Arteriviruses
Eric J. Snijder
Marjolein Kikkert
History and Classification of Arteriviruses
The family Arteriviridae68 was established in 1996 and currently comprises the following four enveloped, plus-stranded RNA viruses: equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV) of mice, porcine reproductive and respiratory syndrome virus (PRRSV), and simian hemorrhagic fever virus (SHFV). Three of these (EAV, LDV, and SHFV) were first isolated and characterized about 50 years ago.62,161,189 The porcine arterivirus PRRSV emerged only about 20 years ago,36,227 causing vast epidemics of a previously unknown reproductive and respiratory disease in swine in both Europe (genotype I) and North America (genotype II). Remarkably, the subsequent molecular characterization of PRRSV strains from both continents revealed considerable genetic differences, suggesting that the two PRRSV genotypes evolved separately and are only distantly related to a common ancestor.152,170 PRRSV infection can cause high-mortality disease outbreaks and has developed into the most prevalent disease of swine worldwide. Recently, a large outbreak of highly virulent PRRSV affected the Asian pig industry, causing enormous economic losses.194,246
In general, the consequences of arterivirus infection can range from an asymptomatic, persistent or acute infection to abortion or lethal hemorrhagic fever.199,226 EAV is capable of inducing a variety of symptoms, including necrosis of the small muscular arteries from which the name of the family prototype EAV was derived. The name of the mouse arterivirus LDV is derived from the increase in the level of lactate dehydrogenase (LDH) caused by LDV infection.161 The virus, which has been used extensively as an in vivo research model, is able to escape immune surveillance and establish a largely asymptomatic persistent infection.22,155 SHFV was isolated from outbreaks of fatal hemorrhagic fever in macaque colonies189 that were probably caused by inadvertent transmission by humans from African monkeys to macaques.
The unification of the previously unclassified arteriviruses was the direct result of the sequence analysis of their genomes, which revealed an intriguing relationship with coronaviruses and toroviruses (discussed in Chapter 28). Despite striking differences in genome size and virion structure, the genome organization and expression strategy of these viruses were found to be comparable and their replicase genes were postulated to share common ancestry54 (e-Fig. 29.1). One of the most prominent features of their genome expression strategy, the generation of a nested set of subgenomic (sg) messenger RNAs (mRNAs), provided the basis for the name Nidovirales
(L. nidus = nest) that was given to the novel virus order comprising the arterivirus and coronavirus families in 1996. Subsequently, the order was further expanded with the invertebrate virus family Roniviridae and the genus Bafinivirus, which contains fish nidoviruses.83 Most recently, the isolation of the first insect nidoviruses (proposed family name Mesoniviridae) was reported,140,249 yet again expanding the exceptional host range of the order Nidovirales. Furthermore, on the basis of its partial genome sequence, a novel nidovirus isolated from Australian possums (wobbly possum disease virus) appears to represent yet another nidovirus lineage, which is relatively closely related to the Arteriviridae.66a
(L. nidus = nest) that was given to the novel virus order comprising the arterivirus and coronavirus families in 1996. Subsequently, the order was further expanded with the invertebrate virus family Roniviridae and the genus Bafinivirus, which contains fish nidoviruses.83 Most recently, the isolation of the first insect nidoviruses (proposed family name Mesoniviridae) was reported,140,249 yet again expanding the exceptional host range of the order Nidovirales. Furthermore, on the basis of its partial genome sequence, a novel nidovirus isolated from Australian possums (wobbly possum disease virus) appears to represent yet another nidovirus lineage, which is relatively closely related to the Arteriviridae.66a
Nidoviruses represent a distinct lineage among plus-strand RNA viruses (e-Fig. 29.1). The complex evolutionary relationship between arteriviruses and nidoviruses with a much larger genome has been reviewed extensively elsewhere.83 Related replicase genes and replication strategies have been combined with seemingly unrelated sets of structural protein genes. RNA recombination likely was an important factor in these evolutionary events and was also invoked to explain some internal rearrangements of arterivirus genomes.52,81,103
Virion Structure
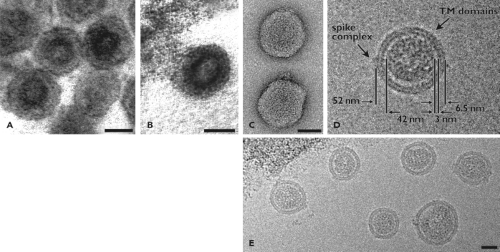
Arteriviruses have been observed as spherical particles, 50 to 60 nm in diameter, and possess a relatively smooth, mostly featureless surface, which is likely explained by the small ectodomains of the two major envelope proteins (Fig. 29.1; see20,68,175,182 and references therein). The nucleocapsid structure has long been assumed to be isometric, but recent cryo-electron tomography studies of PRRSV revealed a rather pleomorphic and “disorganized” core structure (average diameter 39 nm). These findings are clearly incompatible with an icosahedral core and suggest a resemblance to the nucleocapsid structure proposed for coronaviruses, a helical coil, or an even more loosely organized filamentous structure61,182
The buoyant density of arteriviruses is 1.13 to 1.17 g/cm3 in sucrose, and their sedimentation coefficient ranges from 214S to 230S. Virions are highly unstable in solutions containing low concentrations of nonionic detergents or at a pH other than 6.0 to 7.5, and quickly lose their infectivity when stored at temperatures higher than 4°C.
The arterivirus nucleocapsid structure (Fig. 29.2, e-Fig. 29.2) is composed of the 12.7 to 15.7 kb RNA genome and the nucleocapsid protein (N). The crystal structure for the capsid-forming C-terminal domain of PRRSV N59; e-Fig. 29.2B) suggested that it represents a new class of viral capsid–forming domains, a hypothesis further supported by cryo-EM studies.
Based on studies with EAV and PRRSV, the lipid bilayer that surrounds the nucleocapsid is now presumed to contain seven envelope proteins (Table 29.1, Fig. 29.2), an unusually large number compared to other plus-stranded RNA viruses. In this chapter, we refer to the glycoproteins as “GPx”, where x indicates the number of the corresponding open reading frame
(ORF) in the genome (Table 29.1, Figs. 29.2 and 29.3). For simplicity, the GP encoded by ORF2a (PRRSV) or ORF2b (EAV/LDV) will be called GP2. Among arterivirus envelope proteins two major and five minor species are discriminated. The two major species, the nonglycosylated triple-spanning membrane protein M and GP5, form a disulfide-linked heterodimer57,69,174 (Fig. 29.2). By separately knocking out the expression of each of the structural proteins, it was established that all major and minor structural proteins are required for the production of infectious progeny,131,232 with the possible exception of the recently discovered ORF5a protein.75,94
(ORF) in the genome (Table 29.1, Figs. 29.2 and 29.3). For simplicity, the GP encoded by ORF2a (PRRSV) or ORF2b (EAV/LDV) will be called GP2. Among arterivirus envelope proteins two major and five minor species are discriminated. The two major species, the nonglycosylated triple-spanning membrane protein M and GP5, form a disulfide-linked heterodimer57,69,174 (Fig. 29.2). By separately knocking out the expression of each of the structural proteins, it was established that all major and minor structural proteins are required for the production of infectious progeny,131,232 with the possible exception of the recently discovered ORF5a protein.75,94
Table 29.1 Molecular Properties of Arteriviruses | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Studies of GP2, GP3, and GP4, which form a heterotrimer in the virion230,232 (Fig. 29.2), have further highlighted their importance. Knockout mutants for minor structural protein genes produced noninfectious subviral particles consisting of GP5, M, N, and the genome RNA.231,232,243 When one of the
components of the GP2-GP3-GP4 trimer or the small nonglycosylated envelope protein (E)176 was lacking, the incorporation of the three minor GPs into virions was blocked.231 Taken together, these data indicate that the basic protein scaffold of the arterivirus particle consists of the three major structural polypeptides, N, M, and GP5. Whether the incorporation of (genome) RNA is essential for the formation of the nucleocapsid structure, and which RNA sequences/structures specifically interact with N, remains to be established.
components of the GP2-GP3-GP4 trimer or the small nonglycosylated envelope protein (E)176 was lacking, the incorporation of the three minor GPs into virions was blocked.231 Taken together, these data indicate that the basic protein scaffold of the arterivirus particle consists of the three major structural polypeptides, N, M, and GP5. Whether the incorporation of (genome) RNA is essential for the formation of the nucleocapsid structure, and which RNA sequences/structures specifically interact with N, remains to be established.
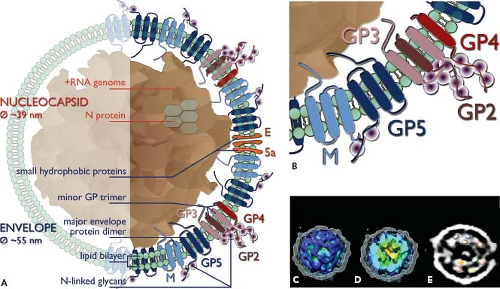 Figure 29.2. Arterivirus structure. A: The presumed location and topology of the envelope proteins GP2 to GP5, E, and M, the recently identified open reading frame (ORF)5a protein, and the N protein are shown (see also Table 29.1 and Fig. 29.3). The major envelope proteins GP5 and M form a disulfide-linked heterodimer. The minor glycoproteins GP2, GP3, and GP4 form a disulfide-linked heterotrimer. Seen Panel B for a close-up. In addition, also GP2-GP4 dimers (not depicted) have been identified in equine arteritis virus (EAV) particles. It should be noticed that not all proteins depicted here have been identified in all four arterivirus particles. C–E: Cryo-electron microscopy–based tomographic reconstruction of a porcine reproductive and respiratory syndrome virus (PRRSV) particle,182 revealing that the virion core is not solid, but consists of a two-layered shell that surrounds a hollow central cavity. C: Cutaway view of the internal core, obtained by peeling away the envelope (shown in mesh representation). The core, which is separated from the envelope by a 3-nm gap, appears disorganized and to consist of density strands that are bundled together into a ball. The data suggest a model for the core in which two layers of N dimers form a linked chain (see also Panel E). The core is shown as an isosurface, colored by the radius from the center of the particle (from red to blue). D: The core has been cut open to show the internal structure, including the central density (red-orange) typically seen in the tomograms. E: A 6.3-nm thick slab through the center of one PRRSV particle tomogram, with several copies of the crystal structure of the dimeric C-terminal domain of N, rendered at a comparable resolution and superimposed on the oblong densities in the core. (See also e-Fig. 29.2). (C–E from Spilman MS, Welbon C, Nelson E, et al. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J GenVirol 2009;90:527–535, with permission.) |
Genome Structure and Organization
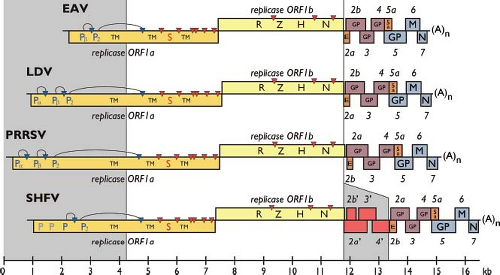
The arterivirus genome is a plus-stranded, 3′-polyadenylated RNA molecule, likely containing a cap structure at its 5′ end.165 Full-length genomic sequences (see also Table 29.1) have been obtained for European and North American isolates of EAV, a large number of European, North American, and Asian PRRSV isolates, two LDV strains, and three SHFV isolates. The arterivirus replicase gene consists of the large ORFs 1a and 1b and roughly occupies the 5′ three-fourths of the polycistronic genome (Fig. 29.3). In contrast to the more conserved ORF1b region, the size of ORF1a is variable (encoding between 1,727 [EAV] and about 2,500 amino acids [PRRSV]), which largely explains the genome size differences encountered among arteriviruses. The region downstream of the replicase gene contains 8 to 11 relatively small genes, most of which have both 5′- and 3′-terminal sequences that overlap with neighboring genes. These genes encode mostly (or exclusively) structural proteins and are translated from sg mRNAs (see below). Their organization is
generally well conserved in the arterivirus genome. An exception is the region downstream of the SHFV replicase gene, which contains four additional ORFs, comprising about 1.6 kb, which may have arisen from the duplication of ORFs 2a to 4.68,81
generally well conserved in the arterivirus genome. An exception is the region downstream of the SHFV replicase gene, which contains four additional ORFs, comprising about 1.6 kb, which may have arisen from the duplication of ORFs 2a to 4.68,81
The Arterivirus Replication Cycle
Attachment and Entry
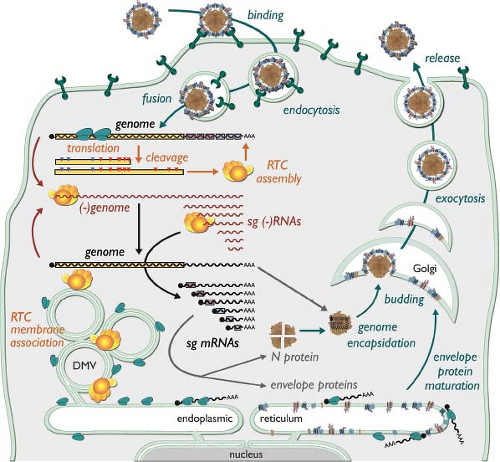
The entry of PRRSV and EAV requires a low pH, suggesting that it occurs via the standard endocytic route101,137,141 (Fig. 29.4). Clathrin heavy-chain knockdown suppressed EAV infection141 and electron microscopy revealed arterivirus particles contained in relatively small vesicles that appeared to be clathrin coated.100,101
The host factors required for arterivirus entry have been studied in detail only for PRRSV (e-Fig. 29.3). Several viral and cellular players have been implicated in binding, entry, and uncoating, although their exact roles remain to be defined in more detail (for recent reviews see 203,224). Sialoadhesin (or sialic acid-binding immunoglobulin [Ig]-like lectin 1 [CD169]; 216), a macrophage-restricted membrane protein, mediates the internalization of the virus by porcine alveolar macrophages (PAMs), the primary target cells of PRRSV.64,66 In addition, glycosaminoglycans (heparan sulfate) on the cell surface51 and sialic acids on the virion surface50 were implicated in the initial binding step. It is believed that the virus initially binds heparin-like molecules on the cell surface and that subsequently internalization via clathrin-mediated endocytosis is triggered by the interaction of CD169 with sialic acids on the ectodomains of the GP5/M dimer.204 Expression of porcine CD169 in nonsusceptible cell lines can mediate PRRSV internalization, but not disassembly and productive infection,216 indicating that additional factors must be required for successful infection. This notion is further supported by the fact that MARC-145 cells, which are commonly used to grow PRRSV, do not express CD169 on their surface.64 In particular CD163, a member of the scavenger receptor cysteine-rich (SRCR) family, was implicated in the early stages of PRRSV infection.26 Although normally a macrophage-specific antigen, CD163 is aberrantly expressed on MARC-145 cells, possibly explaining their unique susceptibility to PRRSV infection among nonengineered cell lines. Expression of CD163 from various species rendered a variety of nonpermissive cell lines susceptible to PRRSV infection, in the absence
of detectable CD169 expression. It was postulated that in PAM, CD169 and CD163 work together, with the former serving as receptor for internalization and the latter playing a key role in virus uncoating and genome release, which are thought to occur in association with the early endosome, following its acidification211 (e-Fig. 29.3).
of detectable CD169 expression. It was postulated that in PAM, CD169 and CD163 work together, with the former serving as receptor for internalization and the latter playing a key role in virus uncoating and genome release, which are thought to occur in association with the early endosome, following its acidification211 (e-Fig. 29.3).
Which virion proteins direct the fusion between viral envelope and endosomal membrane remains one of the key questions to be addressed. A role for the minor glycoproteins in arterivirus receptor recognition and tropism has not been rigorously excluded.44 In fact, recent data show that a chimeric PRRSV carrying E, GP2, GP3, and GP4 of EAV acquired the broad tropism for cultured cells that is typical of the latter virus.193 These findings are in line with the previously observed phenotypes of recombinant viruses in which the GP5 or M ectodomain was replaced, which did not result in an altered tropism in cell culture.60,219 These studies, together with the identification of several other host factors as potential “PRRSV entry mediators”,203,224 illustrate that several questions and controversies regarding arterivirus entry remain to be addressed.
Genome Translation and Replication
The arterivirus replication cycle (Fig. 29.4) is presumed to be entirely cytoplasmic, despite the fact that at least two viral proteins are (in part) targeted to the nucleus (see below). The
incoming genome is translated into the two large replicase polyproteins pp1a (1,727 to 2,502 amino acids) and pp1ab (3,175 to 3,959 amino acids), which comprise all functions required for viral RNA synthesis.131 Despite the relatively large 5′ nontranslated region (NTR), translation presumably initiates following “conventional” ribosomal scanning of the genomic 5′ NTR.206 ORF1b translation requires a −1 ribosomal frame shift (estimated efficiency of 15% to 20%) just before ORF1a translation is terminated54 (Fig. 29.3). The ORF1a/1b overlap region contains two signals that are assumed to promote this event: a so-called “slippery” sequence, which is the actual ribosomal frame shift site, and a downstream RNA pseudoknot structure.
incoming genome is translated into the two large replicase polyproteins pp1a (1,727 to 2,502 amino acids) and pp1ab (3,175 to 3,959 amino acids), which comprise all functions required for viral RNA synthesis.131 Despite the relatively large 5′ nontranslated region (NTR), translation presumably initiates following “conventional” ribosomal scanning of the genomic 5′ NTR.206 ORF1b translation requires a −1 ribosomal frame shift (estimated efficiency of 15% to 20%) just before ORF1a translation is terminated54 (Fig. 29.3). The ORF1a/1b overlap region contains two signals that are assumed to promote this event: a so-called “slippery” sequence, which is the actual ribosomal frame shift site, and a downstream RNA pseudoknot structure.
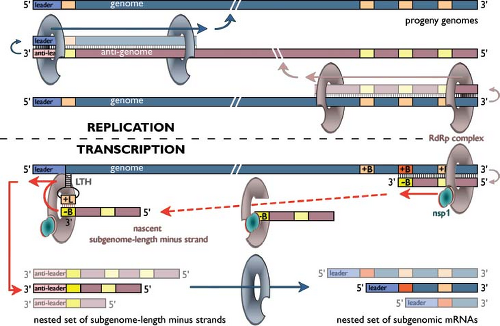 Figure 29.5. Arterivirus RNA synthesis. Model for arterivirus (and coronavirus) replication and transcription146,166,167,168 using a hypothetical arterivirus genome encoding three subgenomic messenger RNAs (mRNAs). The top half of the scheme depicts the replication of the genome by the viral RNA-dependent RNA polymerase (RdRp) complex, which requires a full-length minus-strand intermediate (anti-genome). The bottom half illustrates how minus-strand RNA synthesis can be interrupted at a body transcription-regulating sequence (TRS) (+B), after which the nascent minus strand, having a body TRS complement (-B) at its 3′ end, is redirected to the leader TRS (+L) near the 5′-end of the genome. This +L sequence is thought to be “presented” for base pairing by a viral RNA structure, the leader TRS hairpin (LTH), that is critical for subgenomic RNA synthesis. Guided by a base-pairing interaction between the complementary –B and +L sequences, RNA synthesis is resumed to add the complement of the genomic leader sequence (anti-leader) to each nascent subgenome-length minus strand. Subsequently, the subgenome-length minus strand RNAs each serve as template to produce one of the subgenomic mRNAs. The RdRp complexes engaged in replication and transcription may be (partially) different. For example, in the equine arteritis virus (EAV) model, nsp1 has been identified as a regulatory factor that is dispensable for replication but required to regulate the accumulation levels of the different subgenomic RNAs, most likely by controlling a step during minus strand RNA synthesis. See text for more details. |
Following proteolytic processing of the replicase polyproteins, a complex for viral RNA synthesis is formed that generates a genome-length minus strand (or “anti-genome”), the template for genome replication. In addition, a complex transcription mechanism operates to produce complementary nested sets of sg-length minus-strand RNAs and sg mRNAs55 (see below and Fig. 29.5). The RNA signals involved in arterivirus genome replication remain to be studied in detail. The coding regions of the genomes are flanked by 5′ and 3′ NTRs of 156 to 221 and 59 to 117 nucleotides, respectively. However, natural and synthetic defective interfering RNAs of EAV invariably require at least 300 nucleotides from both genome termini for efficient replication, indicating that replication signals extend into the coding sequences.130,198 Likewise, in the case of PRRSV, a so-called “kissing interaction” between the loop sequences of RNA hairpin structures in the 3′ NTR and the N protein gene was found to be crucial for viral RNA synthesis.218
Using a combination of approaches, detailed RNA secondary structure models were developed for the EAV 5′ and 3′ NTRs. In the 5′ NTR, a region involved in translation, replication, and transcription,205 (e-Fig. 29.4B), one domain in particular was found to be crucial for sg RNA production (see below). This so-called “leader TRS [transcription-regulating sequence] hairpin” (LTH) is potentially conserved in the 5′ NTR of all arteriviruses (e-Fig 29.4C). The importance of the other structural features of the EAV 5′ NTR and, for example, their involvement in RNA–protein interactions, remains to be investigated because few of these elements are conserved in other arteriviruses.205 A possible exception is EAV hairpin C (termed SL2 in PRRSV; 115) that was reported to be crucial for PRRSV replication and subgenomic RNA synthesis in particular (e-Fig. 29.4C).
The 3′ NTR of the arterivirus genome does not contain obviously conserved primary sequences. For EAV, the 3′-terminal CC motif immediately upstream of the poly(A) tail plays a critical role in viral RNA synthesis.15 Furthermore, a stem-loop structure near the 3′-terminus of the EAV genome is also required for RNA synthesis14 (e-Fig. 29.5A) and its loop was implicated in an essential pseudoknot interaction with an upstream stem-loop structure residing in the N protein gene.15 This conformation was predicted to be conserved in all arteriviruses and proposed to constitute a molecular switch that could regulate the specificity or timing of viral (minus strand) RNA synthesis (e-Fig. 29.5B).
Synthesis and Translation of Subgenomic mRNAs
One of the hallmarks of the replication cycle of arteriviruses (and other nidoviruses) is the synthesis of a 3′-co-terminal nested set of sg mRNAs (Fig. 29.5) from which the genes in the 3′ end of the genome are expressed. In the case of arteriviruses, all these genes encode structural proteins. Arterivirus sg mRNAs also have a common 5′ end, the so-called “leader sequence,” which is derived from the 5′ end of the genome.55 This property is shared with coronaviruses, but—remarkably—not with some other nidoviruses (toroviruses and roniviruses; for reviews, see 83,146,168). Supported by the presumed common ancestry of the arterivirus and coronavirus replicase genes, leader-to-body fusion during arterivirus sg RNA synthesis was proposed to rely on a mechanism of discontinuous RNA synthesis similar to that previously proposed for coronaviruses. In both virus groups, short conserved TRSs are present at the 3′ end of the leader sequence (“leader TRS”) and at the 5′ end of each of the transcription units specifying a sg mRNA “body” (“body TRS”; reviewed in 146,168,175). The observation that arterivirus-infected cells contain a nested set of sg-length minus-strand RNAs, complementary to the sg mRNAs, is another important parallel with coronaviruses.30,53
With the exception of the smallest species, the arterivirus sg mRNAs are structurally polycistronic, but most of them are assumed to be functionally monocistronic. Notable exceptions are mRNAs 2 and 5 (in EAV, LDV, and PRRSV; Fig. 29.3), which are functionally bicistronic transcripts from which the partially overlapping gene sets E/GP2 and ORF5a/GP5 are expressed.75,94,176 The mRNAs tentatively numbered 4 and 6 are thought to be used to translate the corresponding SHFV gene sets, and also mRNA2 of this virus was proposed to be functionally bicistronic.81
A substantial number of models for coronavirus and arterivirus sg mRNA synthesis have been proposed and reviewed extensively (see also Chapter 28; 146,168,175, and references therein). The detection of sg-length minus strands indicated that the discontinuous step in sg RNA synthesis likely occurs during minus-strand RNA synthesis. This concept was subsequently supported by data from biochemical and genetic studies with coronaviruses and arteriviruses and resulted in a model (Fig. 29.5; e-Fig. 29.4) in which discontinuous extension of minus-strand RNA synthesis yields sg-length minus-strand templates for sg mRNA synthesis.166,168
Direct proof for base-pairing between leader TRS and antibody TRS was obtained from reverse genetics studies using an EAV infectious complementary DNA (cDNA) clone.147,214 The mechanism by which the transcriptase is translocated between the body and leader TRS in the genomic template, a step that may resemble copy-choice RNA recombination,19,147,214 remains to be elucidated. Arterivirus sg RNAs are produced in nonequimolar, but relatively constant amounts, thus providing a mechanism to regulate the expression of the various structural protein genes. EAV reverse genetics studies have rigorously demonstrated that transcription depends on duplex formation between leader TRS and anti-body TRS and that—in general—the relative amount of sg mRNA correlates with the calculated stability of this duplex.147,148,214 Sequences flanking the body TRS, the relative order and/or location of body TRSs in the genome, and possibly also higher order RNA structure were also shown or postulated to influence transcription.145 Structural studies on the 5′-proximal part of the EAV genome205 placed the leader TRS in a single-stranded loop of the structure referred to as the “leader TRS hairpin” (or LTH; e-Fig. 29.4B,C), which was characterized as a critical player in transcription.206
At the protein level, transcription-specific functions have been attributed to several replicase subunits, in particular nonstructural protein 1 (nsp1) and nsp10, for which mutations were described that resulted in the (near) complete inactivation of sg mRNA synthesis.138,196,198,208 EAV nsp1 controls the accumulation levels of viral genome and individual sg mRNAs in the infected cell by determining the levels at which the minus-strand templates for each of these molecules are produced.138 An N-terminal zinc finger (ZF) domain was implicated in this function, but also other nsp1 domains appear to be important. Mutagenesis of nsp1 triggered the evolution of numerous nsp1 pseudorevertants with compensatory mutations that invariably rescued both balanced EAV mRNA accumulation and efficient virus production.138 In the case of PRRSV, where nsp1 is internally cleaved into nsp1α and nsp1β, the ZF-containing nsp1α subunit is presumed to fulfill a similar role in transcription regulation.102
Arterivirus Proteinases and Posttranslational Processing of the Replicase
The proteolytic maturation of the arterivirus pp1a and pp1ab replicase polyproteins involves the rapid autoproteolytic release of three or two N-terminal nsps and the subsequent cleavage of the remaining part of both polyproteins by the viral nsp4 “main protease.” The posttranslational processing of the
replicase polyproteins has been studied most extensively for EAV (see 202,248, and references therein; e-Fig. 29.6A,B) for which pp1a and pp1ab are cleaved 8 and 11 times, respectively, by three ORF1a-encoded proteinases (see below). In combination with the ORF1a/1b ribosomal frame shift, this yields 13 processing end products (named nonstructural protein [nsp] 1 to 12, including nsp7α and nsp7β; Fig. 29.3; e-Fig. 29.6E). Of these, nsp1-8 are generated from ORF1a, whereas nsp10-12 are entirely ORF1b-encoded and nsp9, due to the ribosomal frame shift consisting of a small, ORF1a-encoded N-terminal domain (identical to nsp8) and a large C-terminal part that is encoded by ORF1b and includes the viral RNA-dependent RNA polymerase (RdRp) domain. EAV reverse genetics studies with cleavage site mutants underscored the critical importance of replicase polyprotein processing for virus replication.202,209 The nsp3-8 region of pp1a (and likely also pp1ab) is subject to two alternative processing cascades, with the “major pathway” requiring an interaction with nsp2 as a cofactor, to mediate cleavage of the nsp4/5 site222 (e-Fig. 29.6D).
replicase polyproteins has been studied most extensively for EAV (see 202,248, and references therein; e-Fig. 29.6A,B) for which pp1a and pp1ab are cleaved 8 and 11 times, respectively, by three ORF1a-encoded proteinases (see below). In combination with the ORF1a/1b ribosomal frame shift, this yields 13 processing end products (named nonstructural protein [nsp] 1 to 12, including nsp7α and nsp7β; Fig. 29.3; e-Fig. 29.6E). Of these, nsp1-8 are generated from ORF1a, whereas nsp10-12 are entirely ORF1b-encoded and nsp9, due to the ribosomal frame shift consisting of a small, ORF1a-encoded N-terminal domain (identical to nsp8) and a large C-terminal part that is encoded by ORF1b and includes the viral RNA-dependent RNA polymerase (RdRp) domain. EAV reverse genetics studies with cleavage site mutants underscored the critical importance of replicase polyprotein processing for virus replication.202,209 The nsp3-8 region of pp1a (and likely also pp1ab) is subject to two alternative processing cascades, with the “major pathway” requiring an interaction with nsp2 as a cofactor, to mediate cleavage of the nsp4/5 site222 (e-Fig. 29.6D).
The three EAV proteinase domains in nsp1, nsp2, and nsp474,175,248 (e-Fig. 29.6) and their corresponding cleavage sites are well conserved in the other arteriviruses (Fig. 29.3). EAV nsp1 and nsp2 both contain a papain-like proteinase domain (PLP; formerly referred to as PCP or CP for [papain-like] cysteine protease) that mediates their rapid release from the polyprotein,178 whereas nsp4 includes a chymotrypsin-like serine proteinase (SP), the arterivirus main proteinase.180 PRRSV and LDV, in addition to having homologs of these three EAV proteinases, possess a fourth nonstructural proteinase,52 which mediates the rapid release of an additional N-terminal cleavage product. This PLPα possibly is a duplication of the proteinase (PLPβ) present in the C-terminal domain of EAV nsp1 and appears to have become inactivated in EAV.52 The sequence analysis of the SHFV nsp1 region revealed an even more complex situation, with an array of three potential PLP domains present in the 480-residue region upstream of the (predicted) nsp1/nsp2 junction. The nsp4 SP combines the His-Asp-Ser catalytic triad of classical chymotrypsin-like proteinases with the substrate specificity of the so-called 3C-like cysteine proteinases, a subgroup of chymotrypsin-like enzymes named after the picornavirus 3C proteinases. Specific residues in the substrate-binding region of the SP are assumed to determine its specificity for cleavage sites containing Glu (or sometimes Gln) as the P1 residue and mainly Gly, Ala, or Ser at the P1′ position. Nine such sites were identified in EAV pp1a/pp1ab, and they were all found to be conserved in the other family members.175,202,248
Nsp4 structures have been obtained by x-ray crystallography for both EAV and PRRSV11,195 (e-Fig. 29.7). The protein consists of three domains, with domains I and II forming the typical chymotrypsin-like two-β-barrel fold of the SP. The C-terminal domain III is dispensable for proteolytic activity and may be involved in fine-tuning replicase polyprotein cleavage.200,201 Recent structural studies also elucidated the structures of PRRSV nsp1α186 and nsp1β,238 including their respective PLP domains (e-Fig. 29.7), which—in line with previous studies—were both confirmed to employ a Cys-His tandem as active site residues. Both PLPα and PLPβ appear to act exclusively in cis and the two structures indeed revealed the presence of the C-terminal region of the proteins in the PLP substrate-binding pocket, suggesting an intramolecular cleavage mechanism that would preclude further proteolytic reactions. Both nsp1α and nsp1β of PRRSV have also been implicated in evasion of the host’s immune response (see below), but this was most directly demonstrated for the PLP that resides in the N-terminal domain of the highly variable nsp2 subunit. This PLP2, which possesses both cis and trans cleavage activities,86,179 not only directs the critical cleavage of the nsp2/3 site in pp1a and pp1ab, but is also able to remove ubiquitin (Ub) and Ub-like modifiers like ISG15 from yet-to-be-identified substrates in the infected cell.213 The protein is distantly related to the ovarian tumor domain (OTU) family of deubiquitinating enzymes.76,123,187
Replicase Proteins and the Replication Complex
Although accelerated by research efforts following the emergence of severe acute respiratory syndrome (SARS)-coronavirus, the functional dissection of the complex array of nidovirus nonstructural protein functions is still in its infancy. Even the arterivirus replicase polyproteins are of extraordinary size and complexity, despite their twofold smaller size compared to nidoviruses with larger genomes like coronaviruses and roniviruses. Therefore, future studies will undoubtedly reveal both novel similarities and differences between these two groups. In arteriviruses, with the notable exception of the role of nsp1 in sg mRNA synthesis (see above), the ORF1a-encoded functions mainly appear important for the regulation of replicase gene expression (by proteolytic processing; see above) and formation of the membrane-anchored “scaffold” for the replication/transcription complex. The ORF1b-encoded proteins, on the other hand, appear to be more directly involved in viral RNA synthesis.
Except for the proteins from the nsp1 region,31,197 all replicase subunits localize to the perinuclear region of the infected cell (e-Fig. 29.8A–D; 207), where they are associated with intracellular membranes that are derived from the endoplasmic reticulum (ER). Upon arterivirus infection, these host cell membranes are modified into vesicular double-membrane structures that presumably carry the viral replication complex (e-Fig. 29.9A–F).149,177,212 The formation of closely paired membranes and double membrane vesicles (DMVs) is a typical feature of arterivirus-infected cells described many years ago.18,184,234 Recent electron tomography studies of EAV-infected cells revealed that these structures are in fact interconnected and form a network of modified ER99 (e-Fig. 29.9E). Biochemical and electron microscopy studies have implicated ORF1a-encoded subunits that contain hydrophobic, probable trans-membrane domains (in particular nsp2, nsp3, and nsp5; e-Fig. 29.6C) in the formation of these membrane structures.71,149,158,177,207
Replicase ORF1b is the most conserved part of the arterivirus genome and encodes the core enzymes for viral RNA synthesis—RdRp (nsp9) and helicase (nsp10).54,83,140 Recombinant EAV nsp9 is able to initiate RNA synthesis de novo in the absence of other viral or cellular proteins, but could not utilize sequences derived from the 3′ end of the viral genome as a template,13 suggesting additional requirements for its activity in vivo. The predicted NTP binding and superfamily 1 helicase activities of arterivirus nsp10 were corroborated by in vitro assays with recombinant nsp10. These also revealed the 5′-to-3′ polarity of the unwinding reaction, a property shared with coronaviruses,12,169 although it has not been reconciled with the protein’s presumed role in unwinding local double-stranded
RNA structures that might hinder the RdRp during viral RNA synthesis, which proceeds in the opposite direction. As in all nidoviruses, the helicase is linked to an N-terminal zinc-binding domain that might assist the proper folding of nsp10 and/or mediate interactions of the protein with its substrate RNAs. This domain was also implicated in a remarkable transcription-specific defect.208,210
RNA structures that might hinder the RdRp during viral RNA synthesis, which proceeds in the opposite direction. As in all nidoviruses, the helicase is linked to an N-terminal zinc-binding domain that might assist the proper folding of nsp10 and/or mediate interactions of the protein with its substrate RNAs. This domain was also implicated in a remarkable transcription-specific defect.208,210
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree