Bornaviridae
Christiane Herden
Thomas Briese
W. Ian Lipkin
Jürgen A. Richt
History
The syndrome we know as Borna disease (BD) was first described in European veterinary textbooks in the 1700s331,358 as a disease of farm horses using various names like Hitzige Kopfkrankheit (German; “hot-headed disease”) or Seuchenhafte Gehirn-Rückenmarksentzündung (German; “epidemic encephalomyelitis”). The contemporary name Borna disease was coined after the occurrence of major outbreaks in the years 1894–1896 in the district around the town of Borna in Saxony, Germany.358 The name contains the “classifying” letters “RNA,” a classification that was only justified at the end of the 20th century, when the etiologic agent Borna disease virus (BDV) was identified as an RNA virus. BD predominantly affects horses and sheep but other Equidae, certain farm and zoo animals, or companion animals are occasionally also diagnosed with natural BD (reviewed in67,113,253,262,310).
During the first decades of the 20th century, studies of BD focused primarily on defining the etiology, pathology, and pathophysiology of the disease. Initial evidence for a viral etiology was presented by Zwick358 by reproducing the disease with bacteria-free filtrates of brain homogenates from affected horses. Histopathologic studies demonstrated a nonpurulent encephalomyelitis characterized by massive lymphohistocytic infiltrates affecting the gray, and to a lesser extent, the white matter of the central nervous system (CNS), reactive astrogliosis, and intranuclear eosinophilic, so-called “Joest-Degen,” inclusion bodies (described in detail in88,141,142,294). Pathologic changes were preferentially localized in the limbic system, most likely resulting in the observed behavioral disturbances.135,262 Detailed studies have been performed on the spectrum of susceptible host species and on the manifestations of the infection. Transmission experiments between naturally infected horses and sheep as well as other host species—including rabbits, guinea pigs, rats, chickens, and monkeys—established the infectious nature of the BD agent and confirmed that the same agent afflicted horses and sheep.113,181,205,223,262,358,359
Interest in BD and its causative virus lapsed until the late 1970s/early 1980s, when the optimization of tissue and cell-culture techniques for propagating the agent paved the way for work on the identification of the agent and on mechanisms of its pathogenesis in rabbit, rat, and tree shrew models.177,191,203,308 Milestones in pathogenesis included the adaptation to Lewis and Wistar rats, the demonstration of an age-dependent outcome in experimentally infected rats,128,203,206 and the recognition of T-cell–dependent immunopathology.128,203,204,257,258,313,318 Narayan’s observations of a biphasic disease in adult-infected rats characterized by initial hyperactivity and followed by hypoactivity prompted efforts to determine whether humans were infected with a related agent. Serologic findings suggestive of a potential role for BDV in affective disorders263 intrigued many investigators (including the authors of this chapter), resulting in new efforts to identify the causative agent and explore the pathobiology and epidemiology/epizootiology of BDV infection.
However, the causative agent remained elusive until the late 1990s, when BDV was isolated and classified as a negative, single-stranded, nonsegmented RNA virus, the first
member of the new family Bornaviridae in the order Mononegavirales.27,44,51,176,258,277,281,326 With the advent of reverse genetics systems to produce infectious cDNA clones, detailed molecular analyses of the BDV genome and its gene products, regulation of their expression, and detailed pathogenicity studies using constructs stably expressing fluorescent proteins became possible.1,52,220,282,286,353
member of the new family Bornaviridae in the order Mononegavirales.27,44,51,176,258,277,281,326 With the advent of reverse genetics systems to produce infectious cDNA clones, detailed molecular analyses of the BDV genome and its gene products, regulation of their expression, and detailed pathogenicity studies using constructs stably expressing fluorescent proteins became possible.1,52,220,282,286,353
In the first decade of the 21st century, a novel BDV, now designated avian bornavirus (ABV), was identified in parrots with proventricular dilatation disease (PDD).131,152 PDD is a progressive, variably contagious and often fatal disease of domesticated and wild psittacine birds worldwide. Typical clinical signs such as gastrointestinal (GI) dysfunction and associated wasting with or without neurologic symptoms are caused by nonpurulent inflammation of the enteric, autonomic, and central nervous system (CNS). A viral etiology for PDD has been assumed for over 40 years; recent work provides evidence for the etiologic role of ABV in the development of clinically manifest PDD. ABV has been detected worldwide in many captive parrots but also in other nonpsittacine species.55,112,132,217,343
Until recently, it has been believed that no endogenous nonretroviral viruses exist in animal genomes. Surprisingly, endogenous elements homologous to BDV genes were detected in the genomes of bats, elephants, fish, lemurs, rodents, squirrels, primates, and humans.11,133 Although phylogenetic analyses indicate that bornaviruses infected primates at least 40 million years ago, there is only controversial data to support current infection of humans. Indeed, a recently published multicenter study used a wide range of molecular and serologic methods to analyze well characterized samples from subjects with schizophrenia and affective disorders, finding no evidence for human infection with a virus similar to either of the two currently known bornaviruses, BDV and ABV.134
The Virus
Taxonomy
Although the syndrome known as BD has been described since the 1700s, its causative agent, BDV, eluded characterization until the late 1980s, when the application of a novel technique, subtractive cDNA cloning, yielded the first cDNA clones of the agent.176,326 Thereafter, analysis of concentrated, partially purified virus preparations24,251 led to the identification of BDV as a nonsegmented, negative-strand RNA virus, distantly related to rhabdo-, paramyxo-, and filoviruses.27,44 Identification of distinctive features including nuclear replication and transcription,24,42,258 differential use of transcription initiation and termination signals,27,276 and the use of alternative mRNA splicing27,45,281 resulted in the creation of a new family Bornaviridae within the order Mononegavirales in 1996.243 BDV is the prototype of the family and was its sole member until a new species, ABV, was discovered in 2008.131,152
Morphology and Physical Characteristics
Spherical, enveloped particles of 80 to 100 nm with an electron dense core have been visualized by electron microscopy (EM) in extracts of BDV-infected cultured cells.159,356 ABV viral particles of 83 to 104 nm in diameter have been detected in the brain, eye, or small intestine of ABV-infected birds and detection of comparable particles 25 years ago led to the conclusion that PDD is due to a virus infection.132,185,352 Smaller structures also identified in these extracts presumably represent defective particles. No similar structures have been reported in tissues or fluids from infected mammals.3,40,270 The virion Mr and the S20,ω are not known; partially purified virus has a buoyant density of 1.15 to 1.22 g/cm3 in CsCl, 1.18 to 1.22 g/cm3 in sucrose, and 1.13 g/cm3 in renografin.47,84,216,251,356 Virus infectivity is only marginally affected after 24 hours in serum or by incubation at 37°C. In tissues and cell-free virus preparations the virus can be more stable than in culture extracts, and depending on the mode of desiccation, dried preparations can remain infectious for months (tissue at ambient temperature) to years (brain suspension under vacuum).47,177,179,205,358,360 At 4°C BDV infectivity is stable for more than 3 months. BDV can withstand both alkaline and acidic environments, but is most stable at neutral pH.47,70,114 Heating to 56°C for more than 3 hours inactivates the virus and common disinfection methods are appropriate as BDV is sensitive to organic solvents, detergents, pH below 4, and to UV light.47,63,113,114,203,205,216,358
Genome
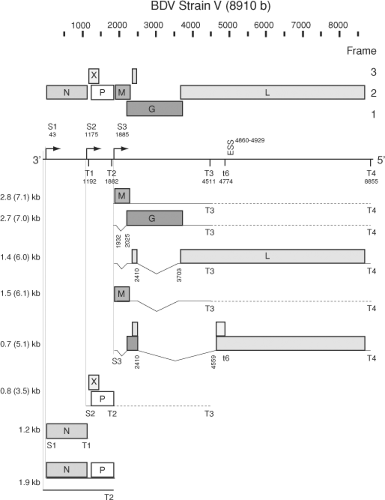
The bornaviral genome is a negative sense, single-stranded, nonsegmented RNA comprising approximately 8,900 nucleotides (nt) that includes six major open reading frames (ORF) with structural proteins in a 3′ and the viral polymerase in a 5′ position.27,131,152,208,230 Short noncoding complementary sequences are found at the termini. Unlike other nonsegmented negative-strand (NNS) RNA viruses, bornaviruses lack specific intergenic regions and instead have mostly overlapping ORFs (Fig. 39.1). The first transcription unit encodes the nucleocapsid protein (N). N exists in two isoforms, p40 (40 kDa) and p38 (38 kDa), that differ in the presence or absence of an amino terminal basic sequence that mediates nuclear localization.156,245 Although nucleocytoplasmic shuttling can be deduced from immunohistochemical and in situ hybridization results for ABV,340,341 there is no proof for the existence of N isoforms in ABV.92,327,328 The viral phosphoprotein (P, p23) and the regulatory X protein (X, p10) are encoded by the second transcription unit.339 The 5′ end of the P ORF overlaps with the 3′ portion of the X ORF in the +1 reading frame, an organization that resembles the P/C/C′ organization of the second gene of vesiculoviruses.167,307 However, BDV X is the first ORF in the RNA transcript and there is no evidence of co-transcriptional mRNA editing, a mechanism used by some paramyxoviruses to regulate expression of multiple reading frames from their second transcription unit.160,321 The first and second transcription units of bornaviruses overlap, because the transcription start S2 is located upstream of the termination/polyadenylation signal T1 (Fig. 39.1).276 The start S3 of the third transcription unit is located 2 nt downstream of T2 and generates multiple transcripts for the matrix protein (M, p16), the type I surface glycoprotein (G, p57), and the L-polymerase (L, p190). The G ORF overlaps the M ORF in the +1 reading frame. The L gene initiates with a short ORF of 6 amino acids (aa) that is spliced to the large 5′ ORF (Fig. 39.1).281,334
Bornaviruses are phylogenetically distinct from other taxa. Sequence divergence between strains of BDV and ABV is less than 20%, while divergence between BDV and ABV is greater than 30%; thus, they are classified as different species.131,152 The only region where bornaviruses have significant sequence
similarity to other known viruses is in the conserved signature motifs of RNA-dependent RNA polymerases (RdRp).234,235 The closest phylogenetic relations exist to rhabdo- and paramyxoviruses,27,44 with the N-terminal half of the L sequence being closer to rhabdoviruses, whereas the C-terminal half is more closely related to paramyxoviruses.
similarity to other known viruses is in the conserved signature motifs of RNA-dependent RNA polymerases (RdRp).234,235 The closest phylogenetic relations exist to rhabdo- and paramyxoviruses,27,44 with the N-terminal half of the L sequence being closer to rhabdoviruses, whereas the C-terminal half is more closely related to paramyxoviruses.
Sequences distantly related to BDV L, M, and N sequences were recently identified in the genomes of several animal species, including bats, elephants, fish, lemurs, rodents, squirrels, primates, and man.11,133 Detailed phylogenetic analyses suggest multiple ancient independent integration events. An intriguing example is a BDV N-related sequence that likely integrated before the separation of marmosets and macaques 40 million years ago.11,133 In some instances, the endogenous Borna-like (EBL) element comprises a complete ORF that includes BDV-like transcription initiation and termination signals. The finding of mRNA transcripts of such EBL elements suggests potential functional roles that may include protection from BDV infection.11,81
Genetic Diversity
BDV isolates reveal a remarkably high degree of genetic stability and homology. Among wild-type and experimentally host-adapted viruses, sequence identity is about 95% at the nucleotide level, and 1.5% to 3% at the predicted amino acid level.18,27,44,123,161,208,230,278,310 Phylogenetic analysis of wild-type and laboratory strains of BDV indicates distinct clusters, which correspond to the different endemic areas in Central Europe.161 Geographical virus clusters exhibited a higher degree of identity to each other than to BDV isolates from distant regions, independent of host species or year of isolation. There is only one highly divergent BDV strain, No/98, which originated from Styria in Austria where Borna disease is not endemic.208
ABV displays remarkable genetic variability in contrast to the high genome conservation of BDV. At present, 7 different genotypes (ABV1, ABV2, ABV3, ABV4, ABV5, ABV6, ABV of canaries) have been identified that share less than 70% sequence identity to any of the described BDV isolates.131,152,217,340,343 The ABV genotypes vary considerably in their gene sequences with
a homology range of 50% to 90% without clustering according to country of origin or avian species.131,132,152,153,259,309,340,343 ABV4 appears to be the most abundant genotype in natural PDD cases but also in healthy carriers. Recently, a distinct ABV genotype has been detected in wild geese and trumpeter swans.55,217
a homology range of 50% to 90% without clustering according to country of origin or avian species.131,132,152,153,259,309,340,343 ABV4 appears to be the most abundant genotype in natural PDD cases but also in healthy carriers. Recently, a distinct ABV genotype has been detected in wild geese and trumpeter swans.55,217
Proteins
Nucleocapsid Protein (N)
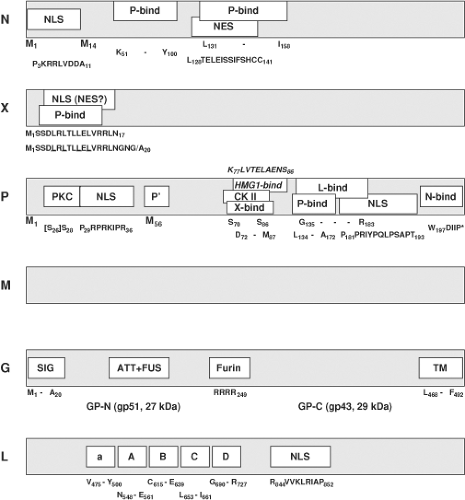
In BDV, N exists as a 40-kDa and 38-kDa isoform;26,156,245 N isoforms have not yet been observed in ABV.92,327,328 The 40-kDa variant (p40) is derived from the full length ORF, while the 38-kDa variant (p38) initiates at a second in-frame AUG, resulting in the lack of 13 aa at the amino terminus (Fig. 39.2). Although an RNA with coding information for p38 has been found that starts downstream of S1,245 it is unknown whether p40 and p38 can both be translated from mRNA transcripts starting at the S1 transcriptional initiation site. The 13 aa amino terminal sequence present in p40 contains a nuclear localization signal (NLS; P3KRRLVDDA11) compatible with the differential cellular distribution of the two isoforms seen in cells transfected with constructs expressing only one of the isoforms; while p40 is primarily nuclear, p38 is primarily cytoplasmic.156,245 However, both, p38 and p40 bind to P. As P contains potent NLS (Fig. 39.2), the in vivo significance of the two N isoforms is unknown; p38 may enter the nucleus through interaction with P. Experimental evidence with expression constructs indicates that p38 can accumulate in the nucleus to levels similar to those of p40; however, p38 alone cannot support transcription/replication of BDV (mini-)genomes.220 Thus, the amino terminal sequence of p40 may have functions in addition to N protein translocation. Both p40 and p38 bind to P through two motifs (K51–Y100, and L131–I158; Fig. 39.2).13,155 p38 appears to regulate cellular levels of free p40 by blocking the respective binding site on P, thus modulating cellular ratios of free p40 to P.284 In addition, p38 and p40 contain a nuclear export signal (NES; L128TELEISSIFSHCC141)155 that overlaps the binding motif for P (Fig. 39.2). It is therefore hypothesized that p38, which lacks the NLS motif, may redistribute to the cytosol after dissociation from bound P, possibly once assembled in ribonucleoprotein (RNP) complexes.155 As indicated by purification experiments and co-localization studies, both p38 and p40, as well as P and M, are included with genomic RNA in the RNP.35,189 Association of genomic RNA with N relies on basic amino acid residues located in a cleft formed between the amino- and carboxy-terminal helical domains of N.129,267 However, binding to N in the multimeric RNP appears not to shield the genomic RNA from enzymatic attack.
Together with P, N constitutes the BDV s-antigen, a complex found in the noninfectious supernatant fluid obtained after high-speed centrifugation of sonicated infected brain tissue or cultured cells. Characterization of the s-antigen provided the
first evidence of protein-protein interactions,8,96,177 and until the development of molecular assays, served as the critical diagnostic marker for infection.211,332,333
first evidence of protein-protein interactions,8,96,177 and until the development of molecular assays, served as the critical diagnostic marker for infection.211,332,333
X Protein (X)
BDV X or p10339 is a nonstructural protein189,288 that, together with P, modulates BDV polymerase activity as a function of the relative abundance of the two proteins.237,288 X effects appear to be differently pronounced in different cell types.353 Although X is not essential for the formation of infectious particles,219 it does perform crucial functions during the BDV life cycle because recombinant BDV constructs carrying a nonfunctional X ORF are not viable.239 Mammalian two-hybrid and co-immunoprecipitation experiments indicate an interaction between X and P,293 and recombinant BDV systems showed that X inhibited BDV RNA replication and transcription through binding to P220,238,285 in the absence of viral M and G.219 The site of X interaction with P has been mapped to the N-terminal motif S3DLRLTLLELVRRL16, with aa 7 through 15 being probably most essential.184,350 X is small (10 kDa) and can be found in the nucleus and cytoplasm. A leucine-rich amino-terminal motif with primary sequence similarity to the NES of cellular and viral export proteins like HIV-1 Rev or PKI (Fig. 39.2, underlined leucines) led to the speculation that X may mediate nucleocytoplasmic shuttling through its interaction with the viral RNP via P. However, X has not been shown to be part of the RNP,35,189,219 and other data suggest that this motif functions as an NLS rather than an NES (R6LTLLELVRRNGN19).351 These experiments also showed that transport of X through the nuclear pore complex is mediated by direct binding to importin-alpha.
Phosphoprotein (P)
P, a cofactor of the L viral polymerase, is phosphorylated at multiple serine residues by two different cellular kinases.289,320 P is phosphorylated predominantly by protein kinase Cε (PKCε) at Ser28 (and Ser26, which is not present in all BDV strains), and to a lesser extent by casein kinase II (CK II) at Ser 70 and Ser 86 (Fig. 39.2). As in other NNS virus phosphoproteins, phosphorylation status may regulate P’s ability to form homo-multimers, bind to other viral proteins, and serve as a transcriptional activator. P interacts with itself, X, N, M, and L as shown by mammalian two-hybrid and co-immunoprecipitation analyses. Regions of interaction of P with P (aa 135–172), with N (aa 197–201),293 with M (aa 1–11),35 and with X (aa 72–87)158 were mapped through analysis of truncation mutants of P. A region of interaction with L (aa 135–183) overlaps that identified for homo-oligomerization; however, the sites are functionally separated as shown by analyses of P mutants that bound to L but had lost the ability to oligomerize.283 These analyses also demonstrated that P-oligomerization is essential for polymerase activity. Overlap also exists between the CK II phosphorylation sites and the region of interaction with X, as well as between the PKCε phosphorylation site(s) and the amino-terminal NLS of P; two NLS have been mapped at the amino- and carboxyl-terminus of P (Fig. 39.2).292,295 Thus, it is conceivable that P phosphorylation may influence nuclear trafficking of P (and possibly of X through its interaction with P). In this context it is intriguing that PKCε is highly concentrated in limbic circuitry,268 as it suggests that PKCε phosphorylation may be important to the limbic distribution of BDV. BDV infection of neurons interferes with synaptic vesicle recycling through blockade of PKC-phosphorylation of myristoylated alanine-rich C kinase substrate (MARCKS) and mammalian uncoordinated-18 (Munc-18). There is speculation that P may contribute to BDV pathogenesis by competing with neuronal substrates for phosphorylation by PKC.241,330
Matrix Protein (M)
Like the BDV s-antigen, a 14.5-kDa protein purified from infected brain homogenate had been linked to the unidentified BD agent prior to molecular characterization of BDV.274 Once genome sequence data became available, microsequencing indicated that this protein is the product of the 16-kDa ORF of BDV (p16).154 Subsequent analyses showed that p16 forms noncovalently linked tetramers and constitutes a nonglycosylated viral matrix protein.165,166 M is a component of the viral RNP and can bind to P. However, in contrast to other NNS RNA virus M proteins, BDV M appears to have no inhibitory effect on polymerase activity.35,189
Glycoprotein (G)
The BDV glycoprotein is a classical type I membrane protein that is generated from the 57-kDa ORF and posttranslationally modified by N-glycosylation to yield a 94-kDa primary product (gp94).252,279 The primary product is processed by cellular furin protease into an amino-terminal GP-N (27 kDa, gp51) and a carboxy-terminal GP-C (29 kDa, gp43) through cleavage after arginine249151,252 (Fig. 39.2). Whereas the gp94 precursor contains only high-mannose glycans, analysis of the cleaved fragments indicated glycan maturation by showing mixtures of high-mannose and complex-type glycans on both GP-N and GP-C.151 Anchored by its transmembrane domain, GP-C is transferred to the cell membrane, while the gp94 precursor predominantly accumulates in the endoplasmic reticulum. The final composition of mature virions is not clear; gp94 as well as GP-C and GP-N have been found in infectious particles,38,68,84 but GP-N and GP-C were also demonstrated in infectious particles that lacked the gp94 precursor after virus purification.151 Computational analyses indicate an arrangement of structural features of BDV G that is co-linear to rhabdoviral G, suggesting that BDV G also belongs to the class III viral fusion proteins.80
RNA-Dependent RNA Polymerase (L)
The large polymerase protein (L) of BDV is the product of alternative splicing, a mechanism unique among NNS RNA viruses.27,281 The generated continuous ORF translates a 190-kDa protein334 that displays motifs characteristic of RNA-dependent RNA polymerases (RdRp). L interacts with P and, analogous to other NNS L-polymerases, it is phosphorylated by cellular kinases (Fig. 39.2).334 Plasmid constructs directing expression of the continuous 190-kDa protein that supported replication of recombinant BDV genomes confirmed the polymerase activity of the protein.187,220,285,286,353 Translocation of L to the nucleus of the infected cell appears to be promoted by an NLS motif (R844VVKLRIAP852; Fig. 39.2) located toward the center of L,335 or in association with BDV P.285
Cycle of Infection
Attachment and Entry
BDV attachment and entry appear to be analogous to the pH-dependent entry via intracellular vesicles described for
rhabdo- and filoviruses, as opposed to the pH-independent surface fusion mechanism used by paramyxoviruses.229,260,269 BDV G binds to one or more still unidentified cellular surface receptor(s).84,279 Receptor interaction of G triggers BDV internalization through energy-dependent, clathrin-mediated endocytosis and subsequent pH-dependent membrane fusion leads to release of the RNP from intracellular vesicles into the cytosol.37,83 Protease inhibitor studies indicate that cleavage of the precursor gp94 is essential for infectivity.252 Whereas the amino-terminal 244 aa of gp94 and/or GP-N are involved in receptor binding, the hydrophobic amino-terminus of GP-C is hypothesized to initiate membrane fusion upon a conformational change induced by acidification in the early to intermediate endosome.37,68,83,221,279 No data are available regarding trafficking of the released nucleocapsid after membrane fusion.
rhabdo- and filoviruses, as opposed to the pH-independent surface fusion mechanism used by paramyxoviruses.229,260,269 BDV G binds to one or more still unidentified cellular surface receptor(s).84,279 Receptor interaction of G triggers BDV internalization through energy-dependent, clathrin-mediated endocytosis and subsequent pH-dependent membrane fusion leads to release of the RNP from intracellular vesicles into the cytosol.37,83 Protease inhibitor studies indicate that cleavage of the precursor gp94 is essential for infectivity.252 Whereas the amino-terminal 244 aa of gp94 and/or GP-N are involved in receptor binding, the hydrophobic amino-terminus of GP-C is hypothesized to initiate membrane fusion upon a conformational change induced by acidification in the early to intermediate endosome.37,68,83,221,279 No data are available regarding trafficking of the released nucleocapsid after membrane fusion.
Despite its predilection for neuronal cell types in vivo, BDV has the capacity to infect a wide variety of cultured neuronal as well as nonneuronal cell types from many species. Only hematopoietic cells264 and mouse cell lines74 have been reported as resistant to infection in vitro. This may indicate potential exploitation of secondary receptors, and has also led to hypotheses about a nonreceptor–mediated cell-to-cell spread of BDV that is supported by in vitro as well as in vivo observations.86,180 More recent studies with a CHO cell line that apparently was resistant to infection by BDV virions, and with furin protease-deficient CHO cells, indicated that BDV can disseminate by G-receptor–independent pathways.38 However, correct G maturation enhanced the proposed receptor independent cell-to-cell spread, and is mandatory for the formation of infectious progeny virions.
Transcription, Replication and Gene Expression
Recombinant virus systems confirm that BDV N, P, and L are essential and sufficient for transcription and replication of the viral genome.187,219,220,285,286,353 As with other negative-strand RNA viruses, genomic RNA packaged by N constitutes the RNP that serves as a template for the associated polymerase complex components L and P.129,189 The BDV X protein, although not part of the incoming RNP complex,189,288 appears to modulate the formation and activity of functional polymerase complexes later in infection by buffering the crucial N-to-P ratio and likely attenuating the enzymatic activity of the polymerase.219,237,337 Furthermore, BDV is unique among known animal NNS RNA viruses in its nuclear location for transcription and replication.24,42 To generate its proteins, BDV uses predominantly polycistronic mRNAs that are transcribed from three transcriptional initiation sites characterized by a CUU consensus sequence and terminate at four AU6 termination/polyadenylation sites (Fig. 39.1).27,276
The first and second transcription units overlap such that the initiation signal S2 lies upstream of the termination signal T1.27,44,240,276 A similar organization was postulated to serve as an attenuation signal for the control of polymerase expression in respiratory syncytial virus.39 However, attenuation appears not to take place in BDV as the two transcription units are found at similar levels,27 in contrast to the usual transcriptional gradient observed in the other viruses of the order. Other factors may modify mRNA levels beyond effects attributable to the typical 3′-to-5′ transcriptional gradient, including the incorporation of regulatory sequences in spliced introns.296
Read-through at termination/polyadenylation signals is a vital feature of BDV transcription, leading to primary, subgenomic RNA transcripts of 0.8, 1.2, 1.9, 2.8, 3.5, and 7.1 kb. The 1.2-kb transcript is the only monocistronic product. It is co-linear with the p40 ORF and directs translation of the p40 and p38 isoforms of BDV N from alternative in-frame AUG codons.156,245 The second transcription unit generates the 0.8-kb transcript that codes for the X and the P protein in overlapping ORFs. There is no evidence of splicing to eliminate the AUG initiating translation of X.27,276,339 Thus, it is likely that P is expressed through a leaky scanning mechanism, possibly analogous to the termination-mediated reinitiation of X translation from the small upstream ORF included in the 0.8-kb transcript. The expression of X may be further modulated by cellular protein interactions with the long 5′-untranslated region (UTR) of this BDV transcript.338 The long 5′-UTR region also includes poorly defined regulatory elements controlling polymerase read-through at the T1 signal.237,240 Interestingly, the region containing the AUG of the upstream small ORF has been found deleted in sequenced psittacine ABV genomes but not in sequences derived from geese.217,259 In BDV, a truncated 16-kDa P′ product of undefined function may result from initiation at a downstream in-frame AUG.157
Read-through at T2 produces an elongated 0.8-kb derivative of 3.5 kb. Two primary transcripts of 2.8 and 7.1 kb are generated from the third transcription unit through differential read-through at T3. The 2.8- and 7.1-kb transcripts include the p16 and p57, or the p16, p57, and pol ORF, respectively; in addition, several secondary transcripts are derived from these RNAs through alternative splicing (Fig. 39.1).
Transcripts of the third transcription unit include two intron sequences, intron-1 and intron-2, that are subject to alternative splicing.281 Whereas the 2.8- and 7.1-kb transcripts direct translation of M, the expression of G likely requires removal of intron-1. Although G may also be expressed in vitro from unspliced transcripts by leaky ribosomal scanning, splicing of intron-1 creates a stop-codon after the 13th aa of the p16 ORF that facilitates translational initiation at the AUG of G.280 Splicing of intron-2 removes almost the complete p57 ORF and fuses a small 17 nt ORF located upstream of the splice donor site to the large downstream ORF that, in the case of the 2.8-kb RNA, terminates at T3, generating a truncated L protein of unknown function (Fig. 39.1). Expression of L is, analogous to G, likely facilitated by splicing of intron-1. In addition, an alternative splice acceptor site, controlled by a downstream exon-splicing suppressor, has been identified at nt 4559.322 Splicing of this potential intron-3 (nt 2410–4559), and transcriptional read-through at t6,27 may result in expression of two additional BDV-specified proteins.43,322 However, the splice acceptor side at nt 4559 is not strictly conserved throughout sequenced BDV isolates. The lack of conservation in BDV No/98230 as well as in ABV isolates indicates that the potential gene products are unlikely to fulfill essential functions in the bornavirus life cycle.
There have been reports of multiple types of BDV 1.9-kb RNA transcripts with unspecified function. Analyses based on RNA circularization and sequencing over the junction indicated the presence of a noncapped RNA complementary to position 1 to 1882 of the BDV genome that interacted with oligo(dT)-beads, but was not fully polyadenylated.276 Such transcripts may represent abortive replication intermediates or subgenomic
RNAs analogous to leader RNAs found in other mononegaviruses. On the other hand, studies employing precipitation with a cap-specific antibody characterized a capped 1.9-kb mRNA with a long poly(A)-tail, extending from S1 to T2240 (Fig. 39.1).
RNAs analogous to leader RNAs found in other mononegaviruses. On the other hand, studies employing precipitation with a cap-specific antibody characterized a capped 1.9-kb mRNA with a long poly(A)-tail, extending from S1 to T2240 (Fig. 39.1).
Initiation of transcription and replication in negative-strand RNA viruses is commonly mediated by sequences located in the UTR. A single transcriptional promoter located in the 3′-UTR of the genome generates the usual transcriptional gradient, and promoter sequences driving genome replication are located in both the complementary 3′- and 5′-genomic termini. However, BDV analyses indicated remarkable terminal heterogeneity.230 Kinetic analyses comparing the genomic termini in acute and persistent BDV infection showed an accumulation of terminal truncations in the course of infection, which in several cases resulted in strongly attenuated replicational and transcriptional promoter activity, possibly contributing to BDV’s persistent lifestyle.261 In addition, rescue of infectious recombinant BDV constructs demonstrated trimmed 5′ genomic termini generated from originally perfectly complementary constructs.286 In this system, when recessed 5′ ends aligned to the 3′ terminus were generated, there was strong attenuation of replication while transcriptional activity appeared to be unaffected. This finding is compatible with the high antigen levels in conjunction with low levels of infectious virus that are observed during persistent BDV infection. The four 5′ terminal bases of genome and antigenome appear to be copied from internal template motifs through backfolding of the termini followed by specific elongation and termination on the template motifs.186 This allows later cleavage of these terminal bases to generate monophosphorylated 5′ termini of progeny strands without the loss of genetic information. Trimming of the termini in BDV may support its persistent infection by escape from innate immune responses through RIG-I–mediated recognition of triphosphorylated genomic termini.97 It is not known whether viral and/or host functions are responsible for the terminal trimming. Further characterization of these unique BDV transcriptional and replicational promoter sequences may help to resolve results obtained in various recombinant test systems.
Assembly and Release
As with other negative-strand RNA viruses, a first step in the production of BDV progeny is packaging of the replicated nascent genomic RNA by N. The 5′-trailer RNA specifically promotes its association with N via basic residues in the cleft between the amino- and carboxy-terminal domains of the protein.129,267 Formation of the RNP complex includes association of P via its carboxy-terminal aa residues, and the inclusion of L is hypothesized to occur via its characterized protein–protein interaction sites (Fig. 39.2). Based on NLS located in N, P, L, and X, and NES in the two N isotypes and possibly X, various hypotheses concerning nucleocytoplasmic shuttling of BDV RNP have been proposed, but experimental confirmation is lacking. Co-localization studies suggest that M is also an integral part of the BDV RNP.35 M, but not X, has been demonstrated in purified RNP,189 a finding consistent with the observation that in recombinant BDV mini-genome systems the expression of M and G, but not X, is required for the formation of infectious BDV-like particles.219 The processing of BDV G by protease digestion appears to be crucial to the assembly of infectious virions.5,38 Distinct packaging signals are not defined.
Epidemiology of BDV Infection
Host Range
BD is reported most commonly in horses and sheep; but disease has also been reported in other Equidae, farm animals (cattle and goats), rabbits, lynx, zoo animals (alpacas, sloths, various monkeys, hippopotamuses), and rarely in companion animals (dogs and cats).21,31,46,54,67,113,138,140,182,192,262,310,344,358 Experimental infections have been achieved in various animal species ranging from chickens to nonhuman primates.4,46,87,113,148,188,201,212,255,262,266,308,314 Infection in horses leads to death 1 to 4 weeks after onset of signs in 80% to 90% of animals.67,89,91,253 In 72% of stables with equine BD cases, only individual animals develop clinically manifested BD. In cattle and sheep, death was noted after 1 to 6 weeks or 1 to 3 weeks in more than 50% of animals, respectively.21,255
Geographic Distribution and Potential Reservoir
Natural BD is endemic in areas of central Europe such as southern and eastern Germany, Switzerland, Liechtenstein, and Austria.30,46,66,113,120,193,344,358 Reports of natural BD outside these endemic areas suggest a wider distribution of the disease.78,98,99,144,147,354 Virus-specific serum antibodies and/or nucleic acids in absence of disease or in association with unusual clinical signs have been reported in animals from different geographic areas, including European countries, such as France, Sweden, Finland, and Italy, as well as Turkey, Israel, Japan, Iran, China, Australia, and the United States.66,78,98,99,144,147,178,253,262,354 However, as some of these data are debated and require more confirmation, further epidemiologic studies are warranted.
A seasonal accumulation of BD cases in April, May, and June—with a significant decrease in late fall and winter—is quite characteristic and argues, in combination with the geographically limited occurrence, for a natural reservoir.66,67,89,113,181,325 Recently, BDV infection has been detected in bicolored shrews (Crocidura leucodon) in endemic areas in Switzerland. These animals had a disseminated virus distribution in the absence of overt disease and represent a potential reservoir for BDV.127,244 Whether other species can serve as reservoir species is currently unknown.
Prevalence and Seroepidemiology
In contrast to the epidemic course of BD at the end of the 19th century, the incidence of BD decreased significantly during recent decades; usually less than 100 horses or sheep are diagnosed with BD per year.66,120,199 However, BDV infections in horses and sheep can be inapparent as indicated by seroepidemiologic surveys in Germany. The average seroprevalence of BDV-specific antibodies in clinically healthy horses in Germany is approximately 11.5%120 and increases in endemic areas up to 22.5%, reaching 50% in stables with a history of clinical BD.91 There is a higher frequency of BD on farms with mixed stock of horse, sheep, and cattle, operating under lower hygiene standards.66 Repeated outbreaks of BD within the same premises have been noted but usually spaced several months or years apart.91,253 The reason for the discrepancy between the high BDV seroprevalence and the low BD incidence remains unknown but may relate to age, immune status, genetic background, virus strain, and/or dose of infection.
Route of Infection and Transmission
There is evidence that nerve endings in the nasal and pharyngeal mucosa represent the most likely natural route of entry.142,188,197 Experimental BDV infection of neonatal rats results in virus persistence and disseminated virus distribution with presence of viral gene products and infectious virus in saliva, urine, and feces.188,197 Such secreta or excreta are important in transmission of other pathogenic viruses (e.g., lymphocytic choriomeningitis virus and hantaviruses). This finding further supports the concept of a natural reservoir of BDV, which is substantiated by stable geographic virus clusters,18,161,325 despite substantial horse movement and trade. This was recently confirmed by a case of natural BD in Great Britain that was traced back to a likely origin of infection in Germany.242 It seems that widespread horse-to-horse or sheep-to-sheep transmissions do not occur.16,66,254,310 The infectious dose for natural infections is unknown.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree