FIGURE 6–1. Size comparison of viruses with other microbes. (Adapted with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
Viruses range in size from 20 to 300 nm in diameter
The basic structure of all viruses places the nucleic acid genome (DNA or RNA) on the inside of a protein shell called a capsid. Some human viruses are further packaged into a lipid membrane, or envelope, which is usually acquired from the cytoplasmic membrane of the infected cell during release from the cell. Viruses that are not enveloped have a defined external capsid and are referred to as naked capsid viruses. The genomes of enveloped viruses form a protein complex and a structure called a nucleocapsid, which is often surrounded by a matrix protein that serves as a bridge between the nucleocapsid and the inside of the viral membrane. Protein or glycoprotein structures called spikes, which often protrude from the surface of virus particles, are involved in the initial contact with receptor on host cells. These basic design features are illustrated schematically in Figure 6–2 as well as in the electron micrographs in Figures 6–3A–C and 6–4.
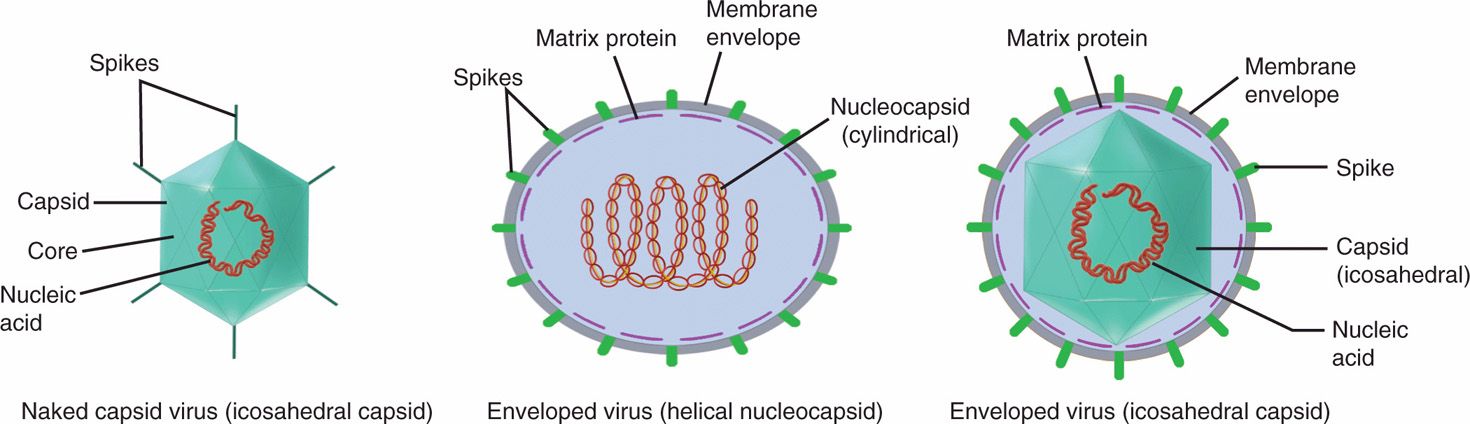
FIGURE 6–2. Schematic drawing of two basic types of virions, naked capsid virus and enveloped virus. In naked capsid virus, the genome is condensed with a defined external capsid (coat protein), whereas enveloped virus has a nucleocapsid or capsid wrapped in a lipid bilayer envelope. (Adapted with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
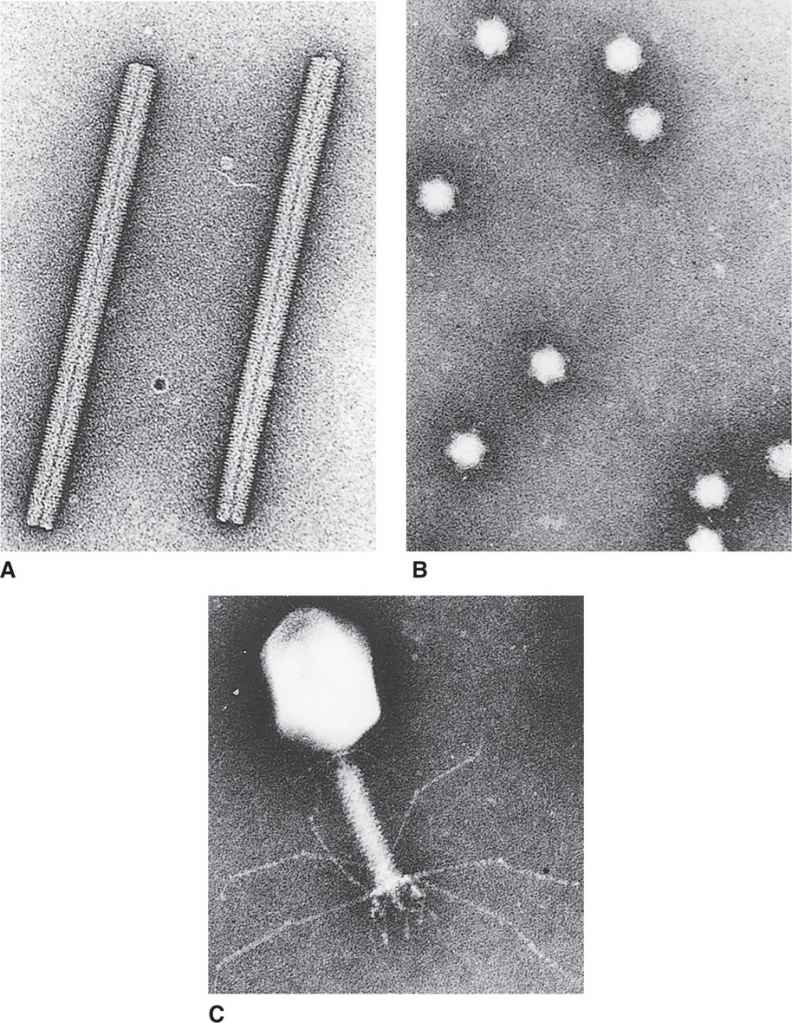
FIGURE 6–3. Three basic virus designs. A. Tobacco mosaic virus. B. Bacteriophage φXI74. C. Bacteriophage T4. (Courtesy of Dr Robley C. Williams.)
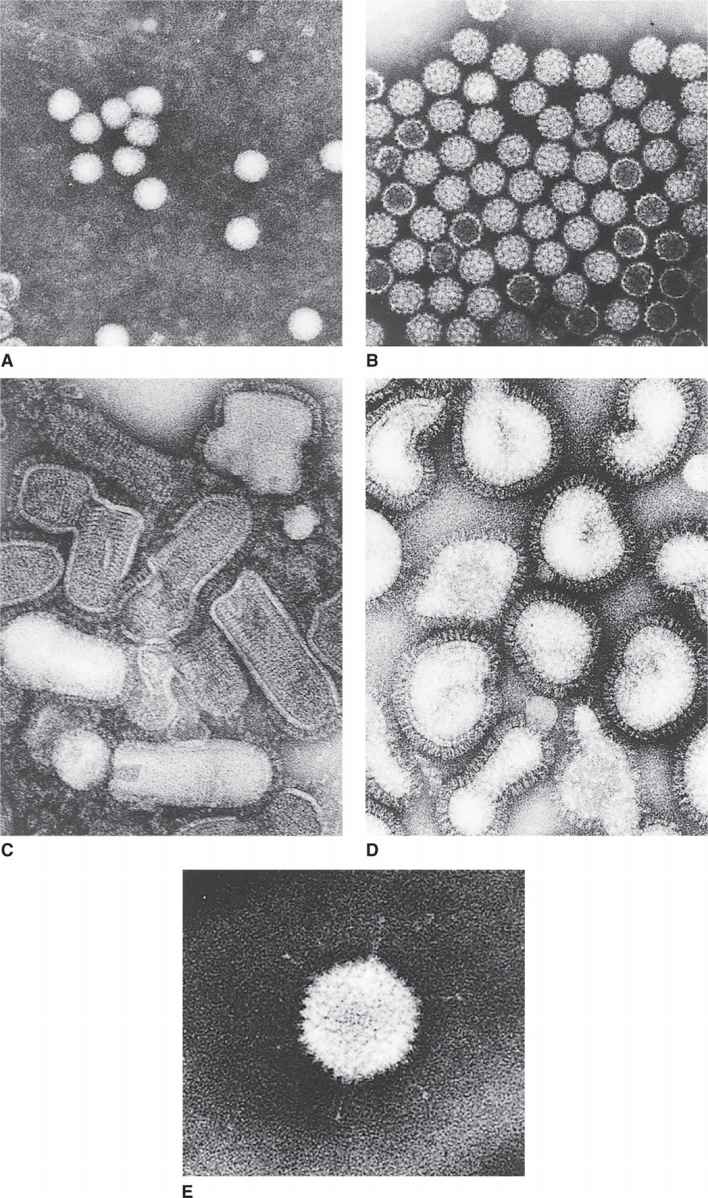
FIGURE 6–4. Representative human/animal viruses. A. Poliovirus. B. Simian virus 40. C. Vesicular stomatitis virus. D. Influenza virus. E. Adenovirus. (Courtesy of Dr. Robley C. Williams.)
Naked capsid viruses have a nucleic acid genome within a protein shell (capsid)
Enveloped viruses have a nucleocapsid (nucleic acid-protein complex) packaged into a lipoprotein envelope
Viruses often have surface protrusions called spikes
The protein shell forming the capsid or the nucleocapsid assumes one of two basic shapes: cylindrical (helical) or spherical (icosahedral). Some of the more complex bacteriophages combine these two basic shapes. Examples of these three structural categories can be seen in the electron micrographs in Figure 6–3.
Two basic shapes: cylindrical (helical) and spherical (icosahedral)
The capsid or envelope of viruses functions (1) to protect the nucleic acid genome from damage during the extracellular passage of the virus from one cell to another, (2) to aid in the process of entry into the cell, and (3) in some cases, to package enzymes essential for the early steps of the infection process.
Outer shell is protective and aids in entry and packaging
In general, the nucleic acid genome of a virus is hundreds of times longer than the longest dimension of the complete virion. It follows that the viral genome must be extensively condensed during the process of virion assembly. For naked capsid viruses, this condensation is achieved by the association of the viral nucleic acid with basic proteins encoded by the virus to form the core of the virus (Figure 6–2). For enveloped viruses, the formation of the nucleocapsid serves to condense the viral nucleic acid genome. The virion may also contain certain virus encoded essential enzymes and/or accessory/regulatory proteins.
Nucleic acid must be condensed during virion assembly
GENOME STRUCTURE
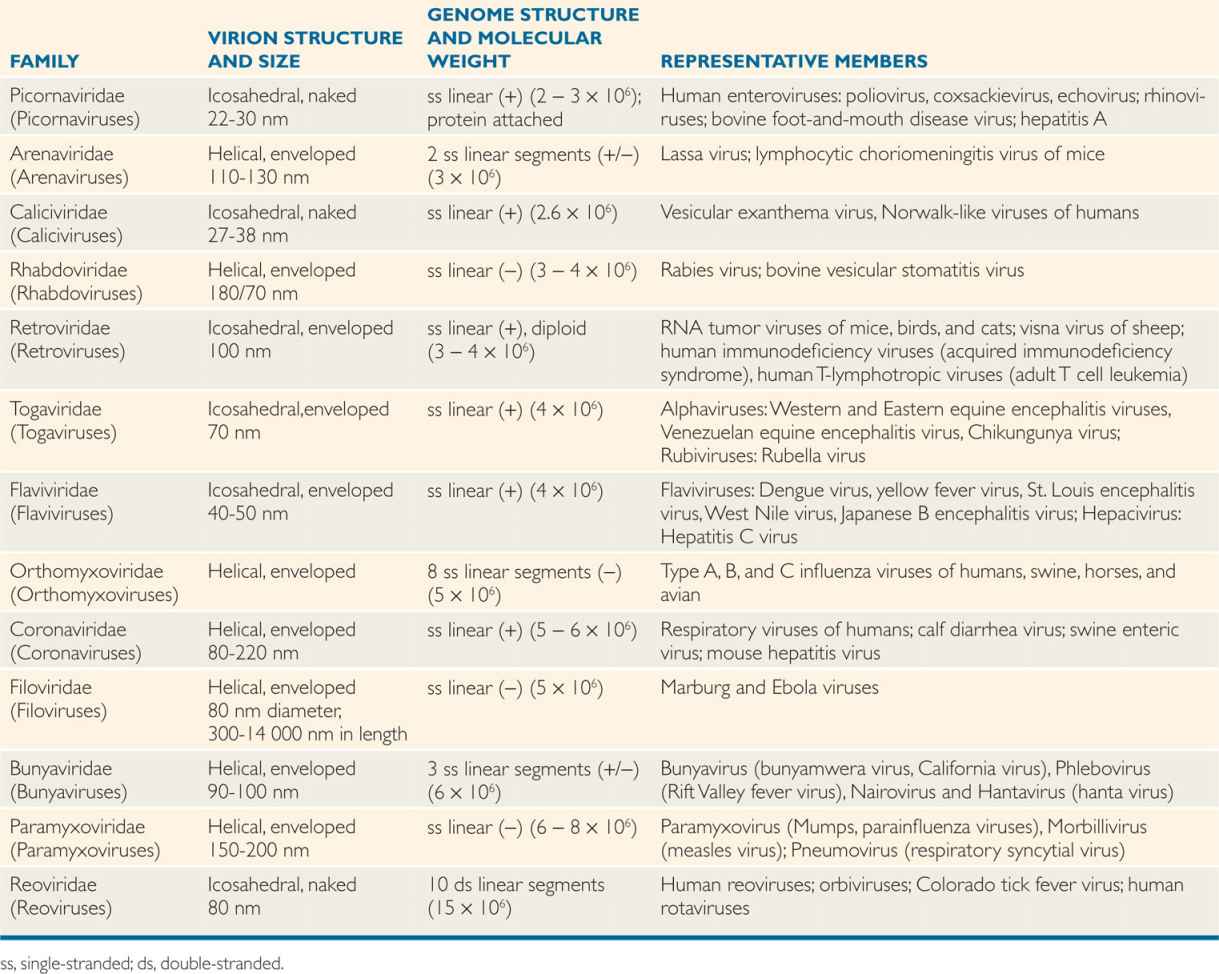
Viral genomes can be made of either RNA or DNA and also can be either single-stranded or double-stranded. The RNA viruses can be either positive sense (indicated by a +) (polarity of mRNA) or negative sense (–) (complementary to or antisense of mRNA), double-stranded (one strand + and the second strand –) or ambisense (both + and – polarity on the same strand). Although the RNA genomes of most viruses are linear, some RNA viruses may also have segmented genomes (several segments or pieces of RNA), with each segment responsible for encoding a protein.
The genomes of viruses can be either RNA or DNA, but not both
DNA or RNA genomes may be single- or double-stranded
RNA genomes could be (+) positive sense, negative (–) sense or ambisense (±)
The DNA genome of viruses can be both linear and circular genomes. Most viruses contain a single copy of their genome, except that retroviruses carry two identical copies of its genome and are, therefore, diploid. A few viral genomes (picornaviruses, hepatitis B virus, and adenoviruses) contain covalently attached protein on the ends of the DNA or RNA chains that are remnants of the replication process. Structural diversity among the viruses is most obvious when the makeup of viral genomes is considered.
Genomes may be linear or circular
Some genomes are segmented
CAPSID STRUCTURE
 Subunit Structure of Capsids
Subunit Structure of Capsids
The capsids or nucleocapsids are virus-encoded specific proteins that protect the genome and confer shapes to viruses. The capsids of all viruses are composed of many copies of one or, at most, several different kinds of protein subunits. This fact follows from two fundamental considerations. First, all viruses code for their own capsid proteins, and even if the entire coding capacity of the genome were to be used to specify a single giant capsid protein, the protein would not be large enough to enclose the nucleic acid genome. Thus, multiple protein copies are needed, and, in fact, the simplest spherical virus contains 60 identical protein subunits. Second, viruses are such highly symmetrical structures that it is not uncommon to visualize naked capsid viruses in the electron microscope as a crystalline array (eg, simian virus 40 in Figure 6–4B). The simplest way to construct a regular symmetrical structure out of irregular protein subunits is to follow the rules of crystallography and form an aggregate involving many identical copies of the subunits, in which each subunit bears the same relation to its neighbors as to every other subunit.
Capsids and nucleocapsids are composed of multiple copies of protein molecule(s) in crystalline array
The presence of many identical protein subunits in viral capsids or the existence of many identical spikes in the membrane of enveloped viruses has important implications for adsorption, hemagglutination, and recognition of viruses by neutralizing antibodies. Two main architectures; cylindrical (helical symmetry) and spherical (icosahedral or cubic symmetry).
 Cylindrical (Helical) Architecture
Cylindrical (Helical) Architecture
A cylindrical shape is the simplest structure for a capsid or a nucleocapsid. The first virus to be crystallized and studied in structural detail was a plant pathogen, tobacco mosaic virus (TMV) (Figure 6–3A). The capsid of TMV is shaped like a rod or a cylinder, with the RNA genome wound in a helix inside it. The capsid is composed of multiple copies of a single kind of protein subunit arranged in a close-packed helix, which places every subunit in the same microenvironment. Because of the helical arrangement of the subunits, viruses that have this type of design are often said to have helical symmetry. Although less is known about the architecture of human viruses with helical symmetry, it is likely that their structures follow the same general pattern as that of TMV. Thus, the nucleocapsids of influenza, measles, mumps, rabies, and poxviruses (Table 6–1) are probably constructed with a helical arrangement of protein subunits in close association with the nucleic acid genome.
TABLE 6–1 Classification of RNA Human Viruses
Cylindrical viruses have capsid protein molecules arranged in a helix
 Spherical (Icosahedral) Architecture
Spherical (Icosahedral) Architecture
The construction of a spherically (icosahedral) shaped virus similarly involves the packing together of many identical subunits, but, in this case, the subunits are placed on the surface of a geometric solid called an icosahedron. An icosahedron has 12 vertices, 30 sides, and 20 triangular faces (Figure 6–5). Because the icosahedron belongs to the symmetry group that crystallographers refer to as cubic, spherically shaped viruses are said to have cubic symmetry. (Note that the term cubic, as used in this context, has nothing to do with the more familiar shape called the cube.)
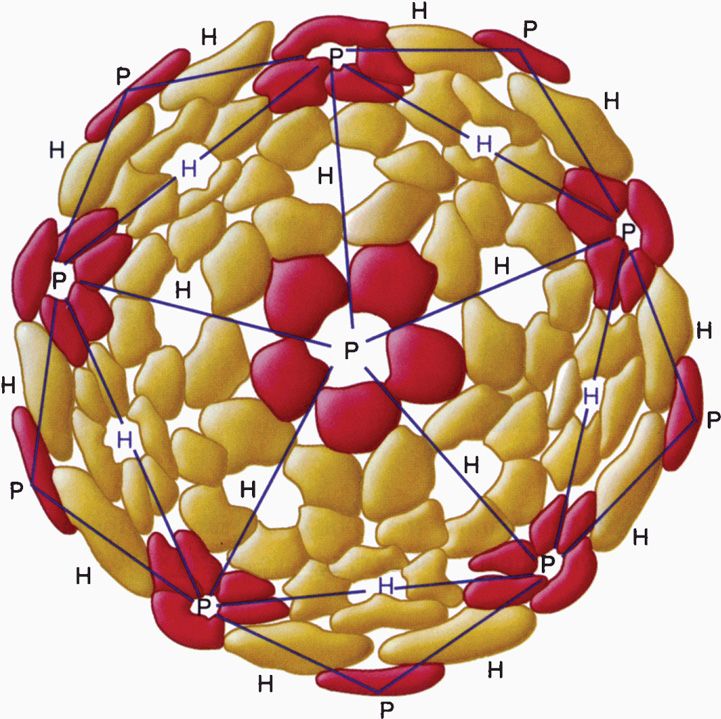
FIGURE 6–5. Diagram of an icosahedron showing 12 vertices, 20 faces, and 30 sides. The colored balls indicate the position of protomers forming a pentamer on the icosahedron. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
Spherical viruses exhibit icosahedral symmetry
When viewed in the electron microscope, many naked capsid viruses and some nucleocapsids appear as spherical particles with a surface topology that makes it appear that they are constructed of identical ball-shaped subunits (Figure 6–4B and E). These visible structures are referred to as morphologic subunits, or capsomeres. A capsomere is generally composed of either five or six individual protein molecules, each one referred to as a structural subunit, or protomer. In the simplest virus with cubic symmetry, five protomers are placed at each one of the 12 vertices of the icosahedron as shown in Figure 6–5 to form a capsomere called a pentamer. In this case, the capsid is composed of 12 pentamers, or a total of 60 protomers. Note that in the case of helical symmetry, this arrangement places every protomer in the same microenvironment as that of every other protomer.
Capsomeres are surface structures composed of five or six protein molecules
To accommodate the larger cavity required by viruses with large genomes, the capsids contain many more protomers. These viruses are based on a variation of the basic icosahedron in which the construction involves a mixture of pentamers and hexamers rather than only pentamers. A detailed description of this higher level of virus structure is beyond the scope of this text. Examples of icosahedral capsids are shown in Figure 6–6.
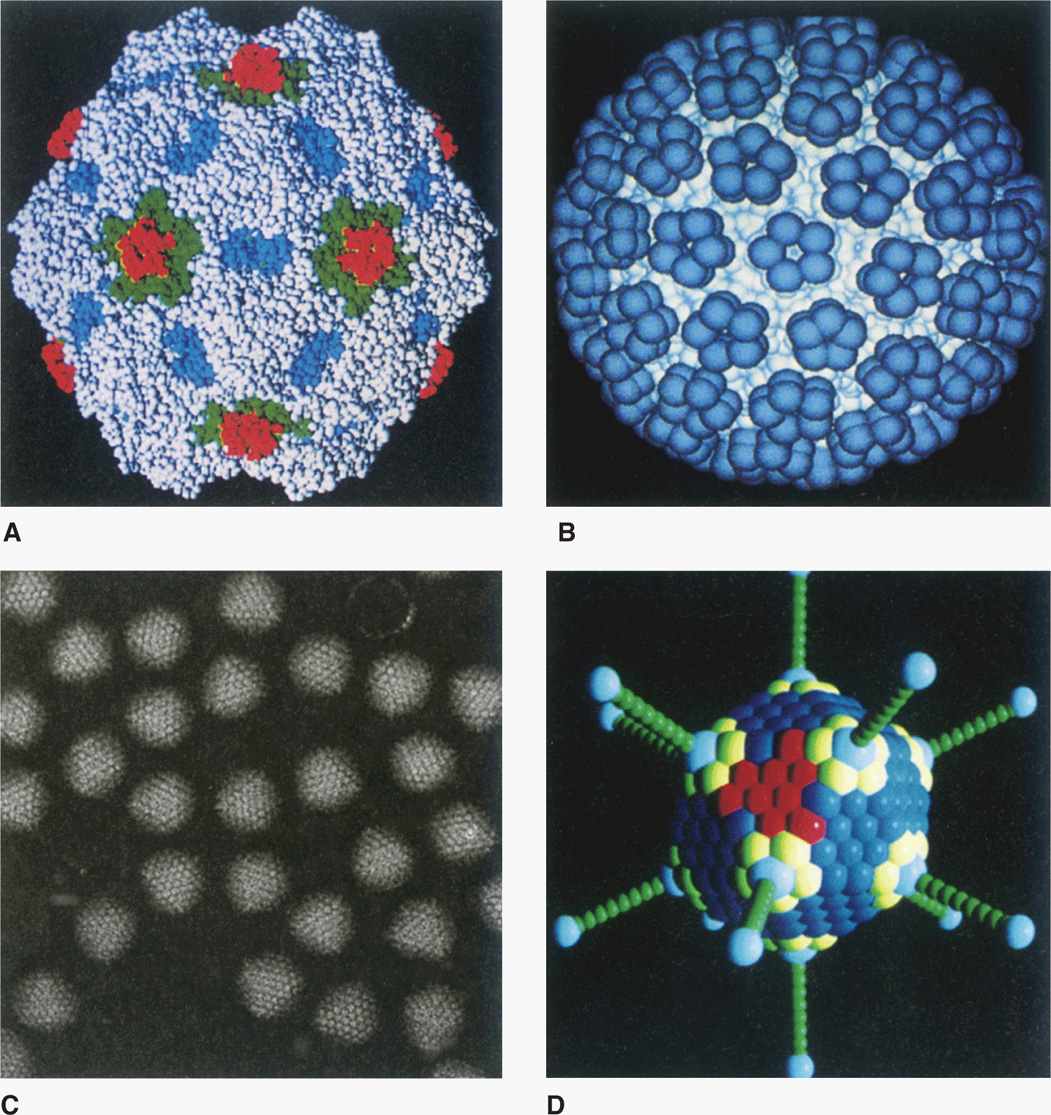
FIGURE 6–6. Examples of icosahedral capsids. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
 Special Surface Structures
Special Surface Structures
Many viruses have structures that protrude from the surface of the virion. In virtually every case, these structures are important for the two earliest steps of infection—adsorption and penetration. The most dramatic example of such a structure is the tail of some bacteriophages (Figure 6–3C), which, acts as a channel for the transfer of the genome into the cell. Other examples of surface structures include the spikes of adenovirus (Figure 6–4E) and the glycoprotein spikes found in the membrane of enveloped viruses (see influenza virus in Figure 6–4D). Even viruses without obvious surface extensions probably contain short projections, which, like the more obvious spikes, are involved in the specific binding of the virus to the cell surface.
Surface structures are important in adsorption and penetration
 Envelope Structure
Envelope Structure
Many human viruses have an outer lipid bilayer membrane that is derived from cellular membranes, mainly the plasma membrane, but also, in some cases, cytoplasmic or nuclear membranes. The viral envelope lipid layer membrane contains virus-encoded glycoproteins called “spikes” or “peplomers” or “viral envelope proteins.” The envelope spikes bind to the receptor on the host cells, help the virus envelope membrane fuse with the cellular membrane of the host cells, and act as principal antigens against which the host mounts immune response for the recognition of the virus. Enveloped viruses have another protein, the matrix protein, which serves as a bridge between nucleocapsid and inner membrane of the envelope (Figure 6–2). Examples of enveloped viruses are shown in Figure 6–7, with both helical (Figure 6–7A and B) and icosahedral or cubic (Figure 6–7C and D) symmetry.
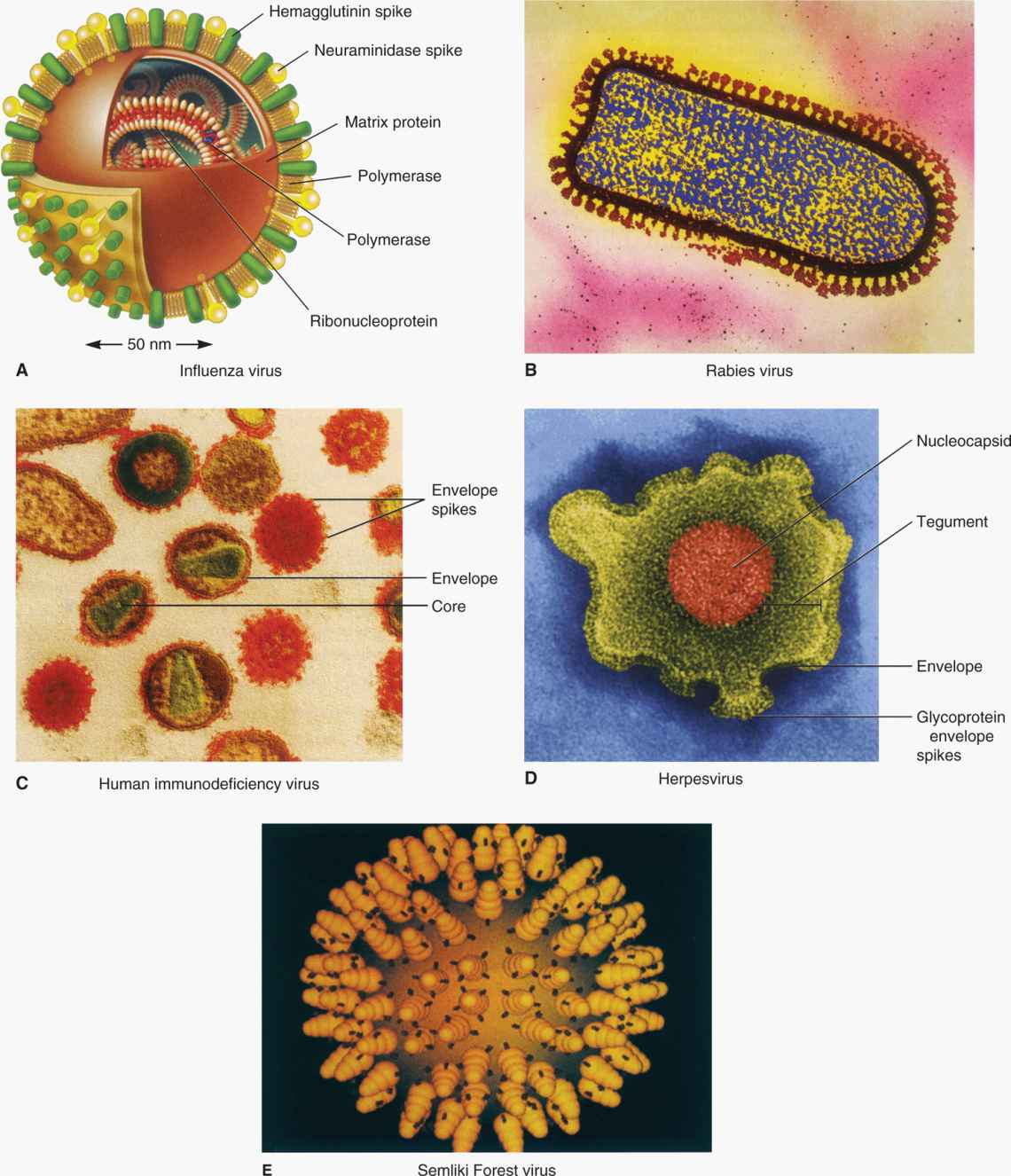
FIGURE 6–7. Examples of enveloped viruses. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
Viral envelopes are lipid bilayer membranes containing virus encoded glycoproteins called spikes or peplomers
Enveloped viruses are more sensitive to detergents, solvents, ethanol, ether, and heat compared with nonenveloped (naked capsid) viruses whose outer coat is capsid protein. Both envelope glycoproteins, and naked capsid viruses’ spikes become antigens after infection and the host mounts both cell-mediated and humoral immune responses for the elimination of virus-infected cells and cell-free virus, respectively. These antigens determine the viral serotypes that are based on antigenic variation and are type-specific such as poliovirus serotypes 1, 2, and 3. Viral serotypes have cross-reactivity but, often, little cross-protection. Viral serotypes arise because of antigenic variations that allow viruses to escape preexisting immune response.
Envelope glycoproteins, like surface proteins of naked capsid viruses, bind to the receptors on host cells for virus entry
Viral serotypes arise due to antigenic variation that have cross-reactivity but, often, little-cross protection
CLASSIFICATION OF VIRUSES
The classification of viruses has evolved at a slower pace than other microorganisms. The International Committee for Taxonomy of Viruses (ICTV) considered various properties, including virions, genome, proteins, envelope, replication, and physical and biologic properties. Based on these properties, virus families are designated with the suffix, -viridae (as in Herpesviridae), virus subfamilies with suffix -virinae (Herpesvirinae), virus genera with suffix -virus (Herpesvirus), and virus species designated by a virus type (herpes simplex virus 1). Tables 6–1, 6–2, and 6–3 present a classification scheme for human RNA and DNA viruses, respectively, which is based solely on their structure. The viruses are arranged in order of increasing genome size. It is important to bear in mind that phylogenetic relationships cannot be inferred from this taxonomic scheme. The tables should not be memorized, but rather used as a reference guide to virus structure. In general, viruses with similar structures exhibit similar replication strategies, as discussed later.
TABLE 6–2 Unclassified RNA Human Viruses
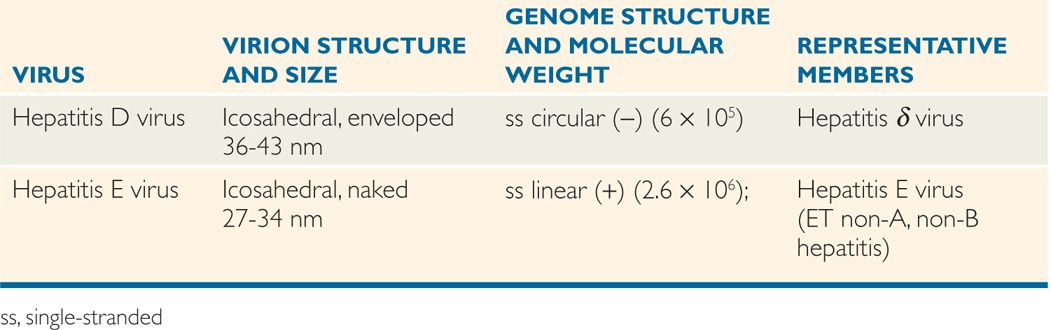
TABLE 6–3 Classification of DNA Human Viruses
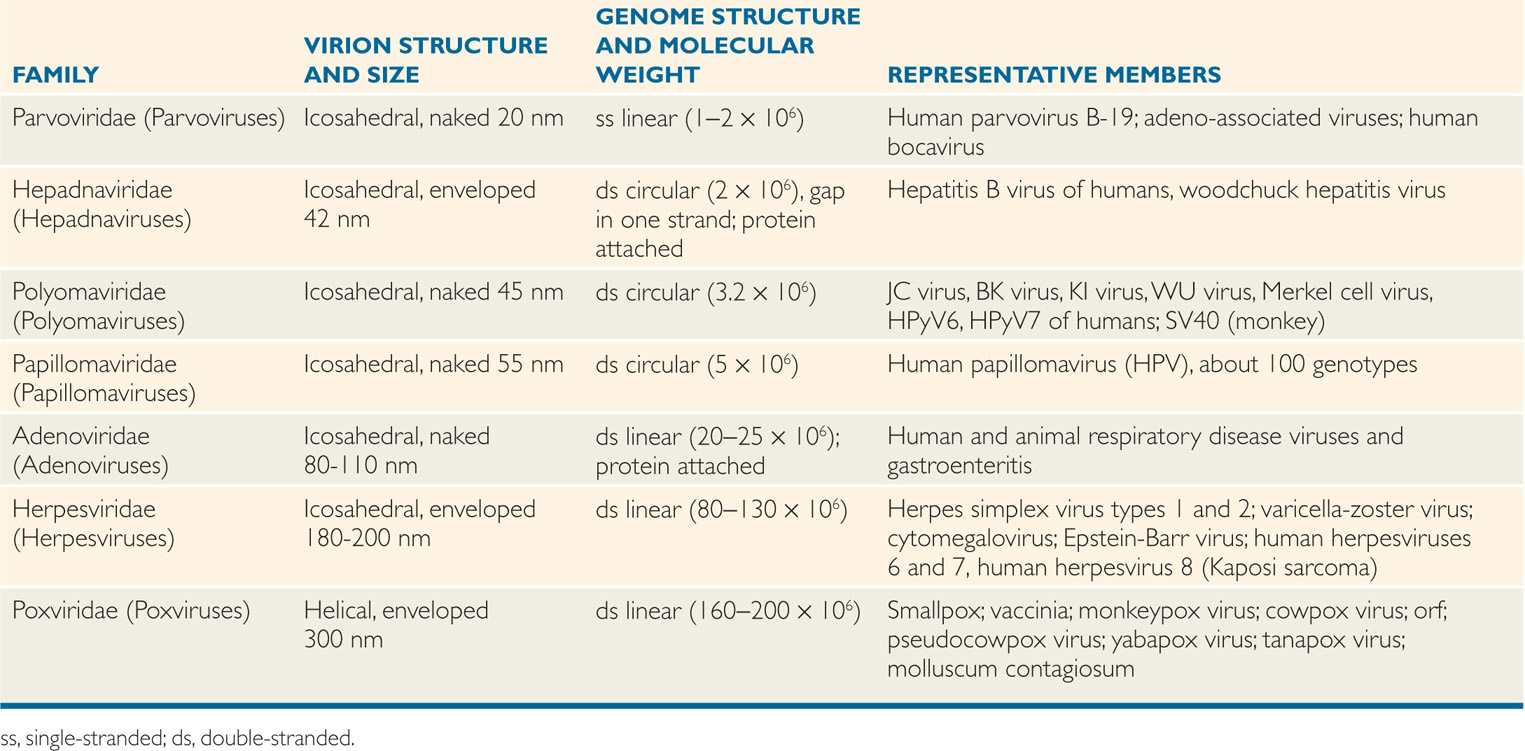
Representative and important bacteriophages are listed along with their properties in Table 6–4. In the chapters that follow, the properties of the well-studied temperate bacteriophage, λ, are described to illustrate the replicative strategies of the more medically important, but less well-studied, β phage of Corynebacterium diphtheriae.
TABLE 6–4 Some Important Bacteriophages
 Virus Replication
Virus Replication
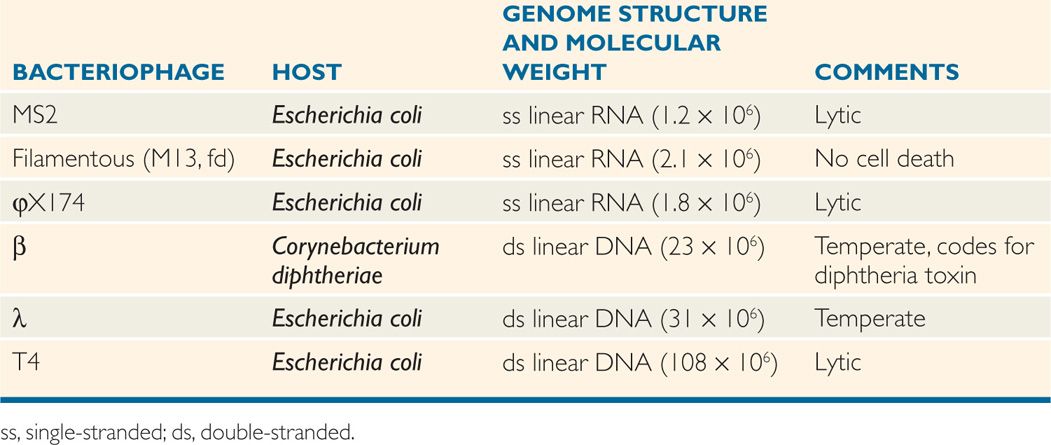
Virus replication cycle typically consists of six discrete phases: (1) adsorption or attachment to the host cell, (2) penetration or entry, (3) uncoating to release the genome, (4) synthetic or virion component production, (5) assembly, and (6) release from the cell. These phases are shown in a general scheme of virus replication cycle in Figure 6–8.
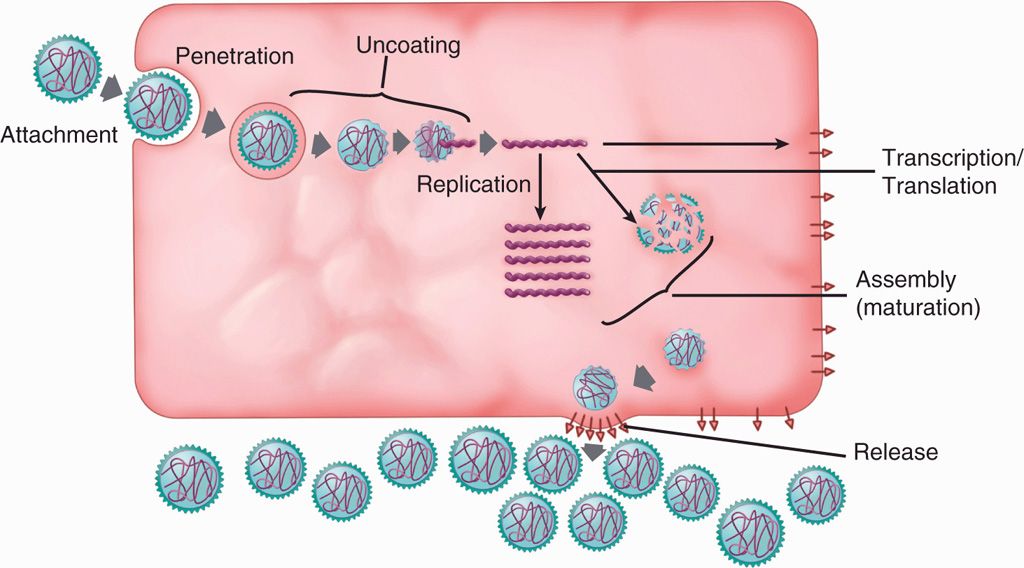
FIGURE 6–8. Virus replication cycle. A general scheme of the six discrete steps of virus replication cycle, including attachment, penetration, uncoating, synthetic phase (transcription, translation and replication), assembly, and release.
This series of events, sometimes with slight variations, describes what is called the productive or lytic response; however, this is not the only possible outcome of a virus infection. Some viruses can also enter into a very different kind of relationship with the host cell in which no new virus is produced, the cell survives and divides, and the viral genetic material persists indefinitely in a latent state. This outcome of an infection is referred to as the nonproductive response. The nonproductive response in the case of bacteriophages is called lysogeny and, in several human and animal viruses under some circumstances, may be associated with oncogenic transformation. (This use of the term transformation is to be distinguished from DNA transformation of bacteria discussed in Chapter 21.)
Viral infections may be productive or nonproductive
Some human viruses can cause oncogenic transformation
Some viruses can also cause a chronic infection where a low level of the virus is produced with little or no damage to the target tissue. Both latent infection and chronic infection are called persistent infection. Virus replication also depends on virus–host cell interaction such as the type of cells it infects—whether permissive or nonpermissive cells. Permissive cells are those that permit production of progeny virus particles and/or viral transformation. However, nonpermissive cells do not allow virus replication, but may allow virus transformation. Some viruses enter cells that do not support virus replication, but some early viral proteins cause cell death; this infection is termed abortive infection.
Some viruses cause chronic or latent infection
Permissive cells allow virus replication and/or viral transformation
Nonpermissive cells do not permit virus replication, but may allow viral transformation
The outcome of an infection depends on the particular virus–host combination and on other factors such as the extracellular environment, multiplicity of infection, and physiology and developmental state of the cell. Viruses that can enter only into a productive relationship are called lytic or virulent viruses. Viruses that can establish either a productive or a nonproductive relationship with their host cells are referred to as temperate viruses. Some temperate viruses can be reactivated or “induced” to leave the latent state and enter into the productive response. Whether induction occurs depends on the particular virus–host combination, the physiology of the cell, and the presence of extracellular stimuli.
Temperate viruses can either replicate or enter a latent state
GROWTH AND ASSAY OF VIRUSES
Viruses are generally propagated in the laboratory by mixing the virus and susceptible cells together and incubating the infected cells until lysis occurs. After lysis, the cells and cell debris are removed by a brief centrifugation, and the resulting supernatant is called a lysate.
The growth of human viruses requires that the host cells be cultivated in the laboratory, mostly in human or animal cell lines (cell derived from tumors or cells transformed by viruses) and, in some cases, in primary cells derived from tissues. To prepare cells for growth in vitro, a tissue is removed from an animal, and the cells are disaggregated using the proteolytic enzyme trypsin. The cell suspension is seeded into a plastic Petri dish in a medium containing a complex mixture of amino acids, vitamins, minerals, and sugars. In addition to these nutritional factors, the growth of animal cells requires components present in animal serum. This method of growing cells is referred to as tissue culture, and the initial cell population is called a primary culture. The cells attach to the bottom of the plastic dish and remain attached as they divide and eventually cover the surface of the dish. When the culture becomes crowded, the cells generally cease dividing and enter a resting state. Propagation can be continued by removing the cells from the primary culture plate using trypsin and reseeding a new plate.
Viruses are cultivated in cell lines or cell cultures derived from animal tissues
Cells taken from a normal (as opposed to cancerous) tissue cannot usually be propagated in this manner indefinitely. Eventually, most of the cells die; a few may survive, and these survivors often develop into a permanent cell line. Cell lines can also be generated directly from tumors or from virus transformed cells. Such cell lines are very useful as host cells for isolating and assaying viruses in the laboratory, but they rarely bear much resemblance to the tissue from which they originated. When cells are taken from a tumor and cultivated in vitro, they display a very different set of growth properties, including long-term survival, reflecting their tumor phenotype.
Permanent cell lines are useful for growing viruses
When a virus is propagated in tissue culture cells, the cellular changes induced by the virus, which usually culminate in cell death, are often characteristic of a particular virus and are referred to as the cytopathic effect of the virus.
Cytopathic effects are characteristic for individual viruses
Viruses are quantitated by a method called the plaque assay (see Plaque Assay under Quantitation of Viruses for a detailed description of the method). Briefly, viruses are mixed with cells on a Petri plate so that each infectious particle gives rise to a zone of lysed or dead cells called a plaque. From the number of plaques on the plate, the titer of infectious particles in the lysate is calculated. Virus titers are expressed as the number of plaque-forming units per milliliter (pfu/mL).
Viruses are quantitated by a plaque assay
ONE-STEP GROWTH EXPERIMENT
The purpose of a one-step growth experiment is to understand various stages of viral infection. To describe an infection in temporal and quantitative terms, it is useful to perform a one-step growth experiment (Figure 6–9). The objective in such an experiment is to infect every cell in a culture so that the whole population proceeds through the infection process in a synchronous fashion. The ratio of infecting plaque-forming units to cells is called the multiplicity of infection (MOI). By infecting at a high MOI (eg, 10, as in Figure 6–9), one can be certain that every cell is infected.
One-step growth experiments are useful in the study of various stages of viral infection
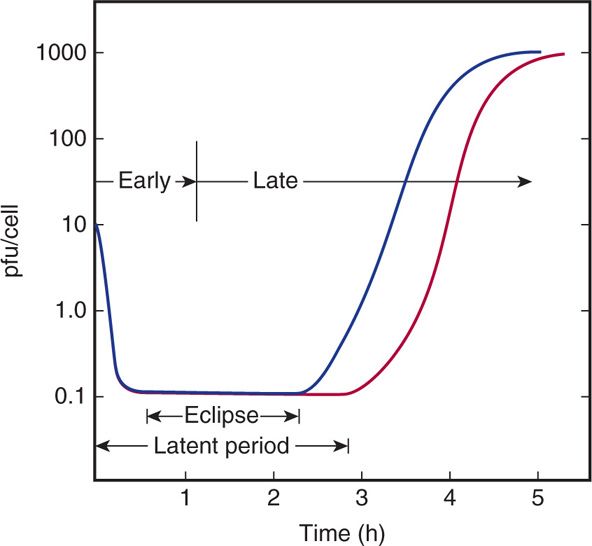
FIGURE 6–9. One-step growth experiment. The purpose of a one-step growth experiment is to understand the various phases of infection that occur following a viral infection. In the graph, blue line measures virus in the culture medium (outside the cell) and red line measures virus inside the cells. pfu, plaque-forming units.
The time course and efficiency of adsorption can be followed by the loss of infectious virus from the medium after removal of the cells (blue line in Figure 6–9). In the example shown, adsorption takes approximately half an hour, and all but 1% of the virus is adsorbed. If samples of the culture containing the infected cells are treated so as to break open the cells before assaying for virus (red line in Figure 6–9), it can be observed that infectious virus initially disappears, because no infectious particles are detectable above the background of unadsorbed virus. The period of infection in which no infectious viruses are found inside the cell is called the eclipse phase and emphasizes that the original virions lose their infectivity soon after entry. Infectivity is lost because, as is discussed later, the virus particles are dismantled as a prelude to their reproduction. Later, infectious virus particles rapidly reappear in increasing numbers and are detected inside the cell prior to their release into the environment (Figure 6–9). The length of time from the beginning of infection until progeny virions are found outside the cells is referred to as the latent period. Latent periods range from 20 minutes to hours for bacteriophages and from a few hours to many days for human viruses.
Shortly after infection, a virus loses its identity (eclipse phase)
Infectious virus reappears at end of eclipse phase inside the cell
The time in the infection at which genome replication begins is typically used to divide the infection operationally into early and late phases. Early viral gene expression is largely restricted to the production of the proteins required for genome replication; later, the proteins synthesized are, primarily, those necessary for construction of the new virus particles.
Proteins for replication are produced early and those for construction of virions are produced late
The average number of plaque-forming units released per infected cell is called the burst size for the infection. In the example shown, the burst size is approximately 1000. Burst sizes range from less than 10 for some relatively inefficient infections to millions for some highly virulent viruses.
VIRUS REPLICATION CYCLE
 Adsorption or Attachment
Adsorption or Attachment
The first step in every viral infection is the attachment or adsorption of the infecting virus particle to the surface of the host cell. A prerequisite for this interaction is a collision between the virion and the cell. Viruses do not have any capacity for locomotion and, therefore, the collision event is simply a random process determined by diffusion. Therefore, similar to any bimolecular reaction, the rate of adsorption is determined by the concentrations of both the virions and the cells.
Only a small percentage of the collisions between a virus and its host cell lead to a successful infection because adsorption is a highly specific reaction that involves protein molecules on the surface of the virion called virion attachment proteins or spikes and certain molecules on the surface of the cell that are called receptors. Typically, 104 to 105 receptors are on the cell surface. Receptors for some bacteriophages are found on pili of bacteria, although most adsorb to receptors found on the bacterial cell wall. Receptors for human viruses are usually glycoproteins located in the plasma membrane of the cell. Table 6–5 lists some of the receptors that have been identified for medically important viruses. It appears that viruses have evolved to make use of a wide variety of surface molecules as receptors, which are normally signaling devices or immune system components. Any attempts to design agents that block viral infections by binding to the receptors for a long time must consider the possibility that the loss of the normal cellular function associated with the receptors would have serious consequences for the host organism.
TABLE 6–5 Examples of Cell Receptors for Human Viruses
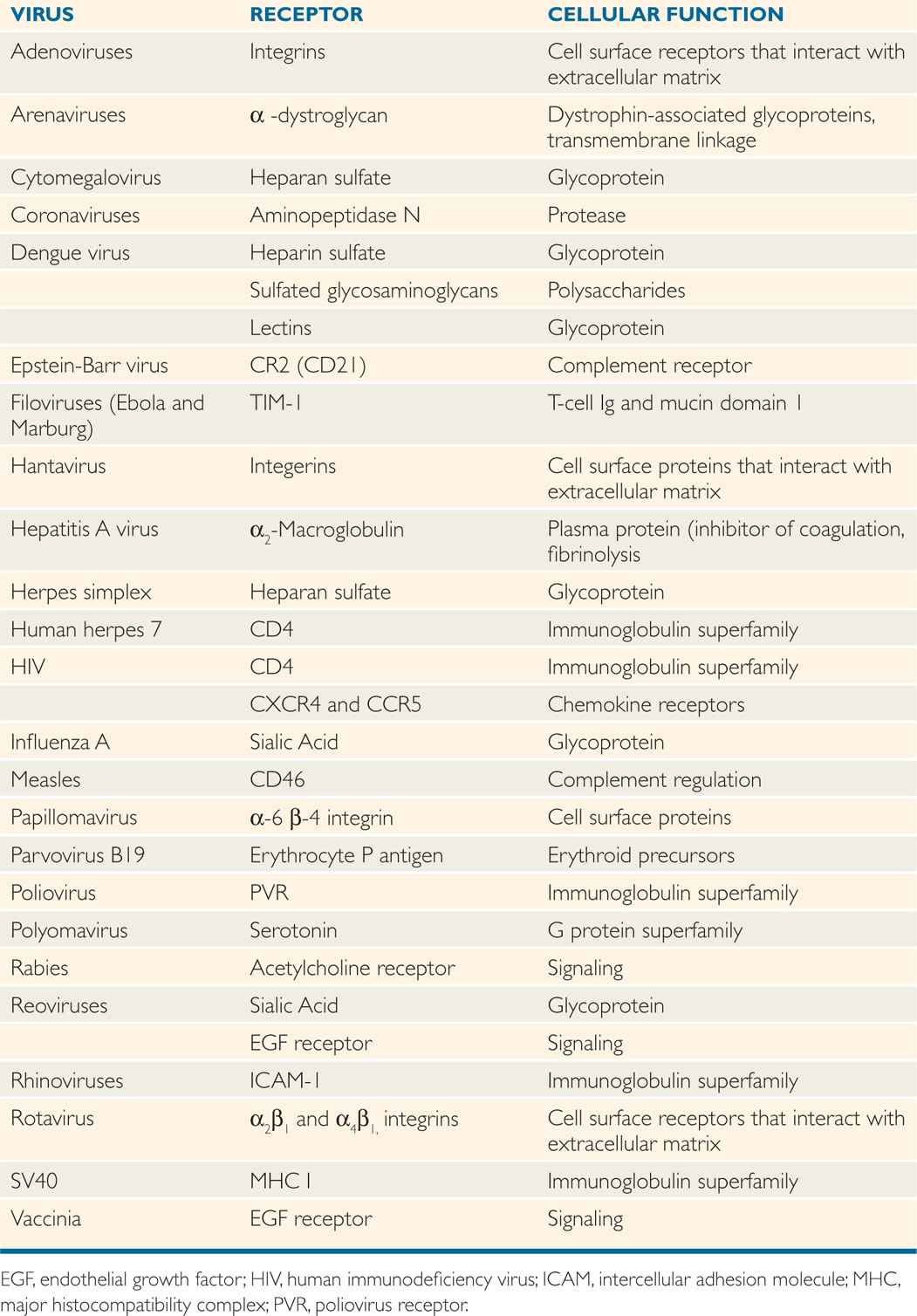
Adsorption involves attachment of viral surface proteins or spikes to the cell surface receptor proteins
For some viruses, two different surface molecules, called coreceptors, are involved in adsorption. Although CD4 was originally thought to be the sole receptor for human immunodeficiency virus type 1 (HIV-1), the discovery of a family of coreceptors that normally function as chemokine receptors (CCR5 and CXCR4) may explain why natural resistance against the virus is found in some individuals with variant forms (Δ32CCR5) of these signaling molecules (discussed in Chapter 18). Although receptors for some human viruses such as influenza viruses are present on lungs cells, these receptors are also found on red blood cells of certain species that are responsible for the phenomena of hemagglutination and hemadsorption discussed later.
Virion attachment proteins are often associated with conspicuous features on the surface of the virion. For example, the virion attachment proteins for the bacteriophages with tails are located at the very end of the tails or the tail fibers (Figure 6–3C). Similarly, the spikes found on adenoviruses (Figure 6–4E) and on virtually all the enveloped human viruses contain the virion attachment proteins.
Viral spikes and phage tails carry attachment proteins
In some cases, a region of the capsid protein serves the function of the attachment protein. For polioviruses, rhinoviruses, and probably other picornaviruses, the region on the capsid that binds to the receptor is found at the bottom of a cleft or trough that is too narrow to allow access to antibodies. This particular arrangement is clearly advantageous to the virus because it precludes the production of antibodies that might directly block receptor recognition.
The repeating subunit structure of capsids and the multiplicity of spikes on enveloped viruses are probably important in determining the strength of the binding of the virus to the cell. The binding between a single virion attachment protein and a single receptor protein is relatively weak, but the combination of many such interactions lead to a strong association between the virion and the cell. The fluid nature of the human cell membrane may facilitate the movement of receptor proteins to allow the clustering that is necessary for these multiple interactions.
Adsorption is enhanced by presence of multiple attachment and receptor proteins
A particular kind of virus is capable of infecting only a limited spectrum of cell types called its host range. Thus, although a few viruses can infect cells from different species, most viruses are limited to a single species. For example, dogs do not contract measles, and humans do not contract distemper. In many cases, human viruses infect only a particular subset of the cells found in their host organism. This kind of tissue tropism is clearly an important determinant of viral pathogenesis. In most cases studied, the specific host range of a virus and its associated tissue tropism are determined at the level of the binding between the cell receptors and virion attachment proteins. Thus, these two protein components must possess complementary surfaces that fit together in much the same way as a substrate fits into the active site of an enzyme. It follows that adsorption occurs only in that percentage of collisions that leads to successful binding between receptors and attachment proteins, and that the inability of a virus to infect a cell type is usually due to the absence of the appropriate receptors on the cell. The exquisite specificity of these interactions is well illustrated by the case of a particular mouse reovirus. It has been found that the tissue tropism—and, therefore, the resultant pathology—are altered by a point mutation that changes a single amino acid in the virion attachment protein. A few cases are known in which the host range of a virus is determined at a step after adsorption and penetration, but these are the exceptions rather than the rule.
Differences in host range and tissue tropism are due to presence or absence of receptors
When a virus particle has penetrated to the inside of a cell, it is essentially hidden from the host immune system. Thus, if protection from a virus infection is to be accomplished at the level of antibody binding to the virions, it must occur before adsorption and prevent the virus from attaching to and penetrating the cell. It is, therefore, not surprising that most neutralizing antibodies—whether elicited as a result of natural infection or vaccination—are specific for virion attachment proteins.
Neutralizing antibodies are often specific for attachment proteins
PENETRATION, ENTRY, AND UNCOATING
The disappearance of infectious virus during the eclipse phase is a direct consequence of the fact that viruses are dismantled before being replicated. As discussed later in the text, the uncoating step may be simultaneous with entry or may occur in a series of steps. Ultimately, the nucleocapsid or core structure must be transported to the site or compartment in the cell where transcription and replication will occur.
Viruses are dismantled before being replicated
 The Bacteriophage Strategy
The Bacteriophage Strategy
The processes of penetration and uncoating are simultaneous for all bacteriophages. Thus, the viral capsids are shed at the surface, and only the nucleic acid genome enters the cell. In some cases, a small number of virion proteins may accompany the genome into the cell, but these are probably tightly associated with the nucleic acid or are essential enzymes needed to initiate the infection.
Bacteriophage capsids are shed, and only the viral genome enters the host cell
Bacteriophages with tails have evolved these special appendages to facilitate the entry of the genome into the cell. The process of penetration and uncoating for bacteriophage T4 is shown schematically in Figure 6–10. The tail fibers extending from the end of the tail are responsible for the attachment of the virion to the cell wall, and, in the next step, the end of the tail itself makes intimate contact with the cell surface. Finally, the DNA of the virus is injected from the head directly into the cell through the hollow tail structure. The process has been likened to the action of a syringe, but the energetics and the nature of the cell surface through which the DNA travels are poorly understood.
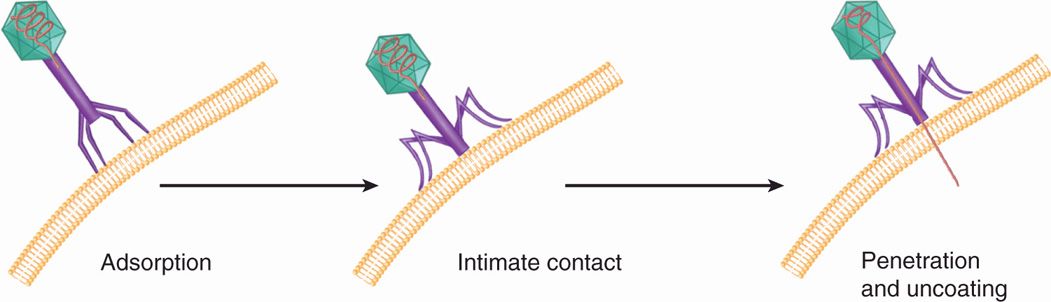
FIGURE 6–10. Bacteriophage entry. Tailed bacteriophages attach to the target cell by their tail fingers followed by injection of genomic material into the cell.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


