11
Antimicrobial Drugs: Resistance
CHAPTER CONTENTS
PRINCIPLES OF ANTIBIOTIC RESISTANCE
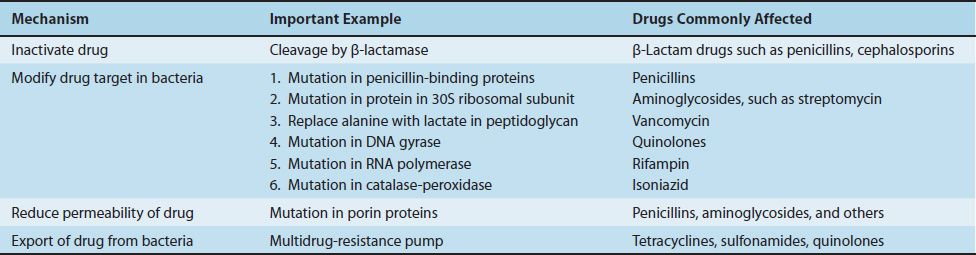
There are four major mechanisms that mediate bacterial resistance to drugs (Table 11–1). (1) Bacteria produce enzymes that inactivate the drug (e.g., β-lactamases can inactivate penicillins and cephalosporins by cleaving the β-lactam ring of the drug). (2) Bacteria synthesize modified targets against which the drug has a reduced effect (e.g., a mutant protein in the 30S ribosomal subunit can result in resistance to streptomycin, and a methylated 23S rRNA can result in resistance to erythromycin). (3) Bacteria reduce permeability to the drug such that an effective intracellular concentration of the drug is not achieved (e.g., changes in porins can reduce the amount of penicillin entering the bacterium). (4) Bacteria actively export drugs using a “multidrug-resistance pump” (MDR pump, or “efflux” pump). The MDR pump imports protons and, in an exchange-type reaction, exports a variety of foreign molecules including certain antibiotics, such as tetracyclines.
TABLE 11–1 Mechanisms of Drug Resistance
Most drug resistance is due to a genetic change in the organism, either a chromosomal mutation or the acquisition of a plasmid or transposon. Nongenetic changes, which are of lesser importance, are discussed on page 90.
The term high-level resistance refers to resistance that cannot be overcome by increasing the dose of the antibiotic. A different antibiotic, usually from another class of drugs, is used. Resistance mediated by enzymes such as β-lactamases often result in high-level resistance, as all the drug is destroyed. Low-level resistance refers to resistance that can be overcome by increasing the dose of the antibiotic. Resistance mediated by mutations in the gene encoding a drug target is often low level, as the altered target can still bind some of the drug but with reduced strength.
To illustrate the use of these terms, strains of Neisseria gonorrhoeae that produce penicillinase cannot be treated successfully with penicillin G. They exhibit high-level resistance, and a different drug such as ceftriaxone must be used. However, strains of N. gonorrhoeae that synthesize altered penicillin-binding proteins exhibit low-level resistance and can be treated successfully with high-dose penicillin G.
Hospital-acquired infections are significantly more likely to be caused by antibiotic-resistant organisms than are community-acquired infections. This is especially true for hospital infections caused by Staphylococcus aureus and enteric gram-negative rods such as Escherichia coli and Pseudomonas aeruginosa. Antibiotic-resistant organisms are common in the hospital setting because widespread antibiotic use in hospitals selects for these organisms. Furthermore, hospital strains are often resistant to multiple antibiotics. This resistance is usually due to the acquisition of plasmids carrying several genes that encode the enzymes that mediate resistance.
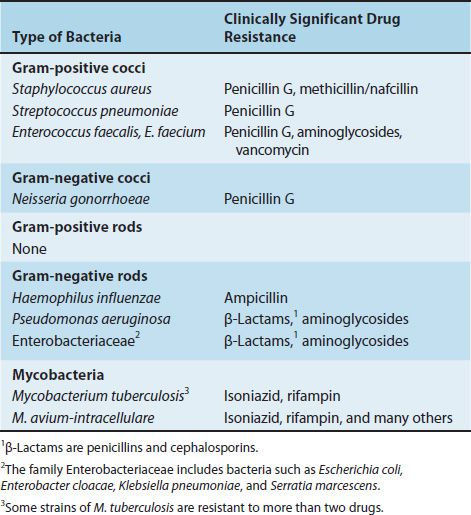
Table 11–2 describes certain medically important bacteria and the main drugs to which they are resistant. Note that although these bacteria are resistant to other drugs as well, for simplicity, only the most characteristic drugs are listed. Some strains of the bacteria listed in Table 11–2 are highly resistant to multiple antibiotics, namely methicillin-resistant S. aureus (MRSA; see Chapter 15), vancomycin-resistant Enterococcus faecium (VRE; see Chapter 15), multidrug-resistant Streptococcus pneumoniae (MDR-SP; see Chapter 15), P. aeruginosa (see Chapter 18), and multidrug-resistant Mycobacterium tuberculosis (MDR-MTB; see Chapter 21).
TABLE 11–2 Medically Important Bacteria That Exhibit Significant Drug Resistance
GENETIC BASIS OF RESISTANCE
Chromosome-Mediated Resistance
Chromosomal resistance is due to a mutation in the gene that codes for either the target of the drug or the transport system in the membrane that controls the uptake of the drug. The frequency of spontaneous mutations usually ranges from 10–7 to 10–9, which is much lower than the frequency of acquisition of resistance plasmids. Therefore, chromosomal resistance is less of a clinical problem than is plasmid-mediated resistance.
The treatment of certain infections with two or more drugs is based on the following principle. If the frequency that a bacterium mutates to become resistant to antibiotic A is 10–7 (1 in 10 million) and the frequency that the same bacterium mutates to become resistant to antibiotic B is 10–8 (1 in 100 million), then the chance that the bacterium will become resistant to both antibiotics (assuming that the antibiotics act by different mechanisms) is the product of the two probabilities, or 10–15. It is therefore highly unlikely that the bacterium will become resistant to both antibiotics. Stated another way, although an organism may be resistant to one antibiotic, it is likely that it will be effectively treated by the other antibiotic.
Plasmid-Mediated Resistance
Plasmid-mediated resistance is very important from a clinical point of view for three reasons:
(1) It occurs in many different species, especially gram-negative rods.
(2) Plasmids frequently mediate resistance to multiple drugs.
(3) Plasmids have a high rate of transfer from one cell to another, usually by conjugation.
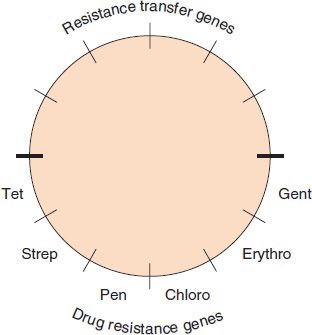
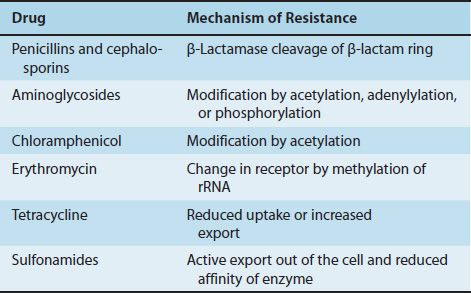
Resistance plasmids (resistance factors, R factors) are extrachromosomal, circular, double-stranded DNA molecules that carry the genes for a variety of enzymes that can degrade antibiotics and modify membrane transport systems (Figure 11–1). Table 11–3 describes the most important mechanisms of resistance for several important drugs.
FIGURE 11–1 Resistance plasmid (R plasmid, R factor). Most resistance plasmids have two sets of genes: (1) resistance transfer genes that encode the sex pilus and other proteins that mediate transfer of the plasmid DNA during conjugation, and (2) drug resistance genes that encode the proteins that mediate drug resistance. The bottom half of the figure depicts (from left to right) the genes that encode resistance to tetracycline, streptomycin, penicillin (β-lactamase), chloramphenicol, erythromycin, and gentamicin.
TABLE 11–3 R-Factor–Mediated Resistance Mechanisms
R factors may carry one antibiotic resistance gene or may carry two or more of these genes. The medical implications of a plasmid carrying more than one resistance gene is twofold: first and most obvious is that a bacterium containing that plasmid can be resistant to more than one class of antibiotics (e.g., penicillins and aminoglycosides) and second, that the use of an antibiotic that selects for an organism resistant to one antibiotic will select for an organism that is resistant to all the antibiotics whose resistance genes are carried by the plasmid. For example, if an organism has the R plasmid depicted in Figure 11–1, then the use of penicillin will select for an organism resistant not only to penicillin, but also to tetracyclines, aminoglycosides (e.g., streptomycin and gentamicin), chloramphenicol, and erythromycin.
In addition to producing drug resistance, R factors have two very important properties: (1) They can replicate independently of the bacterial chromosome; therefore, a cell can contain many copies; and (2) they can be transferred not only to cells of the same species, but also to other species and genera. Note that this conjugal transfer is under the control of the genes of the R plasmid and not of the F (fertility) plasmid, which governs the transfer of the bacterial chromosome (see Chapter 4).
R factors exist in two broad size categories: large plasmids, with molecular weights of about 60 million, and small ones, with molecular weights of about 10 million. The large plasmids are conjugative R factors, which contain the extra DNA to code for the conjugation process. The small R factors are not conjugative and contain only the resistance genes.
In addition to conveying antibiotic resistance, R factors impart two other traits: (1) resistance to metal ions (e.g., they code for an enzyme that reduces mercuric ions to elemental mercury) and (2) resistance to certain bacterial viruses by coding for restriction endonucleases that degrade the DNA of the infecting bacteriophages.
Transposon-Mediated Resistance
Transposons are genes that are transferred either within or between larger pieces of DNA such as the bacterial chromosome and plasmids. A typical drug resistance transposon is composed of three genes flanked on both sides by shorter DNA sequences, usually a series of inverted repeated bases that mediate the interaction of the transposon with the larger DNA (see Figure 2–7). The three genes code for (1) transposase, the enzyme that catalyzes excision and reintegration of the transposon; (2) a repressor that regulates synthesis of the transposase; and (3) the drug resistance gene.
SPECIFIC MECHANISMS OF RESISTANCE
Penicillins & Cephalosporins—There are several mechanisms of resistance to these drugs. Cleavage by β–lactamases (penicillinases and cephalosporinases) is by far the most important (see Figure 10–1). β-Lactamases produced by various organisms have different properties. For example, staphylococcal penicillinase is inducible by penicillin and is secreted into the medium. In contrast, some β-lactamases produced by several gram-negative rods are constitutively produced, are located in the periplasmic space near the peptidoglycan, and are not secreted into the medium. The β-lactamases produced by various gram-negative rods have different specificities: some are more active against cephalosporins, others against penicillins. Clavulanic acid and sulbactam are penicillin analogues that bind strongly to β-lactamases and inactivate them. Combinations of these inhibitors and penicillins (e.g., clavulanic acid and amoxicillin [Augmentin]) can overcome resistance mediated by many but not all β-lactamases.
Extended-spectrum β-lactamases (ESBLs) are produced by several enteric bacteria, notably E. coli, Klebsiella, Enterobacter, and Proteus. ESBLs endow the bacteria with resistance to all penicillins, cephalosporins, and monobactams. However, these bacteria remain sensitive to combinations such as piperacillin/tazobactam. In 2009, a new strain of highly resistant Klebsiella was isolated in India carrying a plasmid that encoded New Delhi metallo-β-lactamase (NDM-1). This plasmid confers high-level resistance to many antibiotics and has spread from Klebsiella to other member of the Enterobacteriaceae. Resistant Enterobacteriaceae carrying NDM-1 have emerged in many countries, including the United States.
Resistance to penicillins can also be due to changes in the penicillin-binding proteins (PBPs) in the bacterial cell membrane. These changes account for both the low-level and high-level resistance exhibited by S. pneumoniae to penicillin G and for the resistance of S. aureus to nafcillin and other β-lactamase–resistant penicillins. The resistance of MRSA to almost all β-lactams is attributed to the presence of PBP2a, which is found particularly in MRSA. The relative resistance of Enterococcus faecalis to penicillins may be due to altered penicillin-binding proteins. Low-level resistance of Neisseria gonorrhoeae to penicillin is attributed to poor permeability to the drug. High-level resistance is due to the presence of a plasmid coding for penicillinase.
Some isolates of S. aureus demonstrate yet another form of resistance, called tolerance, in which growth of the organism is inhibited by penicillin but the organism is not killed. This is attributed to a failure of activation of the autolytic enzymes, murein hydrolases, which degrade the peptidoglycan.
Carbapenems—Resistance to carbapenems, such as imipenem, is caused by carbapenemases that degrade the β-lactam ring. This enzyme endows the organism with resistance to penicillins and cephalosporins as well. Carbapenemases are produced by many enteric gram-negative rods, especially Klebsiella, Escherichia, and Pseudomonas. Carbapenem-resistant strains of Klebsiella pneumoniae are an important cause of hospital-acquired infections and are resistant to almost all known antibiotics.
Vancomycin—Resistance to vancomycin is caused by a change in the peptide component of peptidoglycan from D-alanyl-D-alanine, which is the normal binding site for vancomycin, to D-alanine- D-lactate, to which the drug does not bind. Of the four gene loci mediating vancomycin resistance, VanA is the most important. It is carried by a transposon on a plasmid and provides high-level resistance to both vancomycin and teicoplanin. (Teicoplanin is used in Europe but is not approved in the United States.) The VanA locus encodes those enzymes that synthesize D-alanine-D-lactate as well as several regulatory proteins.
Vancomycin-resistant strains of enterococci (VRE) have been recovered from clinical specimens. Rare isolates of S. aureus that exhibit resistance to vancomycin have also been recovered from patient specimens. Rare isolates of S. pneumoniae that exhibit tolerance to vancomycin have been recovered as well.
Aminoglycosides—Resistance to aminoglycosides occurs by three mechanisms: (1) modification of the drugs by plasmid-encoded phosphorylating, adenylylating, and acetylating enzymes (the most important mechanism); (2) chromosomal mutation (e.g., a mutation in the gene that codes for the target protein in the 30S subunit of the bacterial ribosome; and (3) decreased permeability of the bacterium to the drug.
Tetracyclines—Resistance to tetracyclines is the result of failure of the drug to reach an inhibitory concentration inside the bacteria. This is due to plasmid-encoded processes that either reduce the uptake of the drug or enhance its transport out of the cell.
Chloramphenicol—Resistance to chloramphenicol is due to a plasmid-encoded acetyltransferase that acetylates the drug, thus inactivating it.
Erythromycin—Resistance to erythromycin is due primarily to a plasmid-encoded enzyme that methylates the 23S rRNA, thereby blocking binding of the drug. An efflux pump that reduces the concentration of erythromycin within the bacterium causes low-level resistance to the drug. An esterase produced primarily by enteric gram-negative rods cleaves the macrolide ring, which inactivates the drug.
Sulfonamides—Resistance to sulfonamides is mediated primarily by two mechanisms: (1) a plasmid-encoded transport system that actively exports the drug out of the cell, and (2) a chromosomal mutation in the gene coding for the target enzyme dihydropteroate synthetase, which reduces the binding affinity of the drug.
Trimethoprim—Resistance to trimethoprim is due primarily to mutations in the chromosomal gene that encodes dihydrofolate reductase, the enzyme that reduces dihydrofolate to tetrahydrofolate.
Quinolones—Resistance to quinolones is due primarily to chromosomal mutations that modify the bacterial DNA gyrase.
Rifampin—Resistance to rifampin is due to a chromosomal mutation in the gene encoding the bacterial RNA polymerase, resulting in ineffective binding of the drug. Because resistance occurs at high frequency (10–5), rifampin is not prescribed alone for the treatment of infections. It is used alone for the prevention of certain infections because it is administered for only a short time (see Table 10–5).
Isoniazid—Resistance of M. tuberculosis to isoniazid is due to mutations in the organism’s catalase–peroxidase gene. Catalase or peroxidase enzyme activity is required to synthesize the metabolite of isoniazid that actually inhibits the growth of M. tuberculosis.
Ethambutol—Resistance of M. tuberculosis to ethambutol is due to mutations in the gene that encodes arabinosyl transferase, the enzyme that synthesizes the arabinogalactan in the organism’s cell wall.
Pyrazinamide—Resistance of M. tuberculosis to pyrazinamide (PZA) is due to mutations in the gene that encodes bacterial amidase, the enzyme that converts PZA to the active form of the drug, pyrazinoic acid.
NONGENETIC BASIS OF RESISTANCE
There are several nongenetic reasons for the failure of drugs to inhibit the growth of bacteria:
(1) Bacteria can be walled off within an abscess cavity that the drug cannot penetrate effectively. Surgical drainage is therefore a necessary adjunct to chemotherapy.
(2) Bacteria can be in a resting state (i.e., not growing); they are therefore insensitive to cell wall inhibitors such as penicillins and cephalosporins. Similarly, M. tuberculosis can remain dormant in tissues for many years, during which time it is insensitive to drugs. If host defenses are lowered and the bacteria begin to multiply, they are again susceptible to the drugs, indicating that a genetic change did not occur.
(3) Under certain circumstances, organisms that would ordinarily be killed by penicillin can lose their cell walls, survive as protoplasts, and be insensitive to cell wall–active drugs. Later, if such organisms resynthesize their cell walls, they are fully susceptible to these drugs.
(4) The presence of foreign bodies makes successful antibiotic treatment more difficult. This applies to foreign bodies such as surgical implants and catheters as well as materials that enter the body at the time of penetrating injuries, such as splinters and shrapnel.
(5) Several artifacts can make it appear that the organisms are resistant (e.g., administration of the wrong drug or the wrong dose or failure of the drug to reach the appropriate site in the body). (A good example of the latter is the poor penetration into spinal fluid by several early-generation cephalosporins.) Failure of the patient to take the drug (noncompliance, nonadherence) is another artifact.
SELECTION OF RESISTANT BACTERIA BY OVERUSE & MISUSE OF ANTIBIOTICS
Serious outbreaks of diseases caused by gram-negative rods resistant to multiple antibiotics have occurred in many developing countries. In North America, many hospital-acquired infections are caused by multidrug-resistant organisms. Three main points of overuse and misuse of antibiotics increase the likelihood of these problems by enhancing the selection of resistant mutants:
(1) Some physicians use multiple antibiotics when one would be sufficient, prescribe unnecessarily long courses of antibiotic therapy, use antibiotics in self-limited infections for which they are not needed, and overuse antibiotics for prophylaxis before and after surgery.
(2) In many countries, antibiotics are sold over the counter to the general public; this practice encourages inappropriate and indiscriminate use of the drugs.
(3) Antibiotics are used in animal feed to prevent infections and promote growth. This selects for resistant organisms in the animals and may contribute to the pool of resistant organisms in humans.
ANTIBIOTIC SENSITIVITY TESTING
Minimal Inhibitory Concentration
For many infections, the results of sensitivity testing are important in the choice of antibiotic. These results are commonly reported as the minimal inhibitory concentration (MIC), which is defined as the lowest concentration of drug that inhibits the growth of the organism. The MIC is determined by inoculating the organism isolated from the patient into a series of tubes or cups containing twofold dilutions of the drug (Figure 11–2). After incubation at 35°C for 18 hours, the lowest concentration of drug that prevents visible growth of the organism is the MIC. This provides the physician with a precise concentration of drug to guide the choice of both the drug and the dose.
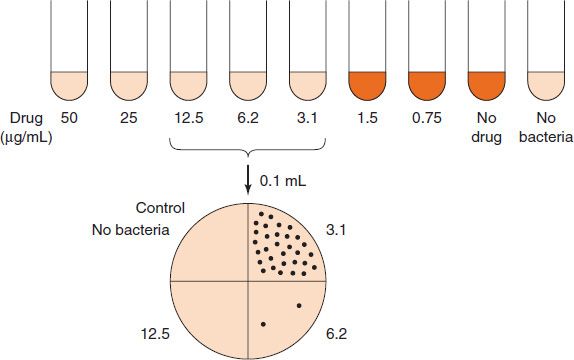
FIGURE 11–2 Determination of minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC). Top: The patient’s organism is added to tubes containing decreasing amounts of the antibiotic. After incubation at 37°C overnight, growth of the bacteria is observed visually. The lowest concentration of drug that inhibits growth (i.e., 3.1 μg/mL) is the MIC. However, at this point, it is not known whether the bacteria have been killed or whether the drug has only inhibited their growth. Bottom: To determine whether that concentration of drug is bactericidal (i.e., to determine its MBC), an aliquot (0.1 mL) from the tubes is plated on an agar plate that does not contain any drug. The concentration of drug that inhibits at least 99.9% of the bacterial colonies (i.e., 6.2 μg/mL) is the MBC.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree