Astroviruses
Ernesto Méndez
Carlos F. Arias
The family Astroviridae includes human and animal astroviruses that show icosahedral morphology; they are nonenveloped and their genome is composed of plus-sense, single-stranded RNA (ssRNA), with three open-reading frames, whose organization distinguishes them from other virus families.
Astroviruses (AstV) have been isolated from a variety of animal species. In most mammals, astrovirus infections are associated with gastroenteritis. In particular, human astroviruses (HAstV) have been found to be the second or third most common cause of viral diarrhea in young children and cause of sporadic gastroenteritis outbreaks. Avian AstV, on the other hand, have been linked with more severe intestinal and extraintestinal manifestations of disease.
History
The term astrovirus was coined by Madeley and Cosgrove in 1975 to describe small, round viruses with a distinctive five- or six-pointed, star-like appearance (astron, star in Greek) of about 28 to 30 nm in diameter.86 They were observed by direct electron microscopy (EM) in the stools of infants hospitalized with diarrhea and in outbreaks of gastroenteritis in newborn nurseries (Fig. 21.1). Subsequently, viral particles of similar size and morphology were identified by EM in association with gastroenteritis in a wide variety of young mammals and birds.
An important milestone was achieved in 1981 when Lee and Kurtz reported the isolation and passage of HAstV in primary cell cultures.82 This achievement led to the recognition of five HAstV serotypes in 1984,79 development of an enzyme immunoassay (EIA) to detect viral antigen in the late 1980s,60 and confirmation of its medical importance.61 The molecular characterization of astrovirus isolates subsequently permitted the recognition of 8 serotypes of HAstV and the design of molecular probes for use as diagnostic tools. Metagenomic approaches have allowed the identification of novel AstV from humans and animal species in recent years.16,33,35,115 The efficient propagation of HAstV in cell lines147 and of turkey isolates in animal models.72 has further advanced our knowledge of the molecular and structural biology, as well as of pathogenesis of these viruses.
Classification
The general organization of the astrovirus genome places the open reading frames (ORF) encoding the nonstructural proteins at the 5′ end, and the ORF encoding the structural proteins at the 3′ end. Distinctive features of this family include the morphology of viruses,116 the lack of an RNA-helicase domain encoded in the genome, and the usage of a ribosomal frameshifting mechanism to translate the RNA-dependent RNA polymerase (RdRp).65
Astroviruses were originally classified into genera and species based only on the host of origin; however, recent characterization of novel AstV has shown that isolates from different animal species can be genetically similar, while genetically
diverse viruses can be isolated from the same animal species.69,85,152 These findings have led to a proposed new classification scheme based on the amino acid sequence of ORF2 (Astroviruses Study Group, 9th Report ICTV, 2010),17 which encodes the capsid polyprotein and represents the most variable region of the genome (see below). Two genera are distinguished within the Astroviridae family: Mamastrovirus and Avastrovirus (Fig. 21.2). Viruses belonging to the genus Mamastrovirus include isolates from a number of mammals, including humans, pigs (PAstV), cats (FeAstV), minks (MAstV), sheep (OAstV), calfs (BoAstV), dogs (CaAstV), bats (BAstV), rats (RAstV), deer (CcAstV), and marine mammals, such as sea lions (CSlAstV) and bottlenose dolphins (BdAstV), among others. This genus includes two genogroups, GI and GII, with 10 and 9 genotype species, respectively. Both genogroups comprise viruses from human and animal origin. Of note, recently identified human viruses are very similar to animal isolates, such as mink and sheep,69 among others. HAstV previously classified within one species that comprised serotypes 1 to 8 (based on their reactivity to hyperimmune sera; HAstV-1 to −8) are now included in the proposed genotype G1 of genogroup I. The genetic diversity found among pig,85 bat,152 and human69 isolates places them into highly divergent groups, suggesting that they have different ancestors that probably emerged during interspecies transmission.85
diverse viruses can be isolated from the same animal species.69,85,152 These findings have led to a proposed new classification scheme based on the amino acid sequence of ORF2 (Astroviruses Study Group, 9th Report ICTV, 2010),17 which encodes the capsid polyprotein and represents the most variable region of the genome (see below). Two genera are distinguished within the Astroviridae family: Mamastrovirus and Avastrovirus (Fig. 21.2). Viruses belonging to the genus Mamastrovirus include isolates from a number of mammals, including humans, pigs (PAstV), cats (FeAstV), minks (MAstV), sheep (OAstV), calfs (BoAstV), dogs (CaAstV), bats (BAstV), rats (RAstV), deer (CcAstV), and marine mammals, such as sea lions (CSlAstV) and bottlenose dolphins (BdAstV), among others. This genus includes two genogroups, GI and GII, with 10 and 9 genotype species, respectively. Both genogroups comprise viruses from human and animal origin. Of note, recently identified human viruses are very similar to animal isolates, such as mink and sheep,69 among others. HAstV previously classified within one species that comprised serotypes 1 to 8 (based on their reactivity to hyperimmune sera; HAstV-1 to −8) are now included in the proposed genotype G1 of genogroup I. The genetic diversity found among pig,85 bat,152 and human69 isolates places them into highly divergent groups, suggesting that they have different ancestors that probably emerged during interspecies transmission.85
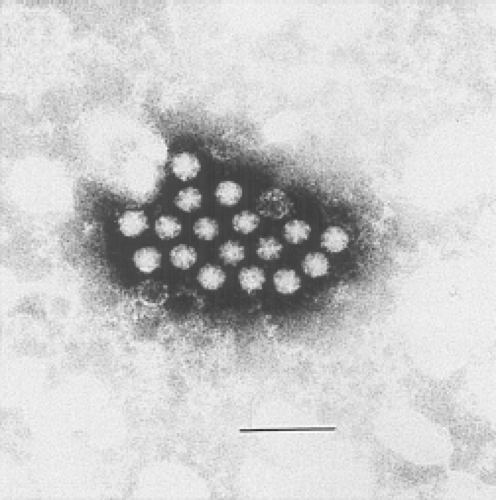 Figure 21.1. Electron micrograph of human astrovirus in a fecal specimen. Bar = 100 nm. (Courtesy of T. W. Lee and J. B. Kurtz.) |
Viruses from the genus Avastrovirus include isolates from turkeys (TAstV), ducks (DAstV), chicken (CAstV), and guinea fowl. This genus includes two proposed species in genogroup GI (GIA, GIB) and one in genogroup II (GIIA). Similarly to canonical HAstV, members of some of these species can be distinguished by serology, indicating the existence of viral serotypes in some, such as in TAstV-2 and ANV.132,134 In general, avastroviruses show higher diversity than mamastroviruses.
Virion Structure and Composition
Virion Structure
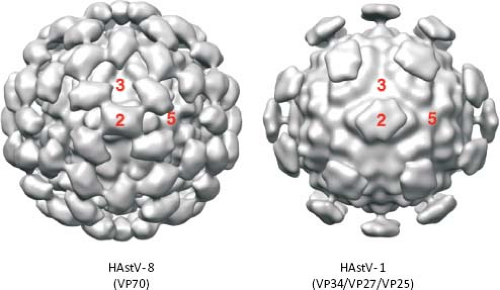
Ultrastructural analysis by EM of human viruses propagated in cell culture in the presence of trypsin revealed icosahedral particles of 41 nm, with spikes protruding from the surface.116 The star-like form of the particles was observed only after alkaline treatment. Recent studies by cryo-EM and image processing of trypsin-treated and untreated HAstV particles confirmed the spiked icosahedral structure of virions (Fig. 21.3) (K. Dryden et al.,
unpublished results). Remarkable differences are observed between the two types of particles. The untreated virus, 46 nm in diameter, contains 180 copies of a single protein of 70 kd arranged in a T = 3 icosahedral symmetry. Two kinds of spikes, localized at two- and fivefold vertices, can be observed in these particles. Two protein layers can be distinguished. The internal layer forms the capsid core and is almost identical to that of treated particles, at the highest resolution reached (23–25 Å); however, the distal layer that forms the spikes shows dramatic changes after trypsin treatment. This treatment results in the cleavage of the 70-kd protein into three polypeptides that is required for virus infectivity (see below). The main difference between the two types of particles is the number of spikes observed, more likely due to disordering of the projections located around the fivefold vertices in the trypsin-treated virions (K. Dryden et al., personal communication).
unpublished results). Remarkable differences are observed between the two types of particles. The untreated virus, 46 nm in diameter, contains 180 copies of a single protein of 70 kd arranged in a T = 3 icosahedral symmetry. Two kinds of spikes, localized at two- and fivefold vertices, can be observed in these particles. Two protein layers can be distinguished. The internal layer forms the capsid core and is almost identical to that of treated particles, at the highest resolution reached (23–25 Å); however, the distal layer that forms the spikes shows dramatic changes after trypsin treatment. This treatment results in the cleavage of the 70-kd protein into three polypeptides that is required for virus infectivity (see below). The main difference between the two types of particles is the number of spikes observed, more likely due to disordering of the projections located around the fivefold vertices in the trypsin-treated virions (K. Dryden et al., personal communication).
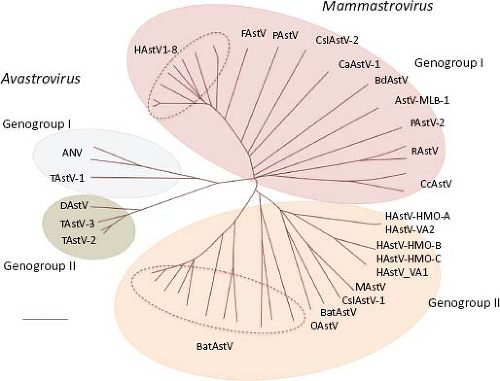 Figure 21.2. The Astroviridae family includes two genera with two genogroups each. Virus species are classified based on the ORF2 amino acid sequence distances. (Classification proposed by the Astroviruses Study Group, 9th Report ICTV, 2010.17). |
Virion Composition
Astrovirus particles are formed by the viral genome surrounded by an icosahedral capsid formed by a single protein of 70 to 90 kd, or by at least three proteins in the range of 25 to 34 kd, depending on the extent of proteolytic processing of the virion.12,93,119 Extracellular particles, released from HAstV-8 infected cells, are formed by protein VP70, which results from the intracellular processing of VP90, the full-length primary protein product encoded in ORF294 (see below). Fully infectious particles obtained by exhaustive treatment of the virus with trypsin are constituted by three proteins, in the range of 32–34, 27–29, and 25–26 kd, depending on the virus strain, with the last two proteins overlapping in sequence.93,119 Thus, proteins of different size (between 24 and 90 kd), probably representing final and intermediate cleavage products, can be found in particles of HAstV.12 The size of some of the proteins identified in animal viruses is similar to that of proteins found in human strains; thus, PAstV contains proteins of 31, 30, and 36 kd,125 and OAstV contains two proteins of approximately 33 kd58; however, PAstV also contains proteins of 39 and 13 kd,125 not usually detected in mature HAstV particles. An isolated report indicates that HAstV particles may contain a small protein of 5.2 kd, but its nature was not established.78
In addition to proteins derived from ORF2, proteins coded by ORF1a have been suggested to be present in viral particles, since antibodies to purified HAstV-1 virions recognize a recombinant protein containing amino acids 757 to 899 of nsp1a.89
Genome Structure and Organization
Astroviruses have an ssRNA genome of positive polarity [ssRNA(+)] (Fig. 21.4) that varies in length from 6.17 kb for the human strain MLB-133 to 7.72 kb for DAstV-2,37
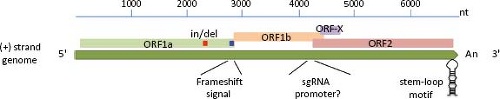
excluding the poly(A) tail at the 3′ end (e-Table 21.1 and e-Fig. 21.1). The RNA extracted from AstV particles, as well as RNA transcribed from a full-length genomic copy of complementary DNA (cDNA) clone, are able to initiate a productive infection in cultured cells,42 although with different efficiencies. The viral genome includes 5′ and 3′ untranslated regions (UTRs), and three ORFs of variable length in different isolates (e-Table 21.1). The two ORFs located towards the 5′ end of the genome, designated ORF1a and ORF1b, encode nonstructural proteins that are presumed to be involved in transcription and replication of the virus genome, based on the sequence motifs they contain (see below). Variation of the ORF1a length in HAstV is mainly due to insertions or deletions (in/del regions) present near the 3′ end of ORF1a.55,144 The third ORF, found at the 3′ end of the genome and designated ORF2, encodes the capsid polyprotein. ORF1a and ORF1b overlap in 10 to 148 nucleotides (nt) in the genome of mammalian viruses, and between 10 and 45 nt in avian viruses. The overlapping region contains an essential signal for translation of the viral RNA polymerase (encoded in ORF1b) through a frameshift mechanism65 (see below).
excluding the poly(A) tail at the 3′ end (e-Table 21.1 and e-Fig. 21.1). The RNA extracted from AstV particles, as well as RNA transcribed from a full-length genomic copy of complementary DNA (cDNA) clone, are able to initiate a productive infection in cultured cells,42 although with different efficiencies. The viral genome includes 5′ and 3′ untranslated regions (UTRs), and three ORFs of variable length in different isolates (e-Table 21.1). The two ORFs located towards the 5′ end of the genome, designated ORF1a and ORF1b, encode nonstructural proteins that are presumed to be involved in transcription and replication of the virus genome, based on the sequence motifs they contain (see below). Variation of the ORF1a length in HAstV is mainly due to insertions or deletions (in/del regions) present near the 3′ end of ORF1a.55,144 The third ORF, found at the 3′ end of the genome and designated ORF2, encodes the capsid polyprotein. ORF1a and ORF1b overlap in 10 to 148 nucleotides (nt) in the genome of mammalian viruses, and between 10 and 45 nt in avian viruses. The overlapping region contains an essential signal for translation of the viral RNA polymerase (encoded in ORF1b) through a frameshift mechanism65 (see below).
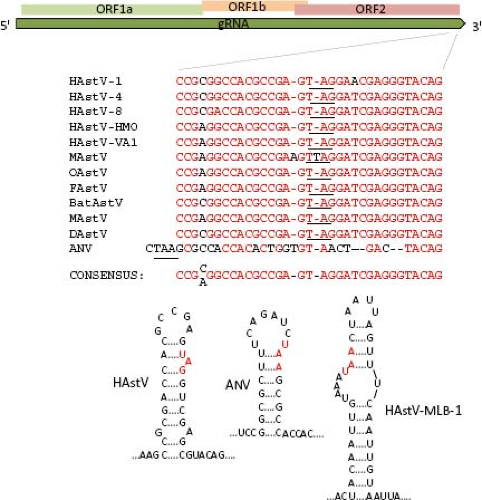
Two positive-sense RNA species have been identified in astrovirus-infected cells: the full-length genomic RNA (gRNA), and a subgenomic RNA (sgRNA) of ∼2.4 kb104 (see below, Transcription/Replication section). Based on the transcription initiation site determined for the sgRNA in HAstV-1 and HAstV-2,103,146 ORF1b and ORF2 overlap in 8 nt; however, the length of this overlapping may vary, and it is not present in DAstV; rather, it is 24 nt apart in this virus.37 The highly conserved sequence around the ORF2 start codon has been suggested to be part of the promoter for synthesis of the sgRNA,68 as described for alphaviruses; however, this region differs in length and sequence between mammalian and avian viruses (Fig. 21.5). This conserved sequence shows partial identity with the 5′ end of the gRNA,68 suggesting that it has an important role for the synthesis of both gRNA and sgRNA. Analysis of recombinant AstV isolated from different species has identified this region as a hot spot for recombination.132,140,148 Of note, the terminal 19 nt of ORF2 and the adjacent 3′-UTR are highly conserved among most
AstV, and similarities have been observed in the sequence and folding of the 3′-UTR of viruses from other families, such as avian infectious bronchitis virus (a coronavirus), a dog norovirus, and equine rhinovirus serotype 2 (a picornavirus),85,101 suggesting that it is relevant for astrovirus genome replication (Fig. 21.6).
AstV, and similarities have been observed in the sequence and folding of the 3′-UTR of viruses from other families, such as avian infectious bronchitis virus (a coronavirus), a dog norovirus, and equine rhinovirus serotype 2 (a picornavirus),85,101 suggesting that it is relevant for astrovirus genome replication (Fig. 21.6).
ORF1a and ORF1b
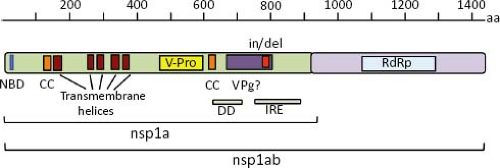
The polypeptide encoded by ORF1a (nsp1a) is 874 to 936 amino acids (aa) in length in most mammalian viruses, although it can be as short as 787 in a human isolate and can reach 1,240 in avian viruses (e-Table 21.1). Five to six helical transmembrane motifs followed by a viral serine protease motif (v-Pro) are predicted in nsp1a68 (Fig. 21.7). The v-Pro has features consistent with trypsin-like proteases, with a serine at the third catalytic amino acid residue.131 A viral protein genome-linked (VPg) encoded downstream of the protease motif has also been predicted based on its similarity with the VPg of calicivirus3; however, its synthesis in astrovirus-infected cells has not been investigated. Two predicted coiled-coil structures are present in nsp1a, one just upstream of the first helical transmembrane motif, and the second one downstream of the protease motif, suggesting that some protein products of nsp1a might form oligomers.68 One region containing insertion/deletions (in/del), located downstream of the VPg motif, was related to the efficiency of HAstV to synthesize viral RNA species and to adaptation to cultured cells55,144; however, it is not known whether the RNA sequence or the encoded protein is involved in modulating those events. ORF1b encodes a polypeptide of 515 to 539 aa that contains motifs of an RdRp, which are similar to RdRp of picornaviruses, caliciviruses, and certain plant viruses.3,65
Regions that could potentially encode an RNA helicase and a methyltransferase have not been identified in the astrovirus genome.65 Given the putative existence of a VPg, the absence of a methyltransferase could be understandable; however, the absence of an RNA helicase domain is unusual for a plus-strand RNA virus with a genome length of more than 6,000 nt, like astrovirus.65 The amino terminus of nsp1a shows similarity with an NTP binding motif of some helicases,3 but lacks other motifs, such as the substrate binding and NTP hydrolysis domains present in these enzymes.135
ORF2
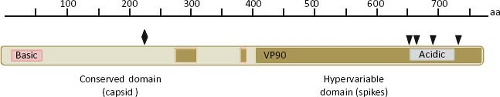
The largest sequence variability in the astrovirus genome is found in ORF2, which codes for the virus structural polyprotein. This polypeptide varies from 672 to 851 aa in length in TastV-1 and a porcine strain, respectively. In general, the ORF2 of avian viruses codes for shorter polyproteins (e-Table 21.1). The N-terminal half of the polyprotein is more conserved than the C-terminal domain, encompasses a region of basic character that is conserved among astroviruses,67,142 and is thought to interact with the genomic RNA in the virion.43 On the other hand, the C-terminal half of the protein shows considerable sequence variability among astroviruses isolated from different species, even among different isolates from the same animal species; the sequence of this region defines the serotypes in HAstV (Fig. 21.8). A delimited region with abundant insertion or deletions among different astroviruses is also present in the carboxy half of the capsid protein.142 The acidic character of a small portion of the protein, located close to the carboxy terminus (residues 649 to 702 for HAstV-8), is highly conserved among all members of the Astroviridae family.92 The last five amino acid residues at the C-terminal end
are conserved, particularly among human and some other mammalian strains.
are conserved, particularly among human and some other mammalian strains.
The conserved domain of the structural protein forms the capsid core of the particle, whereas the hypervariable C-terminal domain forms the spikes of the virion; thus, this last domain is predicted to participate in the early interactions of the virus with the host cell.76
An alternative ORF (named ORF-X) of 91 to 122 codons overlapping ORF2 in a +1 reading frame has been described in all HAstV and some other mammalian viruses36 (Fig. 21.4). Its initiation codon, usually located 41–50 nt downstream of the ORF2 AUG, is placed in a better Kozak’s consensus sequence than that of ORF2, and might be translated through a leaky scanning mechanism. It remains to be determined whether ORF-X is actually translated and, if it is, its significance for virus replication.
Stages of Replication
Attachment and Entry
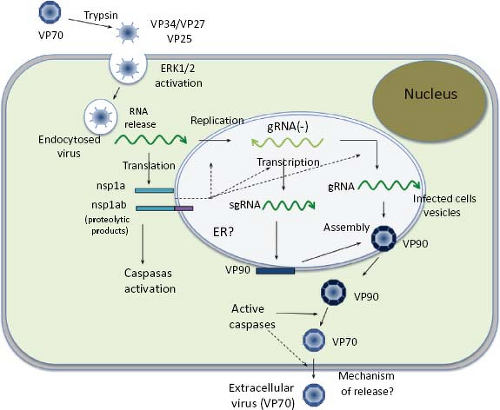
Studies directed toward understanding the early interactions of astroviruses with their host cells have been limited; however, a general view of the replication cycle can be depicted (Fig. 21.9). Cell receptor molecules for these viruses have not been identified. Different HAstV strains show different tropism in cultured cells19; thus, it is likely that their initial interactions with the host cells might be different.
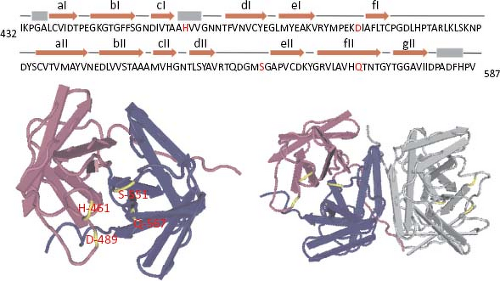
The infectivity of human astroviruses is greatly enhanced (3 to 5 logs) by, and probably requires, the treatment of the virus particles with trypsin.8,93,119 Although the proteolytic pathway of VP70 processing present in the virion has been elucidated for HAstV-8,93 the mechanism by which this treatment enhances virus infectivity is still unknown. Trypsin cleavage of the precursor polyprotein induces drastic structural changes in the particles8 (Fig. 21.3) and the generation in HAstV-8 of three final products: VP34, VP25, and VP27 (see details in the Protein Synthesis and Processing section). VP34 represents the conserved domain and forms the capsid core, while both VP25 and VP27 form the spikes on the virion surface. In spite of its high variability, VP25 contains two conserved structural motifs that have been suggested to be involved in the interaction of the virus with the host cell, one of which includes residues Lys455, Ser554, Thr575, and Glu610, predicted to interact with oligosaccharide moieties that could act as cell receptors or co-receptors (Fig. 21.10) (Dr. Y.J. Tao, unpublished data). Antibodies that recognize the spike proteins VP25 and VP27 neutralize virus infectivity,9,119 probably by blocking virus binding.
Early studies on HAstV entry in HEK-293 Graham cells using endocytosis-blocking agents (ammonium chloride, methylamine, and dansylcadaverine), as well as ultrastructural analysis, indicated that an endocytic, coated pit–dependent pathway is used by the virus to enter cells.30 Recently, a clathrin-dependent endocytosis was confirmed to be a functional pathway for the entry of HAstV-8 into Caco-2 cells (unpublished results).
The interaction of HAstV with the host cell provokes activation of the ERK1/2 signaling pathway within the first 15 minutes after virus attachment.107 Although the mechanism for this activation is unknown, it was independent of virus replication, suggesting that this pathway is triggered during virus binding or entry into the cell. Accordingly, ERK1/2 seems to be required at early times to establish a productive infection,
since inhibitors of this kinase blocked synthesis of viral proteins and RNA and, consequently, reduced virus yield. HAstV also induce an increase in the epithelial barrier permeability that is independent of virus replication, apparently due to an interaction between the capsid protein and the cell surface.105 It has been suggested that HAstV could trigger tight junction instability to reach putative receptors present in the basolateral membrane; however, the fact that the increase in permeability is observed after at least 16 hours of addition of the virus suggests that more than a requirement for infection, it is a consequence of the cellular transduction signal pathways induced by the virus.
since inhibitors of this kinase blocked synthesis of viral proteins and RNA and, consequently, reduced virus yield. HAstV also induce an increase in the epithelial barrier permeability that is independent of virus replication, apparently due to an interaction between the capsid protein and the cell surface.105 It has been suggested that HAstV could trigger tight junction instability to reach putative receptors present in the basolateral membrane; however, the fact that the increase in permeability is observed after at least 16 hours of addition of the virus suggests that more than a requirement for infection, it is a consequence of the cellular transduction signal pathways induced by the virus.
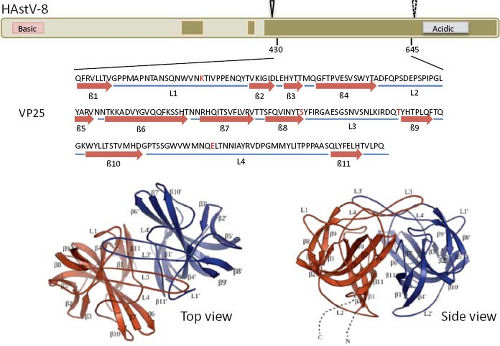 Figure 21.10. Three-dimensional structure of the spike protein VP25. The hypervariable region of HAstV-8 VP90, including residues 430 to 645, crystalized to 1.8 Å resolution as a dimer. The upper diagram shows the relative position of the crystalized protein; the cleavage carried out by trypsin is indicated (arrowheads, as in Fig. 21.15). Amino acid residues Lys455, Ser554, Thr575, and Glu610 (red in the sequence) are strictly conserved in all eight HAstV serotypes, and have been proposed as potentially interacting with cell surface carbohydrate motifs during the early virus–cell interactions. Top and side views of the spike are shown. (Courtesy of Dr. Y. J. Tao, Rice University.) |
Uncoating
The mechanism through which the viral genomic RNA is released from the infecting virus particle into the cytoplasm for translation, the cell site where it occurs, and the cellular and viral factors involved in this event are unknown.
Translation
No detectable changes in cellular protein synthesis are observed upon HAstV infection. As do most cellular mRNAs, astroviral RNA contains a polyA tail at the 3′ end, but the presence of a cap structure at its 5′ end has not been described. Since a VPg has been suggested to be encoded in astrovirus ORF1a,3 this protein could modulate the translation of viral mRNAs by interacting with translation initiation factors, as has been described for other viruses, such as calicivirus.23,29
Nonstructural Polyproteins Synthesis and Processing
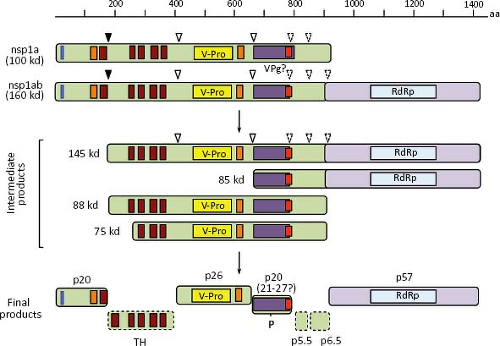
After uncoating, the gRNA is translated into nonstructural proteins that are produced as polyprotein precursors and subsequently proteolytically processed into smaller proteins. ORF1a directs the synthesis of protein nsp1a (of about 100 kd), whereas protein nsp1ab (160 kd) is derived from both ORF1a and ORF1b, through a frameshift mechanism of translation (see below). Proteins nsp1a and nsp1ab are apparently processed co-translationally at their amino terminus, so that the expected full-length proteins are not, or very rarely, observed in HAstV-infected cells (Fig. 21.11). Information regarding processing of the nonstructural polyproteins has been obtained by in vitro translation, transient expression of cDNA clones, and analysis of HAstV-infected cells, using antibodies to different regions of nsp1a and nsp1ab.41,45,95,145 No specific processing of nsp1a and nsp1ab were observed by in vitro translation of cDNA-derived transcripts,45 suggesting a requirement for cellular factors.
In HAstV-1-infected Caco-2 cells, Willcocks et al.145 detected proteins of 74, 34, 20, 6.5, and 5.5 kd with antibodies raised to the predicted C-terminal 298 aa of nsp1a. Proteins of 20 and 74 kd—in addition to products of 88, 27, and 19 kd—were also detected in Caco-2 cells infected with an HAstV-8 strain using antibodies to several regions of nsp1a.95 The 19- and 20-kd proteins likely represent the most N-terminal products of nsp1a, apparently cleaved out by a cellular protease, probably a signalase, because it is produced by in vitro translation experiments only in the presence of microsomes,91 it occurs co-translationally,41,95 and it is independent of the v-Pro activity.41 The 27-kd protein represents the v-Pro and probably spans from around amino acid residue 410 to 654 of nsp1a.41 Proteins of 160, 75, and 38 to 40 kd, and phosphorylated forms of 21 to 27 kd were also detected with antibodies to a synthetic peptide that comprises aa residues 778 to 792 of nsp1a.54
Translation of the RdRp occurs through a ribosomal-1 frameshift mechanism in the overlapping region between ORF1a and ORF1b. The signal that modulates this event has two key features, conserved among all astroviruses: a heptameric sequence (AAAAAAC) and the potential to form a downstream stem-loop structure18 (Fig. 21.12). Translation of heterologous proteins, with the HAstV frameshift signal included, revealed that its efficiency varies between
7% and 28%, depending on the system in which it was evaluated84; however, the efficiency of the frameshift has not been determined in HAstV-infected cells. Antibodies raised to recombinant proteins containing the RdRp motif identified a protein of 57 kd as the final product in Caco-2 cells infected with an HAstV-8 strain.95
7% and 28%, depending on the system in which it was evaluated84; however, the efficiency of the frameshift has not been determined in HAstV-infected cells. Antibodies raised to recombinant proteins containing the RdRp motif identified a protein of 57 kd as the final product in Caco-2 cells infected with an HAstV-8 strain.95
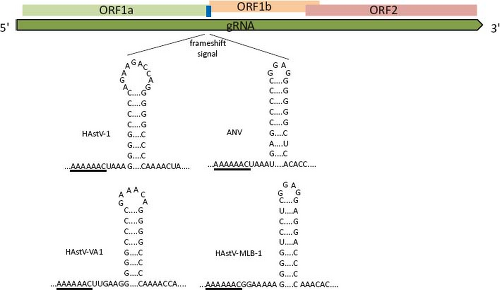 Figure 21.11. The frameshift signal (fss) of astroviruses is composed by a slippery sequence (underlined) and a stem-loop structure. The length and sequence of the loop and of the region between the slippery motif and stem-loop structure are not important for its function,18 as seen for the viruses shown. |
In summary, proteins of 27, 20, 19, 6.5, and 5.5 kd, as well as the phosphorylated 21 to 27 kd polypeptides, seem to represent the final processing products of polyprotein nsp1a, whereas a 57 to 59 kd protein represents the mature protein derived from ORF1b. The proteins observed of 145, 88, 85, 75, and 34 kd most likely represent intermediate products of the nonstructural protein processing (Fig. 21.11). With exception of the cleavage that releases the 20 kd most amino-terminal polypeptide, all other cleavages at nsp1a and nsp1ab polyproteins are believed to depend on the v-Pro activity, although only those identified at around amino acid residues 410 and 655 of nsp1a have been confirmed.41 Crystal analysis of the v-Pro domain (Fig. 21.13) showed that the enzyme contains a basic S1 pocket that recognizes and cleaves acidic residues (Glu and Asp) at position P1.131 Thus, Asp-413, but not Ala-409, and Glu-654 seem to be the actual residues cleaved by v-Pro on nsp1a. The C-terminal region of nsp1a contains many acidic residues that prevent the prediction of cleavage sites downstream of Glu-654 recognized by the viral enzyme to generate the 6.5 and 5.5 kd products observed.
Structural Polyprotein Synthesis and Processing
The structural proteins of HAstV, encoded in ORF2, are synthesized from the sgRNA as a polyprotein precursor of 87 to 90 kd. Studies with HAstV-8 have revealed that the 90-kd primary translation product is intracellularly cleaved at Asp-657 to yield a product of 70 kd (named VP70)7 through intermediates of 75 to 85 kd,94 whose biological relevance, if any, is unknown (Fig. 21.14). Processing of VP90 to VP70 is carried out by cellular enzymes (caspases) that are involved in apoptotic processes,7 and whose activity is triggered during viral infection by an unknown mechanism.7,53 The pan-caspases inhibitor Z-VAD-fmk strongly inhibits this processing, whereas specific inhibitors of caspase activity partially block it. On the contrary, some proapoptotic factors—such as TNF-related apoptosis-inducing ligand (TRAIL) and staurosporine, which triggers different apoptotic pathways—enhance the cleavage of VP90 to VP70.94 It is believed that caspases are important for processing the structural polyprotein precursor in most, if not all, astroviruses since the caspase-recognition motifs present at the carboxy-terminal region of VP90 are highly conserved among different strains. For HAstV-8, caspase-3 and caspase-9 seem particularly important for processing VP90; however, other caspases might also be involved, since VP90 is substrate of caspase-8 and caspase-4 in vitro.7 Also, it cannot be excluded that different caspases are responsible for cleavage of VP90 from different astrovirus strains, since the motifs recognized by caspases in this precursor protein vary among viruses. Of interest, the VP90 to VP70 processing is not required for VP90 to assemble as viral particles, but it is important for the egress of the virus from infected cells7,91 (see below).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree