation of a large epizootic of disease causing abortion and high mortality in sheep.113 Large RVFV epizootics in various areas of sub-Saharan Africa have been noted since that time, and clinically compatible outbreaks have been retrospectively identified as far back as 1912.133 Several decades passed before serologic and molecular similarities to phlebotomine fever viruses were noted. Sandfly fever Sicilian and Naples viruses (SFSV, SFNV) were first isolated from American troops with febrile illness in the Palermo region of Sicily, Italy in 1943 and Naples, Italy in 1944, respectively, by inoculation of acute-phase sera into human volunteers and passage in newborn mice.456 Outbreaks of compatible human illness in the Mediterranean region date back to the time of the Napoleonic Wars, and association of the disease with sandflies was suggested as early as 1905.203
Nairovirus Genus
The nairoviruses are almost exclusively tick-borne viruses, although a few isolations have been made from Culicoides flies and mosquitoes (Table 42.4). Several serogroups exist, but the most important ones are the Crimean-Congo hemorrhagic fever (CCHF) group, which includes CCHF virus (CCHFV) and Hazara virus, and the Nairobi sheep disease (NSD) group, which includes NSD virus (NSDV) and Dugbe virus (DUGV).
Table 42.3 Consensus 3′ and 5′ Terminal Nucleotide Sequences of Genome RNAs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
The Nairovirus genus was named after NSDV, which was originally isolated in Nairobi, Kenya in 1910 by inoculation of sheep with the blood of sheep with acute gastroenteritis.354 The virus is now known to be present throughout various parts of Africa, and possibly India. CCHF was first recognized in the Crimean peninsula in the mid-1940s, when a large outbreak of severe hemorrhagic fever among agricultural workers was identified. The outbreak included more than 200 cases and a case fatality of about 10%.99 Similar disease cases were later reported throughout the European and central Asian republics of the former Soviet Union, Romania, and Bulgaria. However, the virus was first isolated from a patient with a one-day fever in Kisangani, Democratic Republic of Congo, in 1956.494 It was some years before the connection was established, but subsequent serologic studies and virus isolations from Asia and Europe revealed that the viruses from the different outbreaks and geographic regions were essentially the same virus, which then became named CCHFV.82,100
Hantavirus Genus
The discovery of hantaviruses traces back to 1951 to 1953, when United Nations troops were deployed during the border conflict between North and South Korea. More than 3,000 cases of an acute febrile illness were seen among the troops, about one third of which exhibited hemorrhagic manifestations, and an overall mortality of 5% to 10% was seen.129,305 The disease was initially termed Korean hemorrhagic fever but is now referred to as hemorrhagic fever with renal syndrome (HFRS). Despite considerable efforts, it took about 25 years until the field mouse, Apodemus agrarius, was identified as the rodent reservoir, and the virus was eventually isolated. At that
time, all identified viruses of the family Bunyaviridae were known to be arthropod-borne viruses. Therefore, it was a big surprise when this rodent-borne virus was shown to share characteristics of viruses of the family Bunyaviridae. The virus was named Hantaan virus (HTNV), after the Hantaan river close to the location of some of the initial Korean cases, and became the prototype of the Hantavirus genus.305
time, all identified viruses of the family Bunyaviridae were known to be arthropod-borne viruses. Therefore, it was a big surprise when this rodent-borne virus was shown to share characteristics of viruses of the family Bunyaviridae. The virus was named Hantaan virus (HTNV), after the Hantaan river close to the location of some of the initial Korean cases, and became the prototype of the Hantavirus genus.305
Table 42.4 Species in the Family Bunyaviridae | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
During the course of the early studies in Korea, it became clear that HFRS cases were also occurring in urban areas. Years of investigation finally demonstrated that the urban cases in Korea, China, and Japan were associated with infection with Seoul virus, which is hosted by the rats Rattus norvegicus and R. rattus.300,305,514 For more than 50 years, a similar, but generally milder, disease termed nephropathia epidemica (NE) was
described in various parts of Scandinavia.375 Following the isolation of HTNV, the virus was shown to react with sera from patients with convalescent phase NE.375 This led to the discovery of Puumala virus (PUUV) as the cause of this form of HFRS and the bank vole, Clethrionomys glareolus, as the virus host.
described in various parts of Scandinavia.375 Following the isolation of HTNV, the virus was shown to react with sera from patients with convalescent phase NE.375 This led to the discovery of Puumala virus (PUUV) as the cause of this form of HFRS and the bank vole, Clethrionomys glareolus, as the virus host.
The most recent major event in the history of hantaviruses was the discovery of hantavirus pulmonary syndrome (HPS) in the southwestern United States in 1993.373 A cluster of cases was identified in the Four Corners region, which presented with a flu-like illness (i.e., fever, headache, muscle aches, chills, and so on), but rapidly progressed to a more severe respiratory disease with bilateral pulmonary infiltrates, respiratory failure, shock, and death, occurring approximately 2 to 10 days after onset of illness in almost 50% of the cases.257 Within a couple of weeks of the initial outbreak, a newly identified virus, Sin Nombre virus (SNV), was shown to be the cause of HPS, and the rodent reservoir was shown to be the common deer mouse, Peromyscus maniculatus.96,373 Within the next several years, HPS was shown to occur throughout the Americas from Canada to Patagonia, and was found to be caused by least 10 hantaviruses, each associated with a different rodent species. During the study of the hantaviruses associated with HPS and HFRS, many additional hantaviruses not known to be associated with human illness were discovered in a wide variety of rodent hosts (Table 42.4).
In addition to rodents, a number of hantaviruses have been isolated from insectivores. In fact the first hantavirus isolated was Thottapalayam virus, from the Asian house shrew, in Southern India,79 although it was not recognized as a hantavirus until several years later.594 Very recently a number of other insectivore-associated hantaviruses have been identified; interestingly, prevalence in these animals is much higher than in rodents. The insectivore-borne viruses are widely distributed across the globe and display greater genetic diversity than the rodent-borne hantaviruses.434
Tospovirus Genus
The tospoviruses are plant viruses that are transmitted propagatively by thrips.579 At least 13 species of thrips, in the Frankliniella and Thrips genera, are reported to transmit these viruses.384,575 Several of these viruses are now recognized as being of significant agricultural importance.427
The history of the tospoviruses goes back to 1915, with the recognition of spotted wilt of tomatoes in Australia,66 and then the subsequent isolation in 1930 of tomato spotted wilt virus (TSWV),460 from which the genus gained its name.275 By the late 1940s, TSWV infections were greatly reduced in the United States and Western Europe due to pesticide use controlling the onion thrips, Thrips tabaci, which was likely the primary vector at that time. The geographic spread during the 1960s and 1970s of the Western flower thrips, Frankliniella occidentalis, another efficient vector of TSWV, led to a rapid expansion of TSWV.543,575 The virus is now known to be global in distribution, and it is found throughout agricultural areas in warmer climate zones and prevalent in greenhouse cultivations in more temperate areas. In contrast to the other genus members, TSWV has an unusually wide host range, with more than 925 plant species, including 82 botanical families, reported to be susceptible to infection. These include several important crops such as peanuts, peppers, tobacco, potatoes, peas, tomatoes, celery, lettuce, and ornamental flowers, with crop losses amounting to more than a billion dollars.393 There are 21 viruses assigned to the Tospovirus genus, with currently eight recognized species.200,372
Virion Structure
Morphology
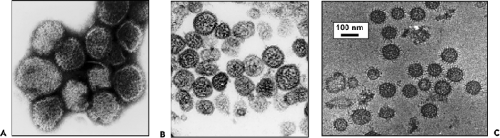
Under the electron microscope, negatively stained bunyaviruses appear spherical or pleomorphic, 80 to 120 nm in diameter, and display surface glycoprotein (GP) projections of 5 to 10 nm, which are embedded in a lipid bilayered envelope approximately 5 to 7 nm thick. (Fig. 42.3). With use of cryoelectron microscopic techniques, which more accurately preserve particle integrity, LACV particles were reported to be 75 to 115 nm in diameter,522 whereas particles of RVFV and Uukuniemi
(UUKV) phleboviruses had mean diameters of 100 nm and 125 nm, respectively,162,386 and Tula and Hantaan hantaviruses had mean diameters of 130 to 135 nm.35,216 The spikes are thought to mostly consist of heterodimers of the two viral GPs, and biochemical analysis of purified LACV particles indicated that the two GP species were equimolar and present at about 650 copies per virion. The GPs interact to form surface morphologic units that vary among viruses in different genera.333 Virions in the Orthobunyavirus genus have surfaces covered with closely packed, knoblike morphologic units with no detectable order. Similarly, no obvious order was found for the small surface structures with central cavities observed on viruses in the Nairovirus genus.333 The appearances of viruses in the Tospovirus genus have been likened to that of the nairoviruses.344 Other than the presence of GP projections, which are observed as a fringe on negatively stained virions, distinctive surface structure has not been noted for these viruses.
(UUKV) phleboviruses had mean diameters of 100 nm and 125 nm, respectively,162,386 and Tula and Hantaan hantaviruses had mean diameters of 130 to 135 nm.35,216 The spikes are thought to mostly consist of heterodimers of the two viral GPs, and biochemical analysis of purified LACV particles indicated that the two GP species were equimolar and present at about 650 copies per virion. The GPs interact to form surface morphologic units that vary among viruses in different genera.333 Virions in the Orthobunyavirus genus have surfaces covered with closely packed, knoblike morphologic units with no detectable order. Similarly, no obvious order was found for the small surface structures with central cavities observed on viruses in the Nairovirus genus.333 The appearances of viruses in the Tospovirus genus have been likened to that of the nairoviruses.344 Other than the presence of GP projections, which are observed as a fringe on negatively stained virions, distinctive surface structure has not been noted for these viruses.
Viruses in the Phlebovirus genus have round, closely packed morphologic units, approximately 10 to 11 nm in diameter, with central cavities approximately 5 nm in diameter.333 Negative staining of glutaraldehyde-fixed particles, freeze-etching techniques, and/or cryoelectron tomography demonstrated that the glycoprotein spikes of UUKV and RVFV are organized on a T-12 icosahedral lattice, an arrangement so far unique to phleboviruses.162,386,557 For UUKV, two pH-dependent conformations of the glycoproteins were observed: tall spikes at pH7 and flat spikes at pH6. It is suggested that at low pH the conformational change exposes hydrophobic regions on the GP, possibly on Gc, that facilitate fusion between the viral and endosomal membrane during viral entry,386 a mechanism observed for other enveloped viruses that enter cells via the endocytic pathway.
The surface structure of viruses in the Hantavirus genus also are distinctly ordered but have a square grid-like appearance (Fig. 42.3).571 Electron cryotomography35,216 confirms that the Gn-Gc spike complex has fourfold symmetry. Each spike is likely composed of four Gn-Gc heterodimers that form lattices on the virion surface. Specific lateral interactions between spike complexes are thought to induce membrane curvature during the budding process.
The interior of virions, as observed by thin-section electron microscopy, has a filamentous or coiled bead appearance, presumably due to the presence of the ribonucleocapsids (Fig. 42.3).122 From single particle reconstructions of tomograms, internally virions contained parallel thread or rod-like structures assumed to be ribonucleoprotein complex (RNP); some were located very close to the membrane suggesting interaction with the cytoplasmic tails of one or both of the glycoproteins.35,216,386,440
Biochemical and Biophysical Properties
The composition and structure of virions has been inferred from biochemical and morphologic studies. An overall chemical content of 2% RNA, 58% protein, 33% lipid, and 7% carbohydrate was estimated for UUKV.380 Sedimentation coefficients of virions range from 400 to 500 S, and their buoyant densities in sucrose are 1.16 to 1.18 g/cm3, and in CsCl, 1.20 to 1.21 g/cm3. Treatment with lipid solvents or nonionic detergents removes the viral envelope and results in loss of infectivity for arthropods and mammals.380 For plants, the envelope is not required for infectivity, as demonstrated in studies with TSWV, for which repeated mechanical passage among plants resulted in a defective virus that was unable to produce enveloped particles but was able to replicate in plant cells.116
Genome Structure and Organization
Viral Genome
Virions contain three single-stranded RNA genome segments designated as large (L), medium (M), and small (S). All three RNA segments of a virus have the same complementary nucleotides at their 3′ and 5′ termini. The terminal nucleotide sequences are highly conserved among viruses within a genus, but differ from those of viruses in other genera (Table 42.3). Base-pairing of the terminal nucleotides is predicted to form stable panhandle structures and noncovalently closed circular RNAs. Direct support for base-pairing comes from electron microscopy of RNA extracted from virions, in which three sizes of circular RNAs were evident.204
The RNA segments are complexed with N to form individual L, M, and S nucleocapsids, which were generally assumed to have helical symmetry,379 but analysis of isolated RVFV nucleocapsids did not show this pattern but rather a string-like appearance.440 Nucleocapsids released by nonionic detergent treatment of virions often also appear as circular structures in electron micrographs,411 suggesting that the complementary RNAs can base-pair even when complexed with protein in an estimated ratio of 4% RNA to 96% protein.431 This hypothesis is supported by data showing cross-linking of the ends of nucleocapsid-enclosed RNAs by treatment with psoralens, which are photoreactive, nucleic acid, cross-linking agents.431
At least one each of the L, M, and S ribonucleocapsids must be contained in a virion for infectivity; however, equal numbers of nucleocapsids may not always be packaged in mature virions, as suggested by various reports of equimolar or nonequimolar ratios of L, M, and S RNAs.49,218 Unequal complements of the ribonucleocapsids may contribute to the size differences of virions observed by electron microscopy.35,522
In addition to ribonucleocapsids containing virion-sense RNA (vRNA), certain viruses in the Phlebovirus and Tospovirus genera encapsidate small amounts of complementary sense (cRNA or antigenome). The phlebovirus UUKV was found to have S but not M segment cRNA, and the tospovirus TSWV had both M and S cRNAs in virions.274,491 For the orthobunyavirus, LACV, S segment cRNA was detected in virions synthesized in insect cells, but not in mammalian cells.431 Interestingly, the phlebovirus RVFV encapsidated all three cRNA gene segments, and, as will be described later (see “S segment products”), at least one of the cRNA genes may have a role in early replication processes.225
Coding Strategies of Viral Genes
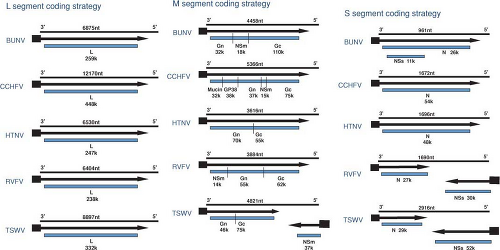
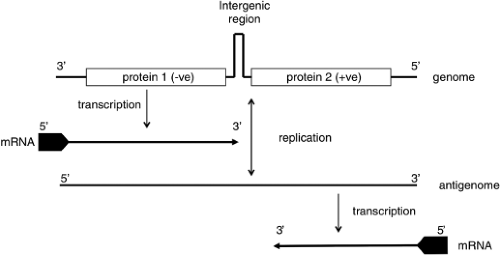
Both similarities and noteworthy differences in the coding strategies of viruses in the family Bunyaviridae are known. Viruses in each genus encode all structural proteins in their cRNA. Certain viruses also encode nonstructural proteins in their cRNA or in their vRNA. Therefore, some viruses in the family Bunyaviridae use only a negative-sense coding strategy and others use a combination of negative-sense and ambisense coding strategies (Fig. 42.2).
S Segment Strategies
Viruses in the Orthobunyavirus genus have smaller S segments than those of viruses in other genera (Table 42.1). Some orthobunyaviruses encode two polypeptides, the nucleocapsid protein (N) and a nonstructural protein (NSs), in ORFs in cRNA48 (Fig. 42.2), whereas others only encode N protein.351 The presence of NSs in virus-infected cells was demonstrated for several members of the genus.140 Only one S segment mRNA species can be found in orthobunyavirus-infected cells84; therefore, N and NSs are generated by alternative start codon recognition by ribosomes.126,144
Hantaviruses and nairoviruses encode larger N proteins than viruses in other genera, (Table 42.2, Fig. 42.2).332,470 Some hantaviruses (e.g., SNV, PUUV, Tula [TULV] and Prospect Hill [PHV] viruses), have ORFs within the N ORF, and an NSs protein has been detected in PUUV- and TULV-infected cells.227 No NSs protein has been detected in nairovirus infected cells. Only one S-segment mRNA, similar in size to the coding region for N, was identified in hantavirus- or nairovirus-infected cells, indicating that transcription termination occurs shortly after the translation stop codon.469,563 Certain hantaviruses (e.g., SNV) have long 3′ noncoding regions (of greater than 700 nucleotides) containing numerous repeated sequences. These repeats may result from polymerase slippage on the vRNA template.505
The ambisense coding strategy of the S segments of phleboviruses and tospoviruses produces N from a subgenomic mRNA that is complementary to vRNA, and NSs from a subgenomic mRNA of the same polarity as vRNA (Fig. 42.2).118,220 Evidence that the mRNA for NSs is copied from cRNA (after genome replication) comes from time-course studies. For the phlebovirus UUKV, N was detected at 4 to 6 hours after infection, whereas NSs did not appear until 8 hours after infection.492,542 Likewise, the mRNA for NSs of the tospovirus TSWV was detected in infected plant cells 15 hours later than for N.507 In addition, studies with the phlebovirus Punta Toro virus (PTV) demonstrated that protein synthesis inhibitors arrest production of NSs mRNA, but not N mRNA.222 These results suggest that protein synthesis must occur before the NSs mRNA can be made. In contrast, studies with the phlebovirus RVFV demonstrated the presence of cRNAs to all three RNA segments in purified virions. Moreover, mRNA for the NSs protein was detected as early as 20 minutes after infection, concomitant with the appearance of mRNA for N, suggesting that the ambisense-encoded NSs mRNA are transcribed from the incoming antiviral sense RNA.225
M Segment Strategies
Sizes of M segments range from ∼3,600 nucleotides to ∼4,900 nucleotides (Table 42.1). All bunyavirus M segments encode the two envelope GPs in a single ORF of cRNA (Fig. 42.2). Previous designations of G1 and G2, which were based on relative migration of the proteins in polyacrylamide gels, have been replaced by designations of Gn and Gc, referring to the amino-terminal or carboxy-terminal coding of the proteins.294 As described below, it is becoming increasingly clearer that the functions of the Gn and Gc proteins are conserved among the five genera.
Some viruses encode NSm proteins, and others do not. Except for the tospoviruses, which use an ambisense strategy to generate NSm from a subgenomic mRNA, a single mRNA, nearly equivalent in size to the cRNA ORF has been detected in virus-infected cells. For tospoviruses, separate, subgenomic messages for the Gn-Gc precursor and for NSm were identified.273,295 NSm is readily detected in infected plants and is the only M segment nonstructural protein in the Bunyaviridae family to have a clearly defined role (i.e., it is a movement protein [see “M Segment Products,” below]). Another possible role for NSm for some viruses may be as a virulence factor. For example, a genetically engineered RVFV lacking NSm induced more extensive apoptosis than did one with NSm and the expression of NSm significantly inhibited the cleavage of caspase 8 and 9 induced by staurosporine, indicating that the NSm protein suppresses apoptosis.582
L Segment Strategies
The L segments of hantaviruses, orthobunyaviruses, and phleboviruses are of similar size, (∼6,500 nucleotides), whereas those of tospoviruses and nairoviruses are considerably larger (∼9,000 and 12,000 nucleotides, respectively; Table 42.1). All L segments of viruses in the family use conventional negative-sense coding strategies (Fig. 42.2). For each L segment described thus far, there are fewer than 200 nucleotides of total noncoding information, and there is no evidence for additional coding regions in either the cRNA or vRNA.117,141,143,209,360,465
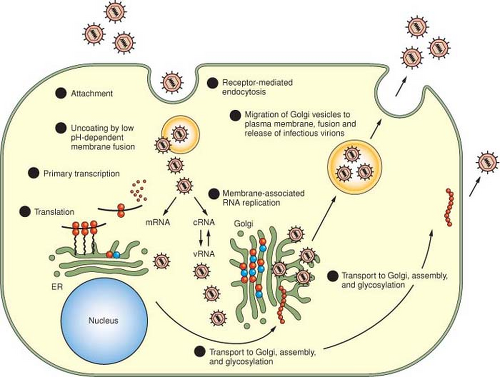
Stages of Replication
The principal stages of the replication process for viruses in the Bunyaviridae are illustrated in Figure 42.4 and are summarized in the following:
Attachment, mediated by an interaction of viral proteins and host receptors
Entry, by receptor-mediated endocytosis
Uncoating, by acidification of endocytic vesicles, and fusion of viral membranes with endosomal membranes
Primary transcription of viral-complementary mRNA species from genome templates using host-cell–derived primers and the virion-associated polymerase
Translation of L, M, and S mRNAs
co-translational cleavage of M-segment polyprotein and postranslational cleavage of precursors for some viruses
dimerization of Gn and Gc in the endoplasmic reticulum (ER)
Membrane-associated RNA replication
synthesis and encapsidation of cRNA to serve as templates for vRNA or, for ambisense genes, templates for subgenomic mRNA
genome replication
Morphogenesis
localization of N in budding compartments
transport of dimerized Gn and Gc to the Golgi
glycosylation
acquisition of modified host membranes, generally by budding into the Golgi cisternae
Fusion of cytoplasmic vesicles containing viruses with the plasma membrane and release of mature virions
more rarely, some viruses in some cell types have been observed to bud directly from the host cell’s plasma membrane
Attachment and Entry
Viral Attachment Proteins and Cellular Receptors
The mechanisms by which members of the family Bunyaviridae gain access to the host cell’s cytoplasm appear similar to those reported for many other enveloped viruses. The first step involves an interaction between cell surface receptors and viral attachment proteins, Gn and/or Gc. The presence of neutralizing and hemagglutination-inhibiting sites on both the Gn and Gc proteins of phleboviruses and hantaviruses27,252 suggests that both proteins may be involved in attachment. Although it is possible that both are directly involved, it is more likely that both are required due to conformational requirements that depend on dimerization of Gn and Gc.
In general, Gc appears to be the primary attachment protein for orthobunyaviruses in mammalian cells, mosquito cells, and mosquitoes.193,261,515 Consistent with this view, expression of M-segment products of three orthobunyaviruses demonstrated that Gc, but not Gn, could effect attachment and entry when tested in a cell-to-cell fusion assay and a pseudotype transduction assay.419 However, the amino terminal half of BUNV Gc ectodomain is not required for infection of cultured mammalian cells,483 and this region can be replaced by green fluorescent protein (GFP) allowing the creation of viable viruses expressing chimeric Gc-GFP in their virions.486
For tospoviruses, the envelope GPs are required only for infection of their arthropod vectors, as evidenced by the ability of envelope-deficient mutants to replicate in plants after
mechanical transmission, but their inability to infect thrips.445 In addition, analysis of reassortant viruses demonstrated that mutations that disrupted the Gn-Gc ORF of TSWV ablated transmissibility by thrips.495 These studies provide indirect evidence that the L-polymerase protein remains associated with ribonucleocapsids and is active despite the absence of intact virions.
mechanical transmission, but their inability to infect thrips.445 In addition, analysis of reassortant viruses demonstrated that mutations that disrupted the Gn-Gc ORF of TSWV ablated transmissibility by thrips.495 These studies provide indirect evidence that the L-polymerase protein remains associated with ribonucleocapsids and is active despite the absence of intact virions.
Although there are no specific reports of Gc being the attachment protein of tospoviruses, functional homology to the Gc proteins of other bunyaviruses was proposed based on amino acid sequence homologies detected for iris yellow spot virus.106 In contrast, however, a study with a soluble, truncated form of TSWV Gn expressed in baculoviruses showed binding of this protein to epithelial cells in the midguts of thrips, suggesting that Gn could mediate attachment in thrips.573 Similar results were reported earlier with the orthobunyavirus LACV, in that virions treated with a protease that cleaved Gc but not Gn exhibited increased binding to the insect vector midgut, but reduced binding to cultured mammalian or mosquito cells.324 Given that both of these systems use a Gn protein in the absence of Gc, it remains to be determined whether Gn functions for attachment when in its native conformation on virions.
Host cell receptors have not been identified for most viruses in the family; however, pathogenic and nonpathogenic hantaviruses were shown to use β3 and β1 integrins, respectively, to enter endothelial cells.170 This finding was suggested to relate to pathogenesis, in that binding of hantaviruses to integrins might disrupt their ability to regulate cell-to-cell adhesion and result in the vascular permeability characteristic of hantaviral diseases.171 In addition, decay accelerating factor (DAF)/CD55 a glycosylphosphatidylinositol (GPI)-anchored protein, has been shown to mediate HTNV and PUUV entry across the apical membrane of polarized epithelial cells,277 and gC1qR/p32, a 32-kD glycoprotein that interacts with complement protein C1q, binds HTNV and mediates infection of cultured A549 lung cells.98
Entry of the Viral Genome into the Host Cytoplasm
Shortly after attachment, viruses in the Phlebovirus and Nairovirus genera were observed in phagocytic vacuoles.145,455 This suggested a mode of viral entry similar to that first described for alphaviruses in which the virus is endocytosed in coated vesicles, and inhibitor studies have confirmed this to be so for the animal-infecting bunyaviruses. Orthobunyaviruses, hantaviruses, and nairoviruses enter by endocytosis into clathrin-coated pits,239,463,490 and HTNV proteins were shown to colocalize with clathrin in confocal immunofluorescence studies.239 In addition, CCHFV requires functional microtubules for infection.489 By contrast, entry of UUKV phlebovirus is predominately in a clathrin-independent manner, and viruses enter Rab5a+ early endosomes and subsequently Rab7+ and LAMP-1+ late endosomes.323 After endocytosis, acidification of the endocytic vesicles is thought to promote a conformational change in Gn and/or Gc that facilitates fusion of the viral and cellular membranes, thereby allowing the viral genome and polymerase access to the cytoplasm. Infection is blocked if cells were treated with ammonium chloride, which prevents acidification.229,419,463,484 Additional indirect support for membrane fusion as a mode of entry came from experiments demonstrating the ability of viruses in the family to mediate syncytia formation at low pH.153,183,229,383,419,484,574
Computational studies revealed that the Gc proteins have characteristics similar to those of class II fusion proteins identified for viruses in the Flaviviridae and Togaviridae families.169 Such proteins have internal fusion domains, as opposed to terminal domains found for the class I fusion proteins. Experimental support for this finding was obtained by demonstrating interaction of the fusion peptide postulated for the Gc protein of the hantavirus, Andes virus (ANDV), with artificial membranes.537 In addition, several lines of evidence indicate that the Gc of orthobunyaviruses is involved in membrane fusion. Protease sensitivity assays, detergent partitioning experiments, and antibody-binding studies suggest that Gc undergoes a conformational change at pH conditions where fusion is observed.182 Mutational analyses of a hydrophobic region (residues 1,066–1,087) of LACV Gc that is well conserved among different orthobunyaviruses106 supported this notion,229,420 and the residues flanking the predicted fusion peptide in BUNV Gc (residues 1,058–1,079) were shown to be structurally critical for the conformational change in Gc that occurs during the fusion process.483 LACV Gc alone, however, when expressed from recombinant vaccinia viruses, could not cause cell fusion, suggesting that an association of the two GPs may be needed for membrane fusion,229 and mutations in the cytoplasmic tail of BUNV Gn severely affected membrane fusion indicating that Gn must also play an important role in the fusion process.484 Likewise for HTNV, both Gn and Gc were required to achieve surface expression and cell-to-cell fusion activity.383 Recombinant LACV carrying mutations in the Gc fusion peptide was impaired for growth in both mammalian and insect cells but were still neurotoxic to neuronal cells, implying that the fusion peptide is a determinate of neuroinvasiveness but not necessarily neurovirulence.501
Transcription and Replication
After uncoating of viral genomes, primary transcription of negative-sense vRNA to mRNA is initiated by interaction of the virion-associated L protein, which is an RNA-dependent RNA polymerase (RdRp), and the three viral ribonucleocapsids.55 Studies with hantavirus N protein suggest that N participates in transcription initiation by facilitating dissociation of the RNA panhandle and by subsequently remaining attached to the 5′ terminus of the RNA, thereby freeing the 3′ terminus for RdRp interactions. Furthermore, N was suggested to be important for replication by acting as an RNA chaperone and transiently and continuously unfolding the RNA to allow it to form more stable structures.347 A role for N in both transcription and replication is also evident by analysis of the behavior BUNV N proteins carrying specific mutations, either in a minigenome assay or in the context of virus infection.135,560 Four BUNV temperature sensitive mutants, carrying single amino acid substitutions in N, could be divided into two groups, those that were defective in antigenome synthesis but not mRNA transcription, and those that were replication defective but transcription competent, suggesting that different domains within N are associated with different RNA synthesis activities.135 The mechanism by which residues in N modulate template activity requires elucidation; one possibility is that transcription and replication require different cellular cofactors that bind to distinct regions on the N protein.
Because only L and N are needed for RNA synthesis,127 the location of these proteins must correlate with the site of RNA
synthesis. For nairoviruses and hantaviruses, N (or L and N) proteins were found to localize to the perinuclear region and to have a peripheral association with perinuclear membranes.283 In cells infected with a recombinant BUNV expressing an epitope-tagged L protein immunofluorescence microscopy showed L to have a punctate to reticular staining pattern, with concentration of staining in the perinuclear region, whereas cell fractionation studies showed L to be distributed in both cytosolic and microsomal fractions.481 Together, these data suggest that bunyavirus RNA replication is membrane-associated, similar to that described for flaviviruses.
synthesis. For nairoviruses and hantaviruses, N (or L and N) proteins were found to localize to the perinuclear region and to have a peripheral association with perinuclear membranes.283 In cells infected with a recombinant BUNV expressing an epitope-tagged L protein immunofluorescence microscopy showed L to have a punctate to reticular staining pattern, with concentration of staining in the perinuclear region, whereas cell fractionation studies showed L to be distributed in both cytosolic and microsomal fractions.481 Together, these data suggest that bunyavirus RNA replication is membrane-associated, similar to that described for flaviviruses.
The L Protein
To mediate replication, the RdRp must affect numerous enzymatic functions. For primed synthesis of mRNA from genomic templates and nonprimed synthesis of vRNA from cRNA templates (as will be described), the RdRp performs endonuclease, transcriptase, replicase, and probably RNA helicase activities. Motifs common to polymerases in general are conserved in the bunyaviral RdRps, most notably a catalytic core motif.26,237,282,360 The N-terminal domain of orthobunyavirus L proteins contains a conserved PD-(D/E)xK amino acid nuclease motif and the isolated domain of LACV L exhibited nuclease activity in vitro; furthermore, bioinformatic analyses indicated that viruses in all the other Bunyaviridae genera contain a similar domain.442 The nuclease domain of LACV bears structural similarities to that of the PA subunit of the influenza A virus polymerase, suggesting a common origin of the cap-snatching process of segmented negative sense viruses. None of the other functions has been definitively localized to a region of the RdRp gene or gene product; however, comparison of two nairovirus L proteins with functionally defined regions of other polymerases revealed potential helicase, topoisomerase, and gyrase coding regions as well as an N-terminal cysteine-protease motif typical of the ovarian tumor (OTU) protein superfamily and protease.262 Biochemical and structural analyses demonstrate that in addition to the deubiquitinase activity as shown by cellular OTU proteases, the nairovirus OTU also targets ISG15 modification and thus enhances its role in overcoming host innate immune defenses.5,77,165,230 The OTU domain is not required for the RNA polymerase activity by CCHFV in a minigenome system.43
Genome Promoters
The template for bunyavirus transcription and replication is not naked RNA but RNA in the form of ribonucleocapsid.127,319 The 3′ and 5′ nontranslated complementary nucleotides contain signals for both mRNA synthesis and antigenome synthesis, and thus are the genome promoters. For the phleboviruses RVFV and UUKV, the first 13 or the first 10 nucleotides, respectively, of the 3′ end of the genome are sufficient for RNA synthesis activity.156,424
For the orthobunyavirus BUNV, a minigenome reporter system was used to demonstrate that optimal transcription required exact complementarity at the 3′ and 5′ termini; however, one to three nucleotide deletions could be tolerated at the 3′ terminus, but not the 5′ terminus.268 Although sequence changes at several positions could be tolerated as long as complementarity was maintained, some changes did reduce transcription. Interestingly, mutagenesis experiments indicated that the single exception to complementarity (a U residue at 3′ position 9) was critical for transcription from the genomic promoter, whereas the corresponding 5′ position 9 could be changed without influencing transcription.33 Therefore, the mismatch itself is not important, but the nucleotide is. This nucleotide, however, does not have to be maintained for transcription activity of the antigenomic promoter.33 Together, these data suggest that the structure of the panhandles is more important than the sequence, but that the sequence also plays a role in promoter strength and activity.
To further investigate promoter regions in an authentic replication system, S segments with shortened complementary termini were introduced into a BUNV reverse genetics system, and rescued viruses were screened to determine the minimal terminal complementary region needed for virus production.322 The minimal 3′ and 5′ deletion mutant viable in this system maintained 29 of the 85 untranslated nucleotides on the 3′ end and 112 of the 174 untranslated nucleotides on the 5′ end, whereas those with shorter untranslated regions were not viable. Therefore, although only the complementary regions are needed for promoter activity in vitro, actual virus production also requires some of the unique sequences in the untranslated regions at the 3′ and 5′ termini of BUNV.322
The in vivo importance of intact termini was also suggested by a study in which terminally deleted RNAs were shown to accumulate during the establishment of persistent infections with the hantavirus, Seoul virus (SEOV). It was hypothesized that as deletions accrued, fewer replication-competent genomes were present, leading to a downregulation of the replication processes, and possibly to persistence.342 Similar deletions were found on the termini of ANDV M and L, but not S genes, leading to a speculation that differences observed in the relative abundance of Gn and Gc compared to N could reflect down regulation of M-segment expression from genes without intact termini.389
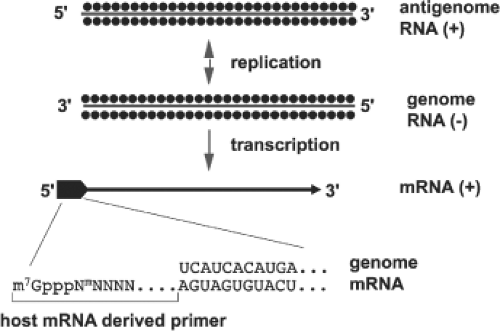
Primed mRNA Synthesis (Cap Snatching)
Like influenza viruses, viruses in the family Bunyaviridae prime mRNA synthesis with capped oligonucleotides that are scavenged from host mRNAs (Fig. 42.5). Cleavage of the capped primers is accomplished by endonuclease activity found in virions and associated with the L protein.398,442 Unlike influenza viruses, which take primers from newly synthesized mRNAs in the host cell’s nucleus, members of the Bunyaviridae family use primers cleaved from cytoplasmic host cell mRNAs. Evidence has been presented that the hantavirus N protein binds to the 5′ cap of cellular mRNAs to protect them from cell-mediated degradation, and that N accumulates in cytoplasmic processing bodies (P bodies) where protected 5′ caps are sequestered, and hence can serve as a pool of primers to initiate mRNA synthesis.346 A result of this mode of mRNA transcription is the presence of 5′ terminal extensions of approximately 10 to 20 heterogeneous nucleotides that are not found in vRNA.47 Studies using anticap antibodies to immunoselect mRNAs192 have provided direct proof for the presence of caps on the scavenged primers. Further evidence for capped extensions of mRNAs was provided by a study of tospoviruses, in which plants were co-infected with TSWV and with a positive-strand RNA virus (alfalfa mosaic virus). The tospovirus acquired 5′ mRNA extensions with nucleotide sequences that matched those of the other virus with a preference for cleaving at an A residue.125 It was suggested that this
preference is due to a need for base-pairing at the ultimate U residue of the gene segments. Consistent with this, later studies indicated that double and triple base complementarity to nucleotides at the ends of the tospovirus gene segments were preferred, even more than the single complementary residue.545 Interestingly, it was also found that newly synthesized mRNAs can serve as cap donors in vitro for tospoviruses, suggesting a “resnatching” mechanism.544
preference is due to a need for base-pairing at the ultimate U residue of the gene segments. Consistent with this, later studies indicated that double and triple base complementarity to nucleotides at the ends of the tospovirus gene segments were preferred, even more than the single complementary residue.545 Interestingly, it was also found that newly synthesized mRNAs can serve as cap donors in vitro for tospoviruses, suggesting a “resnatching” mechanism.544
Other viruses in the family also have nucleotide or nucleotide motif preferences for endonuclease cleavage of capped primers. These preferences vary among the genera, and sometimes among viruses within a genus. A preferred primer sequence, or a favored nucleotide at the site of cleavage due to a need for limited base pairing with the viral genome, appears to be a common feature of primed transcription for bunyaviruses. In one study, the 3′-terminal nucleotides of the scavenged host primers often were similar to the 5′-terminal viral nucleotides.238 It was proposed that after transcription of two or three nucleotides of the nascent mRNA, the viral polymerase might slip backward on the template before further elongation, resulting in a partial reiteration of the 5′-terminal sequence. An extension of this concept, termed “prime and realign,” was proposed for mRNA transcription of hantaviruses.168 According to this model (e-Fig. 42.2), priming by host oligonucleotides with a terminal G residue would initiate transcription by aligning at the third nucleotide of the viral RNA template (C residue). After synthesis of a few oligonucleotides, the nascent RNA could realign by slipping backward two nucleotides on the repeated terminal sequences (AUCAUCAUC) (Table 42.3), such that the G becomes the first nucleotide of the nontemplated 5′ extensions (e-Fig. 42.2). The frequent deletion of one or two of the triplet repeats in hantaviral mRNA supports this sort of slippage mechanism and suggests that sometimes the initial priming might start at the C residue of the third triplet in the conserved sequence rather than at the C of the second triplet.168
Bunyavirus transcription is unique among negative-sense RNA viruses in that functional viral mRNA synthesis requires ongoing protein synthesis,31,400,430,432,433,553 a finding that at face value appears incompatible with the presence of a virion transcriptase. In the absence of protein synthesis, only short transcripts are produced in vivo and in vitro. If the in vitro reaction is supplemented with rabbit reticulocyte lysate, however, full-length RNAs are synthesized. The translational requirement is not at the level of mRNA initiation, but rather during elongation or, more precisely, to prevent the transcriptase from terminating prematurely. A model to account for these observations proposed that in the absence of ribosome binding and protein translation, the nascent mRNA chain and its template can base-pair, thereby preventing progression of the transcriptase enzyme.37 Recently this model (e-Fig. 42.3) has been tested using BUNV model templates containing translational stop codons,31 and the results showed that translation of nascent mRNAs prevented transcription termination. Thus orthobunyaviruses couple transcription and translation, a feature commonly found in prokaryotes, but rare in eukaryotic cells.
Transcription Termination
For gene segments with simple negative-sense coding, synthesis of M- and S-segment mRNAs terminates about 40 to 100 nucleotides before the end of the genome RNA template.109,148,235,399 Although most other negative-strand RNA viruses use a stretch of nucleotides rich in U residues to signal transcription termination and polyadenylation, no such tract has been consistently identified for members of the family Bunyaviridae. This is supported by experimental evidence showing that bunyavirus mRNAs are not 3′ polyadenylated,1,400,542 but many have the potential to form stem-loop structures that are probably involved in enhancing translation of viral mRNAs.53,551
By using a reporter system that involved mutated S segments, the transcription termination signal for the orthobunyavirus BUNV was mapped to a 33-nucleotide region within the 5′ nontranslated region of the S segment.32 Within this region, a 6-nucleotide motif, 3′-GUCGAC-5′ was critical to transcription termination. In addition, changing the nucleotides in this transcription termination signal revealed a second downstream transcription termination signal, which had a 5-nucleotide motif, 3′-UGUCG′-5′, that was also found within the 33-nucleotide region of the upstream site, and that partially overlapped the critical 6-nucleotide motif. The finding that there are no U-rich regions in the transcription termination signal of BUNV is consistent with the absence of poly-A tails on the mRNAs.
Comparison of these S-segment sequences with those of other orthobunyaviruses revealed a high degree of conservation, suggesting that similar motifs also function for transcription termination throughout the genus, at least for S segments. A motif similar to that on the BUNV S segment was observed within the BUNV L, but not the M segment.32 Experimental mapping of BUNV L and M mRNA termination sites has not been reported.
Analysis of the L mRNAs of SNV hantavirus218 and RVFV and Toscana (TOSV) phleboviruses7 showed they were co-terminal with the L vRNA template, suggesting L mRNA synthesis terminates by run-off of the RNA template.
A U-rich motif was proposed as the site of termination for the M mRNA and a C-rich motif (CCCACCC) as termination site for the S mRNA of SNV hantavirus.218 Fine mapping
of M-segment mRNA termination sites for three phleboviruses (RVFV, TOSV, and SFSV) showed that termination occurred immediately following a C-rich domain in the template at a conserved motif 3′-C1-3 GUCG/A-5′.7 Although previously it was thought the C-rich region itself was involved in transcript termination178 deletion of this region in template RNA did not affect specific termination at the identified motif.7
of M-segment mRNA termination sites for three phleboviruses (RVFV, TOSV, and SFSV) showed that termination occurred immediately following a C-rich domain in the template at a conserved motif 3′-C1-3 GUCG/A-5′.7 Although previously it was thought the C-rich region itself was involved in transcript termination178 deletion of this region in template RNA did not affect specific termination at the identified motif.7
The mechanism of transcription termination of the ambisense genes of phleboviruses and tospoviruses was initially thought to involve RNA secondary structure in the intergenic region.103,118,146,187,188,493 However, reanalysis of computer predictions of hairpin formation in these intergenic regions suggest that they are unlikely to form because of their complexity or low energy.7 Detailed mapping of mRNA termination sites for the N and NSs mRNA of RVFV, TOSV, and SFSV phleboviruses showed that the 3′ ends of the mRNAs contained most of the intergenic sequence and indeed overlapped each other.7 Furthermore, termination occurred for both messages at the same motif, 3′-C1-3GUCG/A-5′, as in the M segment. Two copies of this motif are thus present, one in vRNA (genome) and one in the cRNA (antigenome). The conservation of this motif in viral genomes that display considerable sequence diversity suggests a similar mechanism for transcription–termination in the M and S segments. Deletion of either copy of the motif in RVFV S segment did not prevent correct termination of unaffected N or NSs mRNA, but the mutant viruses were attenuated in growth compared to wild-type.7
Genome Replication
The change from primary transcription to replication requires that the RdRp, either acting alone or in concert with undefined viral or cellular factors, must at some point, switch from primed mRNA synthesis to independently initiating transcription at the precise 3′ end of the template to produce a full-length transcript. The processes involved in making that switch from primary transcription to genome replication have not been defined completely for any member of the family. Presumably, some viral or host factor is required to signal a suppression of the transcription termination signal responsible for generation of truncated mRNA and also to prevent the addition of the capped and methylated structures to the 5′ termini of the cRNAs. There is no question that genome replication and subsequent secondary transcription are prevented by translational inhibitors such as cycloheximide. These results indicate that continuous protein synthesis is required for replication of the genome. Although not proven, it is likely that synthesis of N is required for genome replication, as described for other negative-strand RNA viruses such as the rhabdovirus, vesicular stomatitis virus, and the paramyxovirus Sendai virus. For these viruses, encapsidation by N seems to serve as an antitermination signal, thus allowing full-length genome synthesis.
A prime-and-realign model was also postulated for the nonprimed transcription of hantaviral vRNA and cRNA, except that transcription would initiate with pppG alignment at the third nucleotide (C residue) of the template RNA. After synthesis of several nucleotides, polymerase slipping would realign the nascent RNA such that the initial priming G residue would overhang the template. It was further theorized that nucleolytic activity of the L protein might remove the overhanging G, leaving a monophosphorylated U residue at the nascent 5′ end. The presence of the monophosphorylated U on HTNV RNA was experimentally demonstrated168 (e-Fig. 42.2).
Indirect evidence suggesting that a prime-and-realign method of initiation is used by phleboviruses was obtained by using a reconstituted transcription system to study the polymerase recognition sequence at the 3′ termini of the ambisense S segment RNA of RVFV. In those studies, mutational analysis of the terminal nucleotides revealed that the first 13 nucleotides are required for polymerase recognition, but that one of the two terminal dinucleotides (UGUG) could be removed without deleterious effects on transcription.424 These data also suggested that realignment is not a prerequisite for transcription initiation. In contrast, a recent study using the BUNV minigenome system showed that the viral polymerase was able to repair both insertions and deletions in model template RNAs, although this did not appear to involve the prime-and-realign mechanism.559
Schematics of transcription and replication based on information above are presented in Figures 42.5 and 42.6.
Encapsidation Signals
Signals for the encapsidation of the vRNAs and cRNAs of orthobunyaviruses, phleboviruses, nairoviruses, and hantaviruses were found entirely within the noncoding 3′ and 5′ regions of minireplicon reporters.52,154,155,157,321,345,348,382 Mutational
analysis of these terminal regions for BUNV revealed a conserved sequence within nucleotides 20–33 of the 5′ end of vRNA that was critical for packaging.270 Variable packaging efficiencies were noted for the L, M, and S segment–derived minireplicons, and packaging appeared to be independent for each.
analysis of these terminal regions for BUNV revealed a conserved sequence within nucleotides 20–33 of the 5′ end of vRNA that was critical for packaging.270 Variable packaging efficiencies were noted for the L, M, and S segment–derived minireplicons, and packaging appeared to be independent for each.
Host Factors
Beyond the need for host-derived primers for initiating mRNA synthesis, little is known about the role that host factors might play in viral replication. One putative host transcription factor is an ∼40-kD protein isolated from the main insect vector of TSWV, and it was shown to bind to the viral RdRp at its C-terminus and to the viral RNA at its N-terminus.119 Both this protein and reticulocyte lysates were needed for RNA synthesis in vitro, indicating that other cellular factors are also required. Expression of this protein in mammalian cells rendered them permissive to TSWV replication.119 As is the case for a number of other negative-strand RNA viruses, heat-shock protein 90 (Hsp90) appeared essential for La Crosse virus replication in that virus yields were diminished in cells treated with Hsp90 inhibitors like geldanamycin due to destabilization of the viral L protein.105
Translation and Processing of Viral Proteins
Viral polypeptides are synthesized shortly after infection, with S and L mRNAs translated on free ribosomes and M mRNAs translated on membrane-bound ribosomes. Expression products vary among the genera, and even within a genus.
S Segment Products: N and NSs
The N protein is the most abundant viral product in virions and infected cells. N plays several important roles in viral replication. In addition to protecting the RNA from degradation, N also interacts with L and Gn and Gc, although the exact interactions have not been defined. A critical step in N interaction with RNA appears to be the ability of the proteins to oligomerize. Oligomerization characteristics of the N proteins are likely different among the genera, which is probably not surprising, given the size differences of the N proteins, which range from approximately 19kD for orthobunyaviruses to 54kD for hantaviruses and nairoviruses (Table 42.2).
Chemical cross-linking studies indicated that the ribonucleocapsids of the phlebovirus RVFV, either from authentic virions or reconstituted from bacterially expressed protein, contain dimers of N as the basic oligomer.297,440 The crystal structure of RVFV N has been determined to 1.93Å resolution, which revealed that it has a novel all-helical fold without a positively charged surface RNA binding cleft as seen for other negative-sense RNA virus nucleocapsid proteins.440 Extensive ribonuclease digestion of authentic or reconstituted RNAs released heterogenous N-RNA multimers, consistent with the lack of observed helical symmetry. In contrast, hantaviral N proteins form stable trimers, which are postulated to assemble on the viral RNA, followed by further protein–protein interactions that encapsidate the entire RNA.249 The N–N interactions are likely to be electrostatic, with both amino- and carboxy-terminal residues participating.250 The N-terminal residues of hantaviral N proteins are predicted to form a coiled coil domain, providing a trigger for trimerization, and containing charged residues important for intermolecular interactions.9,10,13,562 For TULV, carboxy-terminal amino acids 393 to 398 were shown to be crucial for trimerization, and amino acids 1 to 43 provided a secondary interaction region.249 These data are in agreement with those obtained with other hantaviruses (e.g., SNV and HTNV, for which carboxy-terminal and amino-terminal regions were shown to be involved in the homotypic interactions of N).9,589
Orthobunyaviruses and tospoviruses, in contrast to hantaviruses, do not appear to assemble preformed trimers, but instead multimerize by adding one N protein at a time. This hypothesis comes from chemical cross-linking experiments, which showed that the N protein of the orthobunyavirus BUNV was able to form a range of multimers from dimers to high–molecular-weight structures, with a preponderance of tetramers.308 Deletion of N- or C-terminal portions of N prevented multimerization and also resulted in loss of functionality in a minireplicon system. The data were interpreted to indicate a head-to-head and tail-to-tail multimerization model of individual N proteins. Likewise, for the tospovirus TSWV, analysis of deletion mutants identified binding regions both at the amino- and carboxy-termini of N, and although the type of multimers formed was not examined closely, it appears to be a continuous addition process, like that proposed for the orthobunyaviruses.242,540
In addition to protein–protein interaction, the amino- and carboxy-terminal regions of the tospovirus N were also implicated in RNA binding.447 In contrast, the RNA-binding site of hantaviral N was localized to a central, conserved region of the protein.474 For hantaviruses and bunyaviruses, RNA-binding studies with bacterially expressed, hexahistidine-tagged N proteins indicated that N preferentially interacts with the 5′ end of the vRNA.385,475 In another study, high-affinity binding of hantaviral N to panhandle RNA was demonstrated, and it was suggested that such binding is related specifically to the trimers, which were able to discriminate viral from nonviral RNA, whereas monomer or dimer subunits bound nonspecifically to RNA.348 These data suggest a requirement for multimerization of N for effective RNA binding and encapsidation, and implicate the panhandle structure as the trigger for encapsidation. Hantavirus N is also able to bind 5′ capped mRNAs (to generate a source of primers for transcription initiation; see above), and the cap and vRNA binding sites have been mapped to separate domains within the protein.349 Furthermore, binding to the mRNA cap induces a conformational change in N, and the structurally altered N loaded with the capped primer binds specifically to the conserved 3′ terminus of vRNA.
The S segments of viruses in the Phlebovirus and Tospovirus genera and some viruses in Orthobunyavirus and Hantavirus genera produce NSs proteins as well as N proteins. Sizes of NSs range from 10 kD for orthobunyaviruses and hantaviruses to more than 50 kD for tospoviruses (Fig. 42.2). The NSs protein of the phlebovirus RVFV is phosphorylated and accumulates in the nuclei of infected cells, where it forms fibrillar structures (e-Fig. 42.4).513 The major phosphorylation sites of the RVFV NSs were mapped to serine residues located near the carboxy-terminus of the protein.267 Expression studies revealed that fibrils could form in the absence of other viral proteins; however, if the carboxy-terminal region of NSs was removed, NSs were still transported to the nuclei, but did not coalesce into fibrils.584 Similar fibrillar structures of NSs were observed for some strains of TSWV, but the structures appeared only in the cytoplasm.
Comparing the deduced amino acid sequences of NSs for five different phleboviruses revealed homologies of only 17% to 30%.178 However, comparison of the NSs gene sequences for a number of strains of a single phlebovirus RVFV showed that certain areas were highly conserved.459 These data suggest that there may be a strong evolutionary pressure to maintain distinct portions of the NSs for individual viruses, but that the remainder of the protein can diverge without affecting the function of NSs.
Earlier work suggested that the NSs of phleboviruses and tospoviruses were only synthesized after viral replication; that is, after the ambisense vRNA has been copied to cRNA and mRNA was transcribed from cRNA. Recent work with RVFV, however, indicates that at least for this phlebovirus, virions can encapsidate cRNA and that the incoming cRNA ribonucleocapsids can serve as templates for NSs mRNA.225 The outcome of this is that NSs are present early in infection and might have a role in early replication. The NSs of three orthobunyaviruses all inhibited BUNV RNA synthesis in a dose-dependent manner in a minireplicon system.567 The results were suggested to indicate a function for NSs in regulating the activity of the RdRp. In addition to a possible role for NSs in early replication processes, NSs proteins have other important roles in viral infection, such as interferon antagonism, host-range determination, and gene silencing. These effects will be described in the context of effects on host cells.
M Segment Products: Gn, Gc, and NSm
The viral envelope GPs Gn and Gc are translated from a single mRNA complementary to vRNA. The polyprotein precursor of Gn and Gc is not seen in infected cells, and has been observed only by in vitro translation of RNA transcripts in the absence of microsomal membranes.542 For most viruses, both Gn and Gc are preceded by signal sequences; therefore, cleavage of the polyprotein precursor is likely mediated by host signalase.150 A pentapeptide motif, WAASA, which is highly conserved among hantaviruses, was shown to be the polyprotein cleavage site for HTNV, by mutational analysis of the signal sequence preceding Gc.318
M-segment gene products have a cysteine content of approximately 4% to 7%. Positions of these residues are highly conserved in the M-segment products of related viruses, suggesting that extensive disulfide-bridge formation may occur and that the positions may be crucial for determining correct polypeptide folding. The secondary structure of the proteins is also involved in immunogenicity, as indicated by the finding that neutralizing or protective epitopes are often nonlinear.
All M-segment polyproteins display variable numbers of predicted transmembrane regions, and a hydrophobic sequence at the carboxy-terminus, indicative of a membrane anchor region. Therefore, the M-segment translation products of viruses in the family Bunyaviridae are typical class 1 membrane proteins, with the amino terminus exposed on the surface of the virion and the carboxy-terminus anchored in the membrane.
The Gn and Gc proteins of all viruses in the family possess asparagine-linked oligosaccharides. Examination of the oligosaccharides attached to the Gn and Gc proteins of orthobunyaviruses revealed that Gc has mostly high-mannose glycans, whereas Gn contains both complex and a novel intermediate-type oligosaccharide.326,377,479 In contrast, the GPs of hantaviruses were found to be mostly of the high-mannose type.467 These findings indicate that the proteins are incompletely processed through the Golgi. This is likely related to retention of Gn and Gc in the Golgi, where assembly occurs, and which will be discussed more fully.
Nairoviruses appear unique among viruses in the family in that they have both N-linked and a region of heavily O-linked carbohydrates (a mucin-like domain, as will be described).461
The M-segment gene-product processing events differ among the genera. Hantaviruses produce only Gn and Gc,471 whereas some viruses in the Phlebovirus and Orthobunyavirus genera produce NSm from the same M-segment mRNA as Gn and Gc. The NSm protein of orthobunyaviruses is encoded between the Gn and Gc proteins.149 Its hydropathy profile suggests that it is a membrane-bound protein, and expression studies demonstrated that it localizes to the Golgi along with Gn and Gc.301 Although a function for this protein has not been identified, it was suggested that it might be involved in facilitating virion assembly in the Golgi.301
For the phlebovirus RVFV, a NSm of approximately 14 kD is cleaved from the amino-terminus of the polyprotein translation product (e-Fig. 42.5). Sequence studies revealed five in-frame translation initiation codons (AUG) upstream of the amino-terminus of Gn. Mutational analysis of those codons indicated the fourth and the fifth AUGs to translate Gn and Gc. Whilst NSm was found to originate from the second AUG and a 78-kD polypeptide, representing uncleaved NSm and Gn, was translated from the first AUG.198 Pulse-chase experiments revealed no precursor–product relationship between the 78- and 14-kD proteins; therefore, it appears that use of the first and second AUGs, respectively, is what dictates generation of these proteins.77 Both the 14-kD NSm and the 78-kD polypeptide are found in abundance in RVFV-infected cells,78 suggesting that they might play a role in replication or morphogenesis.
An even larger NSm (∼30 kD) is cleaved from the amino terminus of the M-segment polyprotein of the phlebovirus, PTV (e-Fig. 42.5). In vitro expression studies indicated that both envelope GPs could be produced in the absence of the NSm coding region,335 but other studies to evaluate the use of the 13 potential translation initiation codons present in the NSm coding information have not been reported. No homology between the NSm proteins of PTV and RVFV was apparent.221
In contrast to the mosquito-borne phleboviruses PTV and RVFV, the tick-borne phlebovirus UUKV does not produce an NSm (e-Fig. 42.5). The first (and only) initiation codon preceding sequences of the envelope GPs is located 17 amino acids upstream of the amino terminus of Gn449; hence, the Gn protein of UUKV appears to be analogous to the 78-kD NSm–Gn fusion product produced by RVFV (although there is no obvious sequence homology of these predicted products). Until a function can be assigned to NSm, it is impossible to determine whether UUKV replicates in the absence of such a function or accomplishes whatever function is required without removal of a portion of the amino terminus of Gn.
M-segment polyprotein processing events for nairoviruses are more complicated and appear to differ from those of other viruses in the family (e-Fig. 42.6). Nucleotide sequences of DUGV, CCHFV, and NSDV revealed a coding strategy similar to other viruses in the family, that is, a single ORF.331 Studies on CCHFV demonstrated the polyprotein is co-translationally
cleaved into 140-kD PreGn and 85-kD PreGc precursors and a 15-kD NSm protein.14,44,461,462,555 PreGn is processed to the mature 37-kD Gn by cleavage with a cellular secretory pathway protease, SKI-1, following a consensus motif, RRLL, early in the secretory pathway.461 SKI-1 cleavage generates a subprecursor of the other two polypeptides that is cleaved by a furin-like protease late in the secretory pathway.461 This cleavage site is completely conserved among several CCHFV strains, suggesting that it has importance for viral replication or pathogenicity.461 The amino-terminal portion of this subprecursor is a highly variable (among CCHFV strains) polypeptide with mucin-like characteristics, including numerous serines, threonines, and prolines and predicted extensive O-linked glycosylation. The carboxy-terminal portion of the subprecursor is a 38-kD nonstructural polypeptide. After furin cleavage, secreted proteins GP38, GP85, GP160, and the mucin-like protein are produced; presumably these products reflect extensive O-linked glycosylation differences. Although the PreGc precursor also possesses a SKI-1–like cleavage motif, SKI-1 cleavage could not be demonstrated for this protein, so it is likely that a related enzyme is responsible for the processing to yield mature Gc.461
cleaved into 140-kD PreGn and 85-kD PreGc precursors and a 15-kD NSm protein.14,44,461,462,555 PreGn is processed to the mature 37-kD Gn by cleavage with a cellular secretory pathway protease, SKI-1, following a consensus motif, RRLL, early in the secretory pathway.461 SKI-1 cleavage generates a subprecursor of the other two polypeptides that is cleaved by a furin-like protease late in the secretory pathway.461 This cleavage site is completely conserved among several CCHFV strains, suggesting that it has importance for viral replication or pathogenicity.461 The amino-terminal portion of this subprecursor is a highly variable (among CCHFV strains) polypeptide with mucin-like characteristics, including numerous serines, threonines, and prolines and predicted extensive O-linked glycosylation. The carboxy-terminal portion of the subprecursor is a 38-kD nonstructural polypeptide. After furin cleavage, secreted proteins GP38, GP85, GP160, and the mucin-like protein are produced; presumably these products reflect extensive O-linked glycosylation differences. Although the PreGc precursor also possesses a SKI-1–like cleavage motif, SKI-1 cleavage could not be demonstrated for this protein, so it is likely that a related enzyme is responsible for the processing to yield mature Gc.461
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree