Arenaviridae
Michael J. Buchmeier
Juan-Carlos de la Torre
Clarence J. Peters
History
Arenaviruses cause chronic infections of rodents indigenous to Europe, Africa, the Americas, and perhaps other continents. These asymptomatically infected animals move freely in their natural habitat and may invade human habitation; when humans come in contact with excreted viruses, disease may result. Arenavirus infections of humans are common and in some cases severe; several members of the group are responsible for severe acute infections termed hemorrhagic fevers. The mechanisms that have evolved to ensure chronic infection and intergenerational survival of the virus in rodents are complex and specific to the particular rodent–virus combination, and their elucidation over the past 60 years has led to a deeper understanding of immunology and immunopathology.164
Lymphocytic choriomeningitis virus (LCMV) was the first isolated arenavirus, discovered in 1933 during the study of samples from a St. Louis encephalitis epidemic.185 It was soon found to be a cause of aseptic meningitis206 and to be identical to an agent that chronically infected mouse colonies.222 This launched a series of studies that showed LCMV to be a common human pathogen and a fascinating virus with many lessons for biologists.31,32,164
By the 1960s, several other viruses had been discovered and found to share common morphology,160 serology,192 biochemical features, and a natural history that depended on chronic infection of rodent reservoirs, leading to the recognition of the Arenaviridae family, named after the sandy (Latin, arenosus) appearance of the ribosomes originally seen in thin sections of virions.192 Some of these viruses caused hemorrhagic fevers (Table 43.1), whereas others were apparently not pathogenic for or rarely infected humans (Table 43.2). Since the isolation of Tacaribe virus (TACV),55 new arenaviruses have continued to be discovered in the Americas on the average of one every 3 years, including three lethal new viruses: Guanarito,198 Sabia in 1990,130 and Whitewater Arroyo in 1996.79 As a result of new technologies of deep sequencing and genome discovery, several new arenaviruses have been revealed in the past decade in both the New World (NW) and Old World (OW) branches of the Arenaviridae.
Classification of Viruses within Family
The Arenaviridae consists of a unique genus (Arenavirus) containing 24 unique species currently recognized by the International Committee on Taxonomy of Viruses (ICTV), as well as several newly discovered viruses whose taxonomic status has not yet been determined by the ICTV (see Table 43.1). The original classification of arenaviruses, based mainly on virus antigenic properties, identified two groups: the NW (also referred to as Tacaribe serocomplex) and the OW (also referred to as Lassa-Lymphocytic choriomeningitis serocomplex) including viruses from Africa and the globally distributed prototypic arenavirus, LCMV. Results derived from molecular genetic studies are consistent with findings from comparative serology.
The OW group consists of a single lineage composed of five species (Lassa virus, Mobala virus, Ippy virus, Mopeia virus, and Lymphocytic choriomeningitis virus), and several newly discovered viruses including Kodoko, Morogoro, Dandenong and Lujo,39,119,166 whereas the NW group is composed of four lineages: clades A, B, C, and A/Rec (Fig. 43.1). Clade A contains five species (Flexal virus, Parana virus, Pichinde virus, Pirital virus, and Allpahuayo virus), whose internal genetic relationships are not well established.39 Clade B contains eight species
(Sabia virus, Cupixi virus, Guanarito virus, Amapari virus, Junin virus, Machupo virus, Tacaribe virus, and Chapare virus), which form three well-supported lineages. Clade C is composed of only two established species, Oliveros virus and Latino virus, and a potential third member, Pinhal virus, for which little information is available. Clade A/Rec comprises all North American arenaviruses: Whitewater Arroyo virus, Tamiami virus, Bear Canyon virus, and the newly discovered Skinner Tank and Catarina viruses. As with clade A, intraclade A/Rec relationships are poorly resolved, owing to limited sequencing data and the incongruence of phylogenetic data derived from different genes. The geographic distribution of each arenavirus is determined by the range of its natural specific rodent reservoir, with the exception of TACV, which was isolated from fruit-eating bats.59,84
(Sabia virus, Cupixi virus, Guanarito virus, Amapari virus, Junin virus, Machupo virus, Tacaribe virus, and Chapare virus), which form three well-supported lineages. Clade C is composed of only two established species, Oliveros virus and Latino virus, and a potential third member, Pinhal virus, for which little information is available. Clade A/Rec comprises all North American arenaviruses: Whitewater Arroyo virus, Tamiami virus, Bear Canyon virus, and the newly discovered Skinner Tank and Catarina viruses. As with clade A, intraclade A/Rec relationships are poorly resolved, owing to limited sequencing data and the incongruence of phylogenetic data derived from different genes. The geographic distribution of each arenavirus is determined by the range of its natural specific rodent reservoir, with the exception of TACV, which was isolated from fruit-eating bats.59,84
Table 43.1 Arenaviruses That Are Known to Be Human Pathogens | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Recently, high-throughput sequencing and metagenomic pathogen discovery approaches have led to the identification and isolation of two highly divergent arenaviruses from cases of inclusion body disease, a common infectious disease of captive snakes.215 These viruses show typical ambisense arenavirus genome organization but have a GP2 reminiscent of filoviruses. These findings indicate that arenavirus natural hosts are broader than previously thought and not restricted to mammalians. In addition, significant genetic divergence between these newly identified reptile arenaviruses and known mammalian arenaviruses together with biological differences between reptiles
and mammals have raised new questions about the biology and evolution of arenaviruses.
and mammals have raised new questions about the biology and evolution of arenaviruses.
Table 43.2 Arenaviruses Not Known to Be Significant Human Pathogens | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
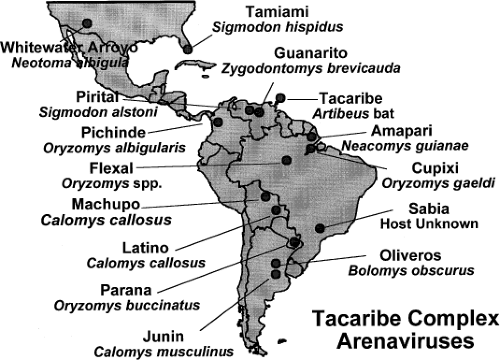 Figure 43.1. Location of American arenaviruses with their natural host. (Courtesy of Dr. S. Nichol, Centers for Disease Control and Prevention.) |
The originally established species demarcation criteria included (a) association with a specific host species or group of species, (b) geographic niche, (c) association with human disease, (d) differences in antigenic cross-reactivity, and (e) significant sequence difference from other species in the genus. However, often limited information and difficulties associated with its analysis regarding these criteria have led more recently to emphasize mainly genetic criteria to define Arenavirus species. This approach, however, faces the problem of how to correctly assign a cutoff value for species demarcation owing to
biased sequence availability. There are many fewer OW than NW arenavirus sequences; however, OW sequences include most complete intraspecies sequences. Therefore, intraspecies diversity is likely underestimated among NW arenaviruses, whereas the interspecies diversity could be underestimated for OW arenaviruses. Notably, sequence comparison of the same genome region for a large number of isolates revealed similar degree of diversity in an OW arenavirus (Lassa virus [LASV]) and two NW arenaviruses (Whitewater Arroyo virus [WWAV] and Pirital virus [PIRV]).39 These findings suggest that accurate classification of a new arenavirus isolate may require to follow the Arenavirus species definition together with comparing the sequence of the new isolate with the larger available set of sequences.
biased sequence availability. There are many fewer OW than NW arenavirus sequences; however, OW sequences include most complete intraspecies sequences. Therefore, intraspecies diversity is likely underestimated among NW arenaviruses, whereas the interspecies diversity could be underestimated for OW arenaviruses. Notably, sequence comparison of the same genome region for a large number of isolates revealed similar degree of diversity in an OW arenavirus (Lassa virus [LASV]) and two NW arenaviruses (Whitewater Arroyo virus [WWAV] and Pirital virus [PIRV]).39 These findings suggest that accurate classification of a new arenavirus isolate may require to follow the Arenavirus species definition together with comparing the sequence of the new isolate with the larger available set of sequences.
Virion Structure
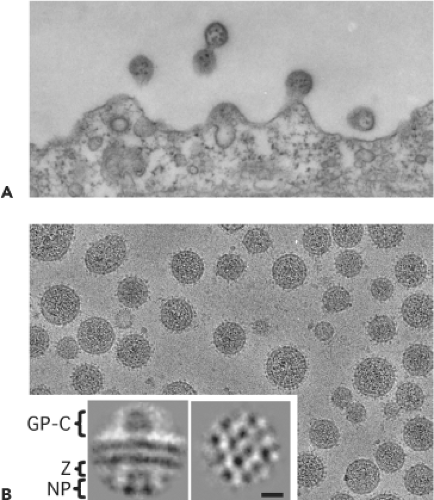
Arenavirus virions are pleomorphic when examined by cryo-electron microscopy, ranging in size from 40 to more than 200 nm in diameter39,103 (Fig. 43.2). Virions contain a surface envelope that is studded with evenly spaced spikes that correspond to glycoprotein (GP) projections consisting of complexes of the viral GPs—GP1 and GP2. The surface GPs are aligned with subjacent Z protein and ribonucleoprotein (RNP) densities, which are packed into a two-dimensional lattice at the inner surface of the viral membrane.103,161 Virions contain the L and S genomic RNAs as helical nucleocapsid (NC) structures that are organized into circular configurations, with lengths ranging from 400 to 1,300 nm.240 The L and S genomic RNA species are not present in equimolar amounts within virions (L:S ratios ∼1:2), and low levels of both L and S antigenomic RNA species are also present within virions. In addition, it has been documented that host ribosomes can be incorporated into virions, although the biological significance of this remains to be determined.28,159
Genome Structure and Organization
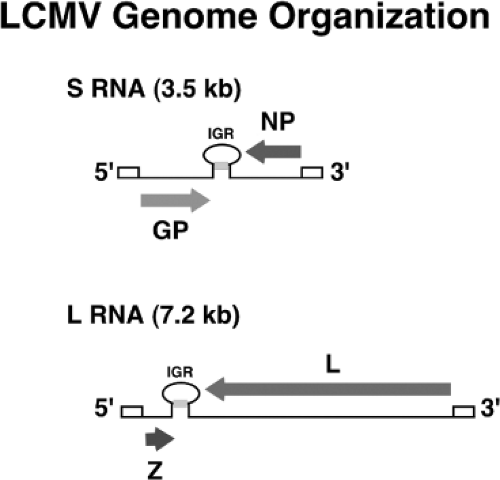
Arenaviruses are enveloped viruses with a bisegmented negative-sense single-stranded RNA genome and a life cycle restricted to the cell cytoplasm. Individual arenaviruses exhibit some variability in the lengths of the two genomic RNA segment, L (ca 7.2 kb) and S (ca 3.5 kb); however, their overall organization is well conserved across the virus family.30 Arenavirus coding strategy has unique features compared to other NS RNA viruses. Each arenavirus genome segment uses an ambisense coding strategy to direct the synthesis of two polypeptides in opposite orientation, separated by a noncoding intergenic region (IGR) with a predicted folding of a stable hairpin structure (Fig. 43.3). The S RNA encodes the viral glycoprotein precursor (GPC; ca 75 kDa) and the nucleoprotein (NP; ca 63 kDa),
whereas the L RNA encodes the viral RNA–dependent RNA polymerase (RdRp, or L polymerase) (ca 200 kDa) and a small (ca 11 kDa) RING finger protein (Z) that functions as a matrix protein.30 This situation—where in one region the S and L RNA are negative sense and in a second, nonoverlapping region they are pseudo–positive sense—led investigators to coin the term ambisense to describe the arenavirus coding strategy. The qualifier pseudo is used because there is no evidence that the genomic S and L RNAs can function as messenger RNAs (mRNAs) and be directly translated into GPC and Z proteins, respectively.
whereas the L RNA encodes the viral RNA–dependent RNA polymerase (RdRp, or L polymerase) (ca 200 kDa) and a small (ca 11 kDa) RING finger protein (Z) that functions as a matrix protein.30 This situation—where in one region the S and L RNA are negative sense and in a second, nonoverlapping region they are pseudo–positive sense—led investigators to coin the term ambisense to describe the arenavirus coding strategy. The qualifier pseudo is used because there is no evidence that the genomic S and L RNAs can function as messenger RNAs (mRNAs) and be directly translated into GPC and Z proteins, respectively.
Arenaviruses exhibit high degree of sequence conservation at the 3′ end of the L and S RNA segments (17 out of 19 nucleotides [nt] are identical), suggesting that this conserved terminal sequence element constitutes the virus promoter for polymerase entry.30,151 Arenavirus genomes and antigenomes exhibit a high degree of complementarity between their 5′ and 3′ termini, and the 5′ and 3′ ends of both L and S genome segments are predicted to form panhandle structures.30,151,215 This prediction is supported by electron microscopy data showing the existence of circular RNP complexes within arenavirus virion particles.240 This terminal complementarity may reflect the presence at the 5′ ends of cis-acting signals sequences that provide a nucleation site for RNA encapsidation, which is required to generate the NC templates recognized by the virus polymerase. Terminal complementarity may be also a consequence of strong similarities between the genome and antigenome promoters used by the virus polymerases. This terminal complementarity has been proposed to favor the formation of both intra- and intermolecular L and S duplexes that might be part of the replication initiation complex; however, its mechanistic aspects remain undefined.199 For several arenaviruses, an additional nontemplated G residue has been detected on the 5′ end of their genome RNAs.82
Arenavirus IGRs are predicted to fold into a stable hairpin structure. Transcription termination of the S-derived NP and GP occurs at multiple sites within the predicted stem of the IGR, suggesting that a structural motif rather than a sequence-specific signal promotes the release of the arenavirus polymerase from the template RNA.30,151 There are significant differences in sequence and predicted folded structure between the S and L IGR; however, among isolates and strains of the same Arenavirus species, the S and L IGR sequences are highly conserved. Some arenaviruses, including LCMV, LASV, and Pichinde virus (PICV), contain a single predicted stem loop within the S IGR, whereas the S IGR of others, such as TACV and Mopeia virus (MOPV), are predicted to contain two distinct stem loops.151,151,214
Arenavirus Life Cycle
Cell Attachment and Entry
Consistent with a broad host range and cell-type tropism, a highly conserved and widely expressed cell surface receptor for extracellular matrix (ECM) proteins—alpha-dystroglycan (α-DG)—has been identified as a main receptor for LCMV, LASV, and several other arenaviruses.35,115,151 Postranslational modifications of α-DG, including O-mannosylation and LARGE-dependent glycosylation, are critical for α-DG’s function as both an ECM and OW and clade C NW arenavirus receptor.151,190 These findings indicate that similar α-DG–derived glycan structures are recognized by both ECM proteins and surface arenavirus GPs. Intriguingly, recent genome-wide studies have uncovered evidence of positive selection of specific LARGE alleles within the Yoruba people in West Africa where LASV is endemic.195 However, many other arenaviruses appear to use an alternative receptor and human transferrin receptor 1 (Tf R1) was identified as a cellular receptor used for cell entry of NW hemorrhagic fever arenaviruses Junin virus (JUNV) and Machupo virus (MACV).184
Upon receptor binding, arenavirus virions are internalized by smooth-walled vesicles into a low-pH subcellular environment that triggers a pH-dependent membrane fusion step between cell and viral membranes that is mediated by GP2 and results in the release of the virus RNP into the cytoplasm of the cell where viral RNA synthesis, both transcription of replication, take place. Consistent with the use of TfR1 as a primary receptor for cell entry, JUNV was found to follow a clathrin-mediated endocytosis as its main route of entry.138 Nevertheless, although TfR1 is internalized and recycles through early endosomes (pH ∼6), optimal fusion activity of JUNV GP requires a significantly lower (<5.5) pH, suggesting that JUNV may redirect TfR1 from its normal recycling pathway into the late endosomes, a process likely influenced by the multivalent nature of the virus particle. In contrast, cell entry of the OW arenaviruses LCMV and LASV appears to use a rather unique endocytic pathway independent of clathrin, caveolin, dynamin, and actin187,188,191,208,227 (Fig. 43.4). Consistent with these findings, endosomal trafficking of LASV and LCMV was only minimally affected by dominant negative forms of Rab5 and Rab7, which are small guanine triphosphatases (GTPases) involved in vesicular trafficking into early and late, respectively, endosomes. Moreover, LCMV was found to circumvent Rab5/EEA-1 early endosomes. LCMV and LASV GP have a high binding affinity for α-DG and thereby the unusual endocytic pathway followed by these viruses may reflect the natural cellular trafficking of α-DG. Nevertheless, it cannot be ruled out that binding of GP to α-DG could trigger a novel route of entry for dystroglycan that is tailored specifically to benefit virus cell entry as previously documented for other pathogens.
Expression and Replication of the Viral Genome
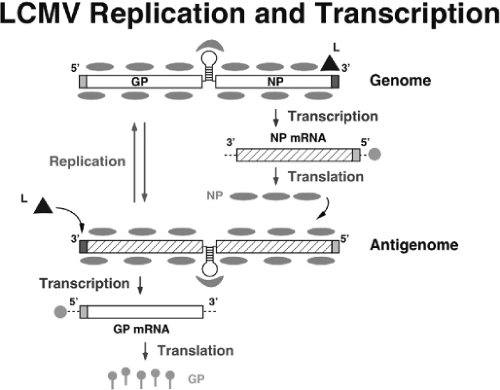
The fusion between viral and cellular membranes releases the viral RNP into the cytoplasm, which is ensued by the onset of viral RNA synthesis. Transcription termination of subgenomic nonpolyadenylated viral mRNAs were mapped to multiple sites within the distal side of the IGR, which suggested that the IGR acts as a bona fide transcription termination signal for the virus polymerase.
The NP and L coding regions are transcribed into a genomic complementary mRNA, whereas the GPC and Z coding regions are not translated directly from genomic RNA, but rather from genomic sense mRNAs that are transcribed using as templates the corresponding antigenome RNA species, which also function as replicative intermediates (Fig. 43.5). LCMV mRNAs have four to five nontemplated nucleotides and a cap structure at their 5′ ends, which are likely obtained from cellular mRNAs via cap-snatching mechanisms whose details remain to be determined. A recently described endonuclease activity associated with the arenavirus L polymerase could play a critical role in this process.157
The NP—the most abundant viral polypeptide both in infected cells and virions—is the main structural element of
the viral RNP and plays an essential role in viral RNA synthesis. Recent evidence indicates that NP exhibits interferon type I (IFN-I) counteracting activity that was mapped to the C-terminus of NP.140,141,143 Two recently determined crystal structures of LASV NP at 1.80183 and 1.590 Å resolution identified distinct N- and C-terminal domains connected by a flexible linker. The N-terminal domain was proposed to contain a potential cap-binding site that could provide the host-derived primers to initiate transcription by the virus polymerase.183 However, analysis of the structure of the N-terminal domain of LASV NP in complex with single-strand RNA (ssRNA) suggest that this originally proposed cap-binding site likely corresponds to a binding site for the viral genome.
the viral RNP and plays an essential role in viral RNA synthesis. Recent evidence indicates that NP exhibits interferon type I (IFN-I) counteracting activity that was mapped to the C-terminus of NP.140,141,143 Two recently determined crystal structures of LASV NP at 1.80183 and 1.590 Å resolution identified distinct N- and C-terminal domains connected by a flexible linker. The N-terminal domain was proposed to contain a potential cap-binding site that could provide the host-derived primers to initiate transcription by the virus polymerase.183 However, analysis of the structure of the N-terminal domain of LASV NP in complex with single-strand RNA (ssRNA) suggest that this originally proposed cap-binding site likely corresponds to a binding site for the viral genome.
The C-terminal domain has a folding that mimics that of the DEDDH family of 3′-5′ exoribonucleases like the one associated with severe acute respiratory syndrome (SARS) coronavirus nsp14 protein.155 Functional studies confirmed the 3′-5′ exoribonuclease activity of LASV NP, which was proposed to
be critical for the anti-interferon (IFN) activity of NP but dispensable for the role of NP on replication and transcription of the viral genome.90 This assertion, however, is difficult to reconcile with the finding that TACV NP, despite containing the conserved DEDDH residues at the active site of the exoribonuclease domain, has a dramatically reduced anti-IFN activity,142 and the finding that an LCMV with a mutant NP lacking the 3′-5′ exoribonuclease had a large decrease in fitness during its replication in IFN-deficient Vero cells.140
be critical for the anti-interferon (IFN) activity of NP but dispensable for the role of NP on replication and transcription of the viral genome.90 This assertion, however, is difficult to reconcile with the finding that TACV NP, despite containing the conserved DEDDH residues at the active site of the exoribonuclease domain, has a dramatically reduced anti-IFN activity,142 and the finding that an LCMV with a mutant NP lacking the 3′-5′ exoribonuclease had a large decrease in fitness during its replication in IFN-deficient Vero cells.140
The arenavirus L protein has the characteristic sequence motifs conserved among the RdRp (L proteins) of NS RNA viruses.181 Detailed sequence analysis and secondary structure predictions done with the LASV L polymerase identified several regions of strong alpha-helical content and a putative coiled-coil domain at the N-terminus.228 Subsequent bioinformatic analysis together with biochemical and minigenome (MG)-based functional studies have shown that LASV L protein is likely organized into three distinct structural domains, and that at specific amino acid positions, LASV L can be split into N- and C-terminal parts that are able to functionally trans-complement each other.27 Notably, the recent electron microscopy characterization of a functional MACV L protein has revealed a core ring domain decorated by appendages, which may reflect a modular organization of the arenavirus polymerase.114
Assembly and Budding
Assembly and cell release of infectious arenavirus progeny requires both Z and GPC, as well as the correct processing by the cellular site 1 protease (S1P) of GPC into GP1 (40–46 kDa) and GP-2 (35 kDa)116,125,178 Oligomers of GP1 and GP2 form the spikes that decorate the virus surface. GP1 located at the top of the spike and held in place by ionic interactions with the N-terminus of the transmembrane GP-2, mediates virus interaction with host cell surface receptors, whereas GP2 exhibits similarities with fusion active domains of other viral envelope proteins.66,80,236 GPC contains a 58-amino-acid signal peptide (SSP) that is expressed as a stable polypeptide in infected cells and it remains associated to the GP complex (GPcx). This SSP has been implicated in different aspects of the trafficking and function of the viral envelope GPs including the GP-2-mediated pH-dependent fusion process.2,203,237,238,239
For most enveloped viruses, a matrix (M) protein is involved in organizing the virion components prior to assembly. Interestingly, arenaviruses do not have an obvious counterpart of M. The arenavirus RING finger protein Z is a structural component of the virion that has no homologue among other known NS RNA viruses. In LCMV-infected cells Z has been shown to interact with several cellular proteins including the promyelocytic leukemia (PML) protein and the eukaryote translation initiation factor 4E (eIF4E), which have been proposed to contribute to the noncytolytic nature of LCMV infection and repression of cap-dependent translation, respectively.54,229 Z has been shown to be the main driving force of arenavirus budding,169,218 a process mediated by the Z late (L) domain motifs: PTAP and PPPY similar to those known to control budding of several other viruses including HIV and Ebola virus, via interaction with specific host cell proteins.78 Consistent with this observation Z exhibited features characteristic of bona fide budding proteins: 1) ability to bud from cells by itself, and 2) substituted efficiently for other L domain. Targeting of Z to the plasma membrane, the location of arenavirus budding, strictly required its myristoylation.171,219 Results derived from cryo-electron microscopy of arenavirus particles161– were also consistent with the role of Z as a functional M protein.
Arenavirus Reverse Genetics: Implications for the Investigation of Arenavirus Molecular and Cell Biology
The generation of an infectious progeny of NS RNA viruses from cloned cDNA, referred to as reverse genetics, has revolutionized the analysis of cis-acting sequences and trans-acting proteins required for virus replication, transcription, maturation and budding. In addition, the possibility to generate predetermined specific mutations within the virus genome and analyze their phenotypic expression in the context of the virus natural infection provides investigator with novel and powerful approaches to investigate cellular and molecular mechanisms underlying virus-host interactions and their implications in viral pathogenesis. In addition, these developments have also paved the way for engineering recombinant viruses for vaccine and gene therapy purposes.
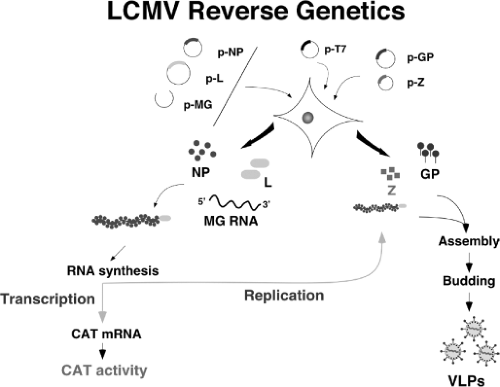
In contrast to positive-strand RNA viruses, deproteinized genomic and antigenomic RNAs of NS RNA viruses cannot function as mRNAs and are not infectious. Generation of biologically active synthetic NS viruses from cDNA requires trans complementation by all viral proteins involved in virus replication and transcription, and polymerases of NS RNA viruses recognize as templates an NC consisting of the genomic RNA tightly encapsidated by the NP, which associated with the virus polymerase proteins forms an RNP complex. These considerations hindered the application of recombinant DNA technology to the genetic analysis of these viruses. However, significant progress has been made in this area and for all NS RNA viruses, short model genomes (aka minigenomes (MG)) could be encapsidated and expressed by plasmid-encoded proteins. Moreover, it has become possible to rescue infectious virus entirely from cloned cDNAs for members of all families of NS RNA viruses.108,162
Arenavirus Minigenome Systems
The first arenavirus MG rescue system was developed for LCMV.122 Subsequently MG systems were developed for LASV87 and the NW arenaviruses PICV,117 TACV,131 and JUNV.3 Results derived from MG-based assays identified NP and L as the minimal viral trans-acting factors required for efficient RNA synthesis mediated by the virus polymerase. For LCMV both genetic and biochemical evidence indicated that oligomerization of L is required for the activity of the arenavirus polymerase.201 Consistent with this finding, biochemical and MG-based functional studies have shown that LASV L protein contains both N-and C-termini sites that mediate L–L interaction. The use of MG-based assays facilitated mutation-function studies involving conserved acidic and basic amino acid residues within the N- and C-termini of LASV L protein uncovered several residues within the N-terminus of L that played a critical role in synthesis of viral mRNA but not in
RNA replication.88,124,201 The recent publication of a 2.13 Å resolution crystal structure and functional characterization of the N-terminal 196 residues (NL1) of the LCMV L protein uncovered an endonuclease functional domain similar to the one found in the N-terminus of the influenza virus PA protein.157 MG-based assays have shown the endonuclease activity of NL1 to be critical for arenavirus transcription.
RNA replication.88,124,201 The recent publication of a 2.13 Å resolution crystal structure and functional characterization of the N-terminal 196 residues (NL1) of the LCMV L protein uncovered an endonuclease functional domain similar to the one found in the N-terminus of the influenza virus PA protein.157 MG-based assays have shown the endonuclease activity of NL1 to be critical for arenavirus transcription.
Mutation-function analysis of the genome 5′- and 3′-termini using the LCMV and LASV MG-based assays indicated that the activity of the arenavirus genomic promoter requires both sequence specificity within the highly conserved 3′-terminal 19 nt of arenavirus genomes and the integrity of the predicted panhandle structure formed via sequence complementarity between the 5′- and 3′-termini of viral genome RNAs.89,157,170 These studies revealed that arenavirus RNA replication and transcription are regulated in a coordinated manner. Likewise, MG-based assays provided direct experimental confirmation that the IGR is a bona fide transcription termination signal,177 and that intracellular levels of NP do not determine the balance between virus RNA replication and transcription68,178,179—a finding conceptually similar to that reported for the paramyxovirus respiratory syncytial virus.68
Generation of Recombinant Arenaviruses
Generation of infectious arenaviruses from cloned complementary DNAs (cDNAs) has been reported for LCMV,73,131,202 PICV,117 and JUNV.3,62,117 Both T7 RNA polymerase (RP)- and pol-I RP-based systems73,202 have been documented to launch intracellular synthesis of S and L genome, or antigenome, RNA species that were subsequently replicated and transcribed by the virus polymerase complex reconstituted intracellularly via plasmid-supplied L and NP (Fig. 43.6). Notably, rescue of PICV and JUNV using a T7RP-based system was efficiently achieved without requiring plasmid-supplied viral NP and L proteins,3,73,117 indicating that T7RP-mediated RNA synthesis produced viral antigenome RNA species that were substrate for encapsidation and replication as well as mRNAs that serve as a template to produce levels of NP and L sufficiently high to facilitate virus rescue. This finding parallels those reported for several other negative sense RNA viruses, including bunyaviruses,20,132 filoviruses,85 and bornaviruses.205
The ability to generate recombinant arenaviruses with predetermined specific mutations and analyze their phenotypic expression in the context of the natural course of infection has opened new opportunities to investigate arenavirus–host
interactions that influence a variable infection outcome, ranging from virus control and clearance by the host defenses to long-term chronic infection associated with subclinical disease, and severe acute disease including hemorrhagic fever. Thus, studies with recombinant lymphocytic choriomeningitis virus (rLCMV)/vesicular stomatitis virus glycoprotein (VSVG) uncovered the arenavirus GP as a viral Achilles’ heal and provided the foundations for a novel strategy to develop safe and effective live attenuated arenavirus vaccines.178,179 Likewise, the use of rLCMV/VSVG facilitated studies aimed at understanding the regulation of CD8+ T-cell function within the infected brain,180 and how viruses can induce organ-specific immune disease in the absence of molecular mimicry and without disruption of self-tolerance.149 Other rLCMVs have been used to investigate pathways of LASV cell entry187 and the role of NP in the inhibition of induction of IFN-I by LCMV.140 Likewise, the use of recombinant arenaviruses where a canonical furin cleavage site substituted for the S1P cleavage site within GPC illustrated the critical role played by the S1P-mediated processing of arenavirus GPC in the virus life cycle.3,189,224 The rescue of attenuated and virulent forms of PICV117,128 or the docile and aggressive strains of LCMV41,60,61 have allowed for the identification of specific genetic determinants of virus virulence. However, despite these successes, the ability to rescue recombinant arenaviruses expressing additional genes of interest posed unexpected difficulties. Approaches successfully employed with other NS RNA viruses including the use of an internal ribosome entry site (IRES), or of the picornavirus self-cleaving 2A sequence, did not work when applied to LCMV.61 This problem was finally overcome by the generation of trisegmented rLCMV (r3LCMV) containing one L and two S segments. For this, each of the two S segments was altered to replace one of the two viral open reading frames (ORFs), GPC or NP, by a gene of interest (GOI).61 The rationale for this approach was that the physical separation of GP and NP into two different S segments would represent a strong selective pressure to select and maintain a virus capable of packaging 1L and 2S segments (Fig. 43.7). Various r3LCMV have been rescued that express one or two additional GOI. Depending on the GOI expressed, with gene size and function being critical parameters, these r3LCMV have shown little or no attenuation and long-term genetic stability in cultured cell. In contrast, the use of r3LCMV to study virus–host interactions in mice has encountered some limitations owing to the observation that even r3LCMV with wild-type growth properties in cultured cells exhibited an attenuated phenotype in the mouse owing to reasons that remain to be determined.
interactions that influence a variable infection outcome, ranging from virus control and clearance by the host defenses to long-term chronic infection associated with subclinical disease, and severe acute disease including hemorrhagic fever. Thus, studies with recombinant lymphocytic choriomeningitis virus (rLCMV)/vesicular stomatitis virus glycoprotein (VSVG) uncovered the arenavirus GP as a viral Achilles’ heal and provided the foundations for a novel strategy to develop safe and effective live attenuated arenavirus vaccines.178,179 Likewise, the use of rLCMV/VSVG facilitated studies aimed at understanding the regulation of CD8+ T-cell function within the infected brain,180 and how viruses can induce organ-specific immune disease in the absence of molecular mimicry and without disruption of self-tolerance.149 Other rLCMVs have been used to investigate pathways of LASV cell entry187 and the role of NP in the inhibition of induction of IFN-I by LCMV.140 Likewise, the use of recombinant arenaviruses where a canonical furin cleavage site substituted for the S1P cleavage site within GPC illustrated the critical role played by the S1P-mediated processing of arenavirus GPC in the virus life cycle.3,189,224 The rescue of attenuated and virulent forms of PICV117,128 or the docile and aggressive strains of LCMV41,60,61 have allowed for the identification of specific genetic determinants of virus virulence. However, despite these successes, the ability to rescue recombinant arenaviruses expressing additional genes of interest posed unexpected difficulties. Approaches successfully employed with other NS RNA viruses including the use of an internal ribosome entry site (IRES), or of the picornavirus self-cleaving 2A sequence, did not work when applied to LCMV.61 This problem was finally overcome by the generation of trisegmented rLCMV (r3LCMV) containing one L and two S segments. For this, each of the two S segments was altered to replace one of the two viral open reading frames (ORFs), GPC or NP, by a gene of interest (GOI).61 The rationale for this approach was that the physical separation of GP and NP into two different S segments would represent a strong selective pressure to select and maintain a virus capable of packaging 1L and 2S segments (Fig. 43.7). Various r3LCMV have been rescued that express one or two additional GOI. Depending on the GOI expressed, with gene size and function being critical parameters, these r3LCMV have shown little or no attenuation and long-term genetic stability in cultured cell. In contrast, the use of r3LCMV to study virus–host interactions in mice has encountered some limitations owing to the observation that even r3LCMV with wild-type growth properties in cultured cells exhibited an attenuated phenotype in the mouse owing to reasons that remain to be determined.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree