FIGURE 4–1. Specimens for the diagnosis of infection. A. Direct specimen. The pathogen is localized in an otherwise sterile site, and a barrier such as the skin must be passed to sample it. This may be done surgically or by needle aspiration as shown. The specimen collected contains only the pathogen. Examples are deep abscess and cerebrospinal fluid. B. Indirect sample. The pathogen is localized as in A, but must pass through a site containing normal flora in order to be collected. The specimen contains the pathogen, but is contaminated with the nonpathogenic flora. The degree of contamination is often related to the skill with which the normal floral site was “bypassed” in specimen collection. Examples are expectorated sputum and voided urine. C. Sample from site with normal flora. The pathogen and nonpathogenic flora are mixed at the site of infection. Both are collected and the nonpathogen is either inhibited by the use of selective culture methods or discounted in interpretation of culture results. Examples are throat and stool.
Quality of the specimen is crucial
 Direct Tissue or Fluid Samples
Direct Tissue or Fluid Samples
Direct specimens (Figure 4–1A) are collected from normally sterile tissues (lung, liver) and body fluids (cerebrospinal fluid, blood). The methods range from needle aspiration of an abscess to surgical biopsy. In general, such collections require the direct involvement of a physician and may carry some risk for the patient. The results are always useful because positive findings are diagnostic and negative findings can exclude infection at the suspected site.
Direct samples give highest quality and risk
 Indirect Samples
Indirect Samples
Indirect samples (Figure 4–1B) are specimens of inflammatory exudates (expectorated sputum, voided urine) that have passed through sites known to be colonized with the resident microbiota. The site of origin is usually sterile in healthy persons; however, some assessment of the probability of contamination with normal flora during collection is necessary in interpretation of the results. This assessment requires knowledge of the potential contaminating flora as well as the probable pathogens to be sought. Indirect samples are usually more convenient for both physician and patient, but carry a higher risk of misinterpretation. For some specimens, such as expectorated sputum, guidelines to assess specimen quality have been developed by correlation of clinical and microbiologic findings.
Bypassing the microbiota requires extra effort
Results require interpretive evaluation of contamination
 Samples from Microbiota Sites
Samples from Microbiota Sites
Frequently, the primary site of infection is in an area known to be colonized with many organisms (pharynx and large intestine) (Figure 4–1C). This is primarily an issue with bacterial diagnosis because they dominate the makeup of the microbiota. In such instances, examinations are selectively made for organisms known to cause infection that are not normally found at the infected site. For example, the enteric pathogens Salmonella, Shigella, and Campylobacter may be selectively sought in a stool specimen or only β-hemolytic streptococci in a throat culture. In these instances, selective media that inhibit growth of the other bacteria are used or, if growing, they are simply ignored. Molecular assays that target the specific pathogens in these specimens are becoming more widely used in place of the selective cultures.
Strict pathogens can be specifically sought
The selection of specimens for viral diagnosis is easier because there is usually little resident viral flora to confuse interpretation. This allows selection guided by knowledge of which sites are most likely to yield the suspected etiologic agent. For example, enteroviruses are the most common viruses involved in acute infection of the central nervous system.
Lack of normal viral flora simplifies interpretation
Specimens that might be expected to yield these agents on culture or in molecular assays include nasopharyngeal or throat swabs, stool, and cerebrospinal fluid.
 Specimen Collection and Transport
Specimen Collection and Transport
The sterile swab is the most convenient and most commonly used tool for specimen collection; however, it provides the poorest conditions for survival, can only absorb a small volume of inflammatory exudate, and is easily contaminated with adjacent microbiota. The worst possible specimen is a dried-out swab; the best is a collection of 5 to 10 mL or more of the infected fluid or tissue. The volume is important because infecting organisms that are present in small numbers may not be detected in a small sample.
Swabs limit volume, survival, and may yield misleading results.
Specimens should be transported to the laboratory as soon after collection as possible because some microorganisms survive only briefly outside the body. For example, unless special transport media are used, isolation rates of the organism that causes gonorrhea (Neisseria gonorrhoeae) are decreased when processing is delayed beyond a few minutes. Likewise, many respiratory viruses survive poorly outside the body. In contrast, some bacteria survive well and may even multiply after the specimen is collected. The growth of enteric Gram-negative rods in specimens awaiting culture may, in fact, compromise specimen interpretation and interfere with the isolation of more fastidious organisms. Significant changes are associated with delays of more than 3 to 4 hours.
Viability may be lost if specimen is delayed
Various transport media have been developed to minimize the effects of the delay between specimen collection and laboratory processing. In general, they are buffered fluid or semisolid media containing minimal nutrients and are designed to prevent drying, maintain a neutral pH, and minimize growth of bacterial contaminants. Other features may be required to meet special requirements, such as an oxygen-free atmosphere for obligate anaerobes or specific (validated) collection-transport systems for molecular assays.
Transport media stabilize conditions and prevent drying
DIRECT EXAMINATION
Of the infectious agents discussed in this book, only some of the parasites are large enough to be seen with the naked eye. Bacteria and fungi can be seen clearly with the light microscope when appropriate methods are used. Individual viruses can be seen only with the electron microscope, although aggregates of viral particles in cells (viral inclusions) may be seen by light microscopy. Various stains are used to visualize and differentiate microorganisms in smears and histologic sections.
All but some parasites require microscopy for visualization
 Light Microscopy
Light Microscopy
Direct examination of stained or unstained preparations by light (bright-field) microscopy (Figure 4–2A) is particularly useful for detection of bacteria, fungi, and parasites. Even the smallest bacteria (1-2 μm wide) can be visualized, although all require staining and some require special lighting techniques. As the resolution limit of the light microscope is near 0.2 μm, the optics must be ideal if small organisms are to be seen clearly by direct microscopy. These conditions may be achieved with a × 100 oil immersion objective, a × 5 to × 10 eyepiece, and optimal lighting.
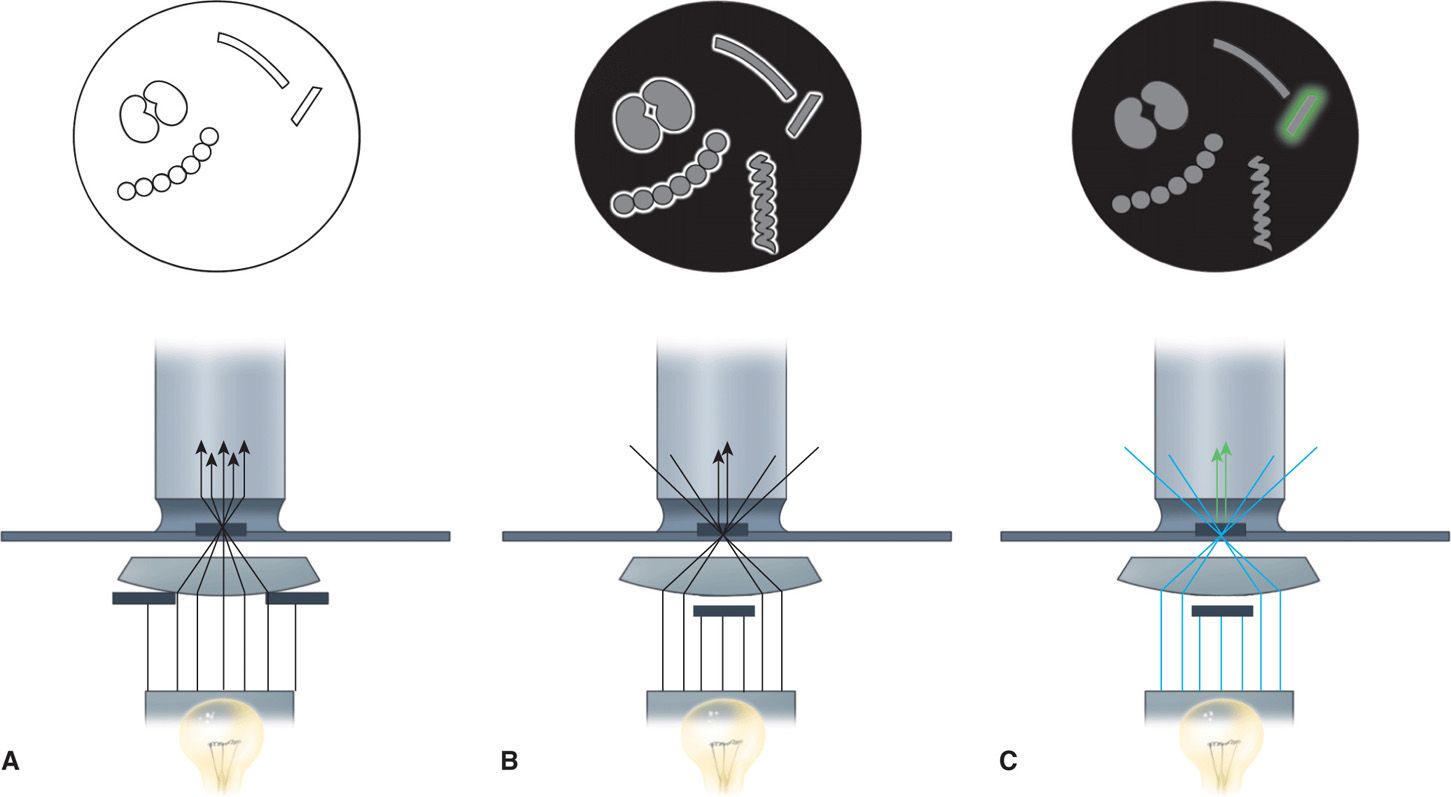
FIGURE 4–2. Bright-field, dark-field, and fluorescence microscopy. A. Bright-field illumination properly aligned. The purpose is to focus light directly on the preparation for optimal visualization against a bright background. B. In dark-field illumination, a black background is created by blocking the central light. Peripheral light is focused so that it will be collected by the objective only when it is reflected from the surfaces of particles (eg, bacteria). The microscopic field shows bright halos around some bacteria and reveals a spirochete too thin to be seen with bright-field illumination. C. Fluorescence microscopy is similar to dark-field microscopy except that the light source is ultraviolet and the organisms are stained with fluorescent compounds. The incident light generates light of a different wavelength, which is seen as a halo (colored in this illustration) around only the organism tagged with fluorescent compounds. For the most common fluorescent compound, the light is green.
Bacteria are visible if optics are maximized
Bacteria may be stained by a variety of dyes, including methylene blue, crystal violet, carbol-fuchsin (red), and safranin (red). The two most important methods, the Gram and acid-fast techniques, use staining, decolorization, and counterstaining in a manner that helps to classify as well as stain the organism.
Bacteria must be stained
The Gram Stain
The differential staining procedure described in 1884 by the Danish physician Hans Christian Gram has proved one of the most useful in microbiology and medicine. The procedure (Figure 4–3A) involves the application of a solution of iodine in potassium iodide to cells previously stained with an acridine dye such as crystal violet. This treatment produces a mordanting action in which purple insoluble complexes are formed with ribonuclear protein in the cell. The difference between Gram-positive and Gram-negative bacteria is in the permeability of the cell wall to these complexes on treatment with mixtures of acetone and alcohol solvents. This extracts the purple iodine-dye complexes from Gram-negative cells, whereas Gram-positive bacteria retain them. An intact cell wall is necessary for a positive reaction, and Gram-positive bacteria may fail to retain the stain if the organisms are old, dead, or damaged by antimicrobial agents. Rarely, a Gram-negative organism (eg, Acinetobacter) will appear Gram positive. The stain is completed by the addition of red counter-stain such as safranin, which is taken up by bacteria that have been decolorized. Thus, cells stained purple are Gram positive, and those stained red are Gram negative. As indicated in Chapter 21, Gram positivity and negativity correspond to major structural differences in the cell wall.
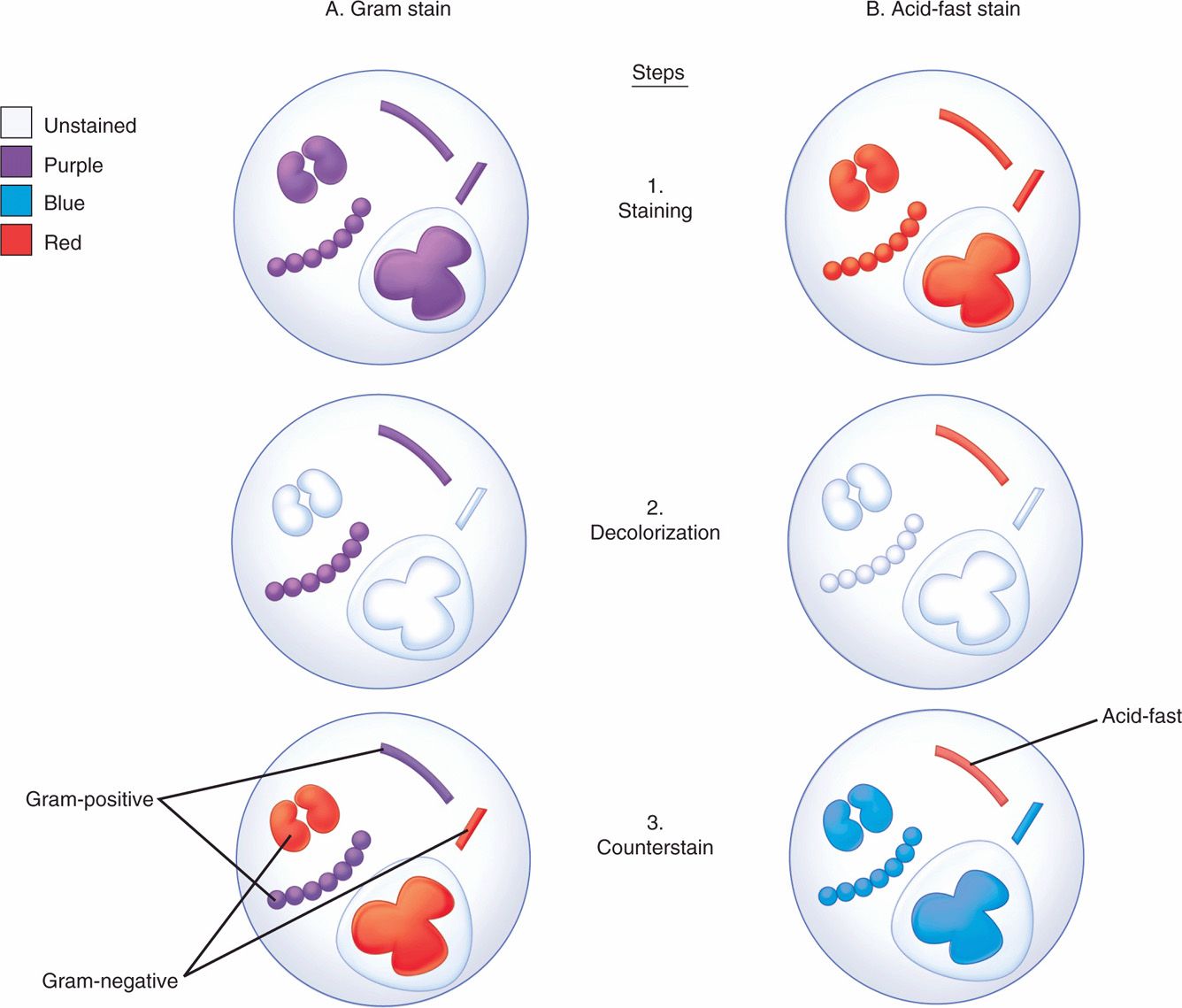
FIGURE 4–3. Gram and acid-fast stains. Four bacteria and a polymorphonuclear neutrophil are shown at each stage. All are initially stained purple by the crystal violet and iodine of the Gram stain (Al) and red by the carbol fuchsin of the acid-fast stain (Bl). After decolorization, Gram-positive and acid-fast organisms retain their original stain. Others are unstained (A2, B2). The safranin of the Gram counterstain stains the Gram-negative bacteria and makes the background red (A3), and the methylene blue leaves a blue background for the contrasting red acid-fast bacillus (B3).
Gram-positive bacteria retain purple iodine-dye complexes
Gram-negative bacteria do not retain complexes when decolorized
In many bacterial infections, the etiologic agents are readily seen on stained Gram smears of pus or fluids. The purple or red bacteria are seen against a Gram-negative (red) background of leukocytes, exudate, and debris (Figures 4–3A and 4–4C). This information, combined with the clinical findings, may guide the management of infection before culture results are available. Interpretation requires considerable experience and knowledge of probable causes, of their morphology and Gram reaction, and of any organisms normally present in health at the infected site.
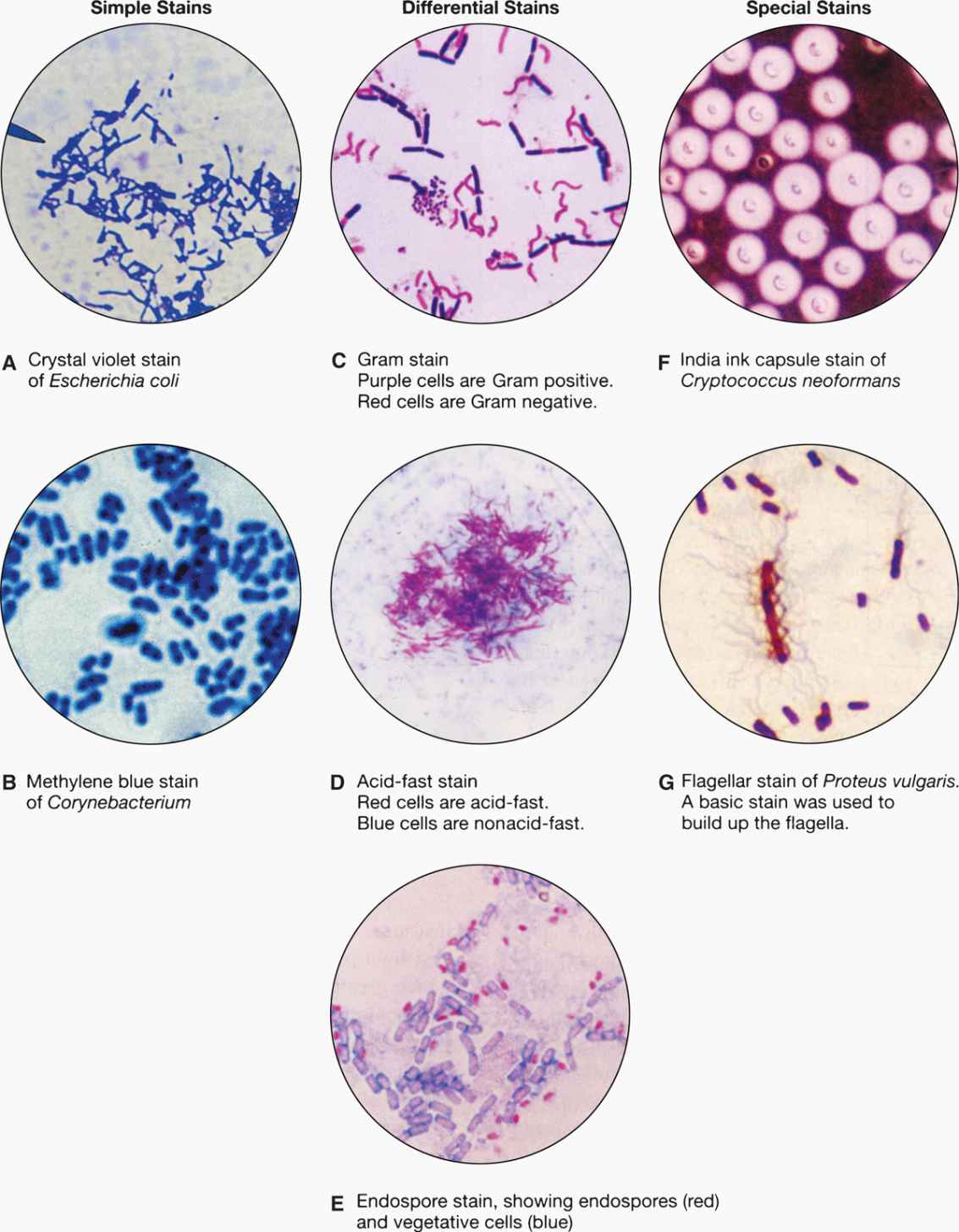
FIGURE 4–4. Types of microbiologic stains. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
Properly decolorized background should be red
Gram reaction plus morphology guide clinical decisions
The Acid-Fast Stain
Acid fastness is a property of the mycobacteria (eg, Mycobacterium tuberculosis) and related organisms. Acid-fast organisms generally stain very poorly with dyes, including those used in the Gram stain. However, they can be stained by prolonged application of more concentrated dyes, by penetrating agents, or by heat treatment. Their unique feature is that when stained, acid-fast bacteria resist decolorization by concentrations of mineral acids and ethanol that remove the same dyes from other bacteria. This combination of weak initial staining and strong retention once stained is related to the high lipid content of the mycobacterial cell wall. Acid-fast stains are completed with a counterstain to provide a contrasting background for viewing the stained bacteria (Figure 4–3B).
Acid-fast bacteria take stains poorly
Once stained, they retain it strongly
In the acid-fast procedure, the slide is flooded with carbol-fuchsin (red) and decolorized with hydrochloric acid in alcohol. When counterstained with methylene blue, acid-fast organisms appear red against a blue background (Figure 4–4B). A variant is the fluorochrome stain, which uses a fluorescent dye (auramine, or an auramine–rhodamine mixture), followed by decolorization with acid–alcohol. Acid-fast organisms retain the fluorescent stain, which allows their visualization by fluorescence microscopy. The fluorochrome stain is more sensitive and allows rapid screening and, therefore, has become the method of choice in most laboratories performing testing for acid-fast organisms.
There are multiple variants of the acid-fast stain
Fungal and Parasitic Stains
The smallest fungi are the size of large bacteria, and all parasitic forms are larger. This allows detection in simple wet mount preparations, often without staining. Fungi in sputum or body fluids can be seen by mixing the specimen with a potassium hydroxide solution (to dissolve debris) and viewing with a medium power lens. The use of simple stains or the fluorescent calcofluor white improves the sensitivity of detection. Another technique is to mix the specimen with India ink, which outlines the fungal cells (Figure 4–4F). Detection of the cysts and eggs of parasites requires a concentration procedure if the specimen is stool, but once done they can be visualized with a simple iodine stain (Figure 4–5).
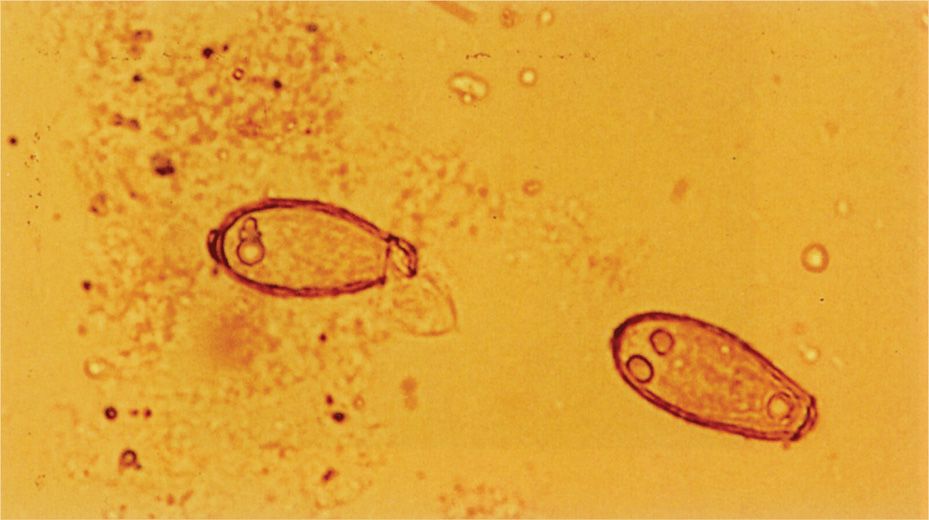
FIGURE 4–5. Iodine-stained parasite eggs. Two eggs of the intestinal fluke Clonorchis sinensis are present in this stool specimen. (Reproduced with permission from Connor DH, Chandler FW, Schwartz DQ, et al: Pathology of Infectious Diseases. Stamford CT: Appleton & Lange, 1997.)
Fungi and parasites visible with simple stains
Dark-field and Fluorescence Microscopy
Some bacteria, such as Treponema pallidum, the cause of syphilis, are too thin to be visualized with the usual bright-field illumination. They can be seen by use of the dark-field technique. With this method, a condenser focuses light diagonally on the specimen in such a way that only light reflected from particulate matter, such as bacteria, reaches the eyepiece (Figure 4–2 B). The angles of incident and reflected light are such that the organisms are surrounded by a bright halo against a black background. This type of illumination is also used in other microscopic techniques, in which a high light contrast is desired, and for observation of fluorescence. Fluorescent compounds, when excited by incident light of one wavelength, emit light of a longer wavelength and thus a different color. When the fluorescent compound is conjugated with an antibody as a probe for detection of a specific antigen, the technique is called immunofluorescence, or fluorescent antibody microscopy (Figure 4–6). The appearance is the same as in dark-field microscopy except that the halo is the emitted color of the fluorescent compound (Figures 4–2C and 4–6 C and D). Immunofluorescence stains are the most commonly used stains for detection of viruses though these are being replaced by the more sensitive molecular assays. For improved safety, most modern fluorescence microscopy systems direct the incident light through the objective from above (epifluorescence).
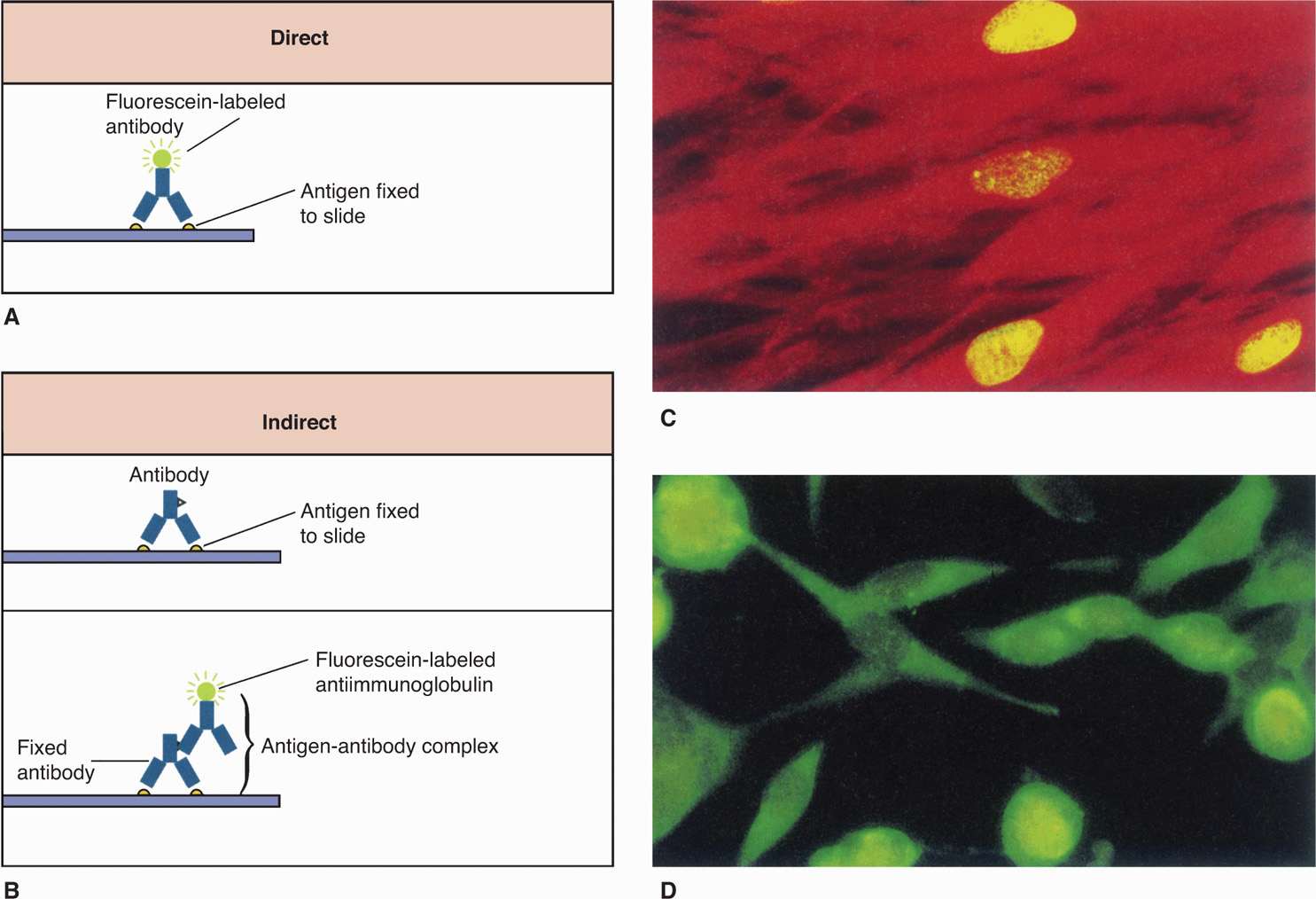
FIGURE 4–6. Direct and indirect immunofluorescence. A. In the direct fluorescent antibody (DFA), the specimen containing antigen is fixed to a slide. Fluorescently labeled antibodies that recognize the antigen are then added, and the specimen is examined with a fluorescence microscope for yellow-green fluorescence. B. The indirect fluorescent antibody technique (IFA) detects antigen on a slide as it reacts with an unlabeled antibody directed against it. The original antigen–antibody complex is detected with a second labeled antibody that recognizes any antibody. C. Three infected nuclei in a cytomegalovirus-positive tissue culture. D. Several infected cells in a herpes simplex virus–positive tissue culture. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
Dark-field creates a halo around organisms too thin to see by bright-field
Fluorescent stains convert dark-field to fluorescence microscopy
 Electron Microscopy
Electron Microscopy
Electron microscopy shows structures by transmission of an electron beam and has 10 to 1000 times the resolving power of light microscopic methods. For practical reasons, its diagnostic application is limited to virology, where, because of the resolution possible at high magnification, it offers results not possible by any other method. Using negative staining techniques for direct examination of fluids and tissues from affected body sites enables visualization of viral particles. In some instances, electron microscopy has been the primary means of discovery of viruses that do not grow in the usual cell culture systems. Molecular assays are replacing electron microscopy for detection of many viruses.
Viruses are visible only by electron microscopy
CULTURE
Growth and identification of the infecting agent in vitro is usually the most sensitive and specific means of diagnosis and is, thus, the method most commonly used. Theoretically, the presence of a single live organism in the specimen can yield a positive result. Most bacteria and fungi can be grown in a variety of artificial media, but strictly intracellular microorganisms (eg, Chlamydia, Rickettsia, and viruses) can be isolated only in cultures of living eukaryotic cells. The culture of some parasites is possible, but used only in highly specialized laboratories.
 Isolation and Identification of Bacteria and Fungi
Isolation and Identification of Bacteria and Fungi
Almost all medically important bacteria can be cultivated outside the host in artificial culture media. A single bacterium placed in the proper culture conditions multiplies to quantities sufficient to be seen by the naked eye. Bacteriologic media are soup-like recipes prepared from digests of animal or vegetable protein supplemented with nutrients such as glucose, yeast extract, serum, or blood to meet the metabolic requirements of the organism. Their chemical composition is complex, and their success depends on matching the nutritional requirements of most heterotrophic living things. The same approaches as well as some of the same culture media used for bacteria are also used for fungi.
Bacteria grow in soup-like media
Growth in media prepared in the fluid state (broths) is apparent when bacterial numbers are sufficient to produce turbidity or macroscopic clumps. Turbidity results from reflection of transmitted light by the bacteria; depending on the size of the organism, more than 106 bacteria per milliliter of broth are required. The addition of a gelling agent to a broth medium allows its preparation in solid form as plates in Petri dishes. The universal gelling agent for diagnostic bacteriology is agar—a polysaccharide extracted from seaweed. Agar has the convenient property of becoming liquid at approximately 95°C but not returning to the solid gel state until cooled to less than 50°C. This allows the addition of a heat-labile substance such as blood to the medium before it sets. At temperatures used in the diagnostic laboratory (37°C or lower), broth–agar exists as a smooth, solid, nutrient gel. This medium, usually termed “agar,” may be qualified with a description of any supplement (eg, blood agar).
Large numbers of bacteria in broth produce turbidity
Agar is a convenient gelling agent for solid media
A useful feature of agar plates is that the bacteria can be separated by spreading a small sample of the specimen over the surface. Bacterial cells that are well separated from others grow as isolated colonies, often reaching 2 to 3 mm in diameter after overnight incubation. This allows isolation of bacteria in pure culture because the colony is assumed to arise from a single organism (Figure 4–7). Colonies vary greatly in size, shape, texture, color, and other features called colonial morphology. Colonies from different species or genera often differ substantially, whereas those derived from the same strain are usually consistent. Differences in colonial morphology are very useful for separating bacteria in mixtures and as clues to their identity. Some examples of colonial morphology are shown in Figure 4–8.
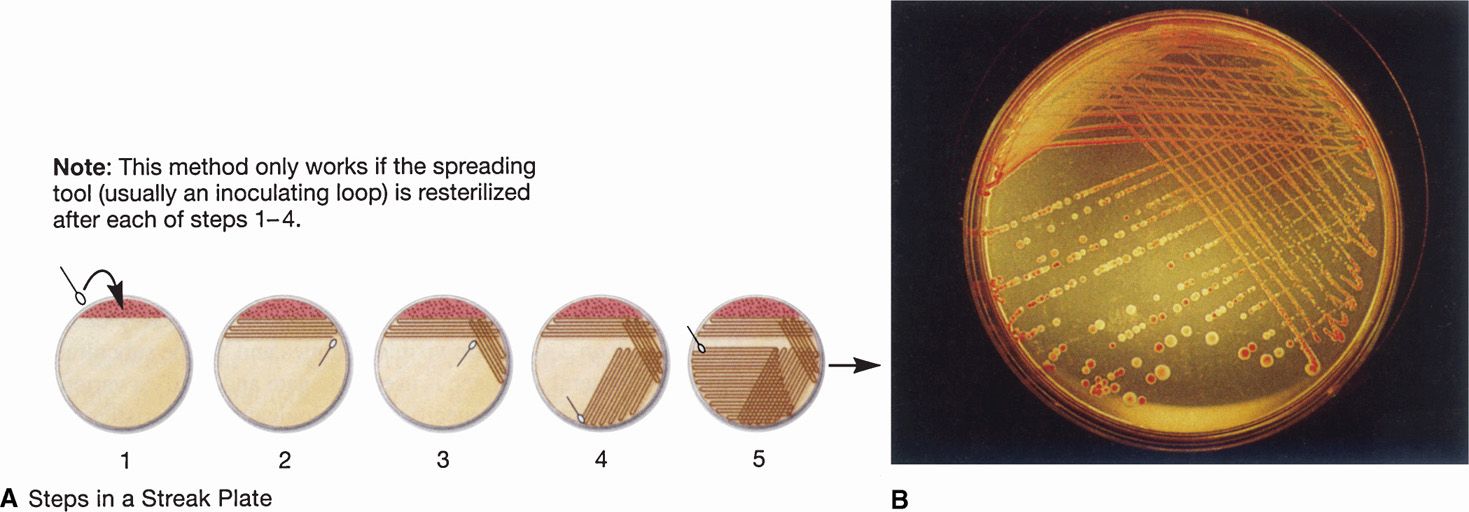
FIGURE 4–7. Bacteriologic plate streaking. Plate streaking is essentially a dilution procedure. A. (1) The specimen is placed on the plate with a swab, loop, or pipette and evenly spread over approximately part of plate surface with a sterilized bacteriologic loop (2-5). The loop is flamed to remove residual bacteria, and a series of overlapping streaks are made flaming the loop between each one. B. After overnight incubation, heavy growth is seen in the primary areas followed by isolated colonies. More than one organism is present because both a red and a clear colony are seen. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
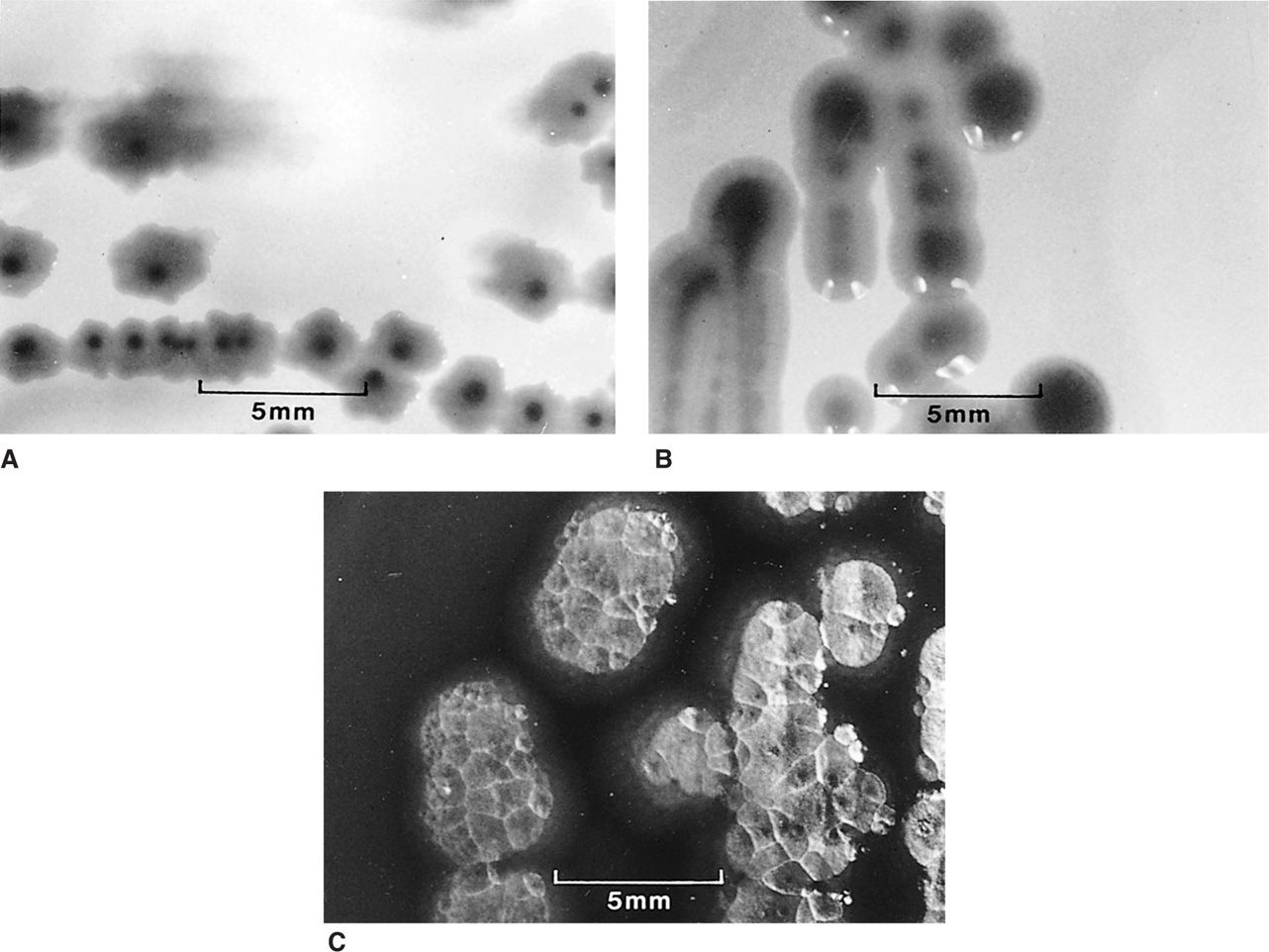
FIGURE 4–8. Bacterial colonial morphology. The colonies formed on agar plates by three different Gram-negative bacilli are shown at the same magnification. Each is typical for its species, but variations are common. A. Escherichia coli colonies are flat with an irregular scalloped edge. B. Klebsiella pneumoniae colonies with a smooth entire edge and a raised glistening surface. C. Pseudomonas aeruginosa colonies with an irregular reflective surface, suggesting hammered metal.
Bacteria may be separated in isolated colonies on agar plates
Colonies may have consistent and characteristic features
New methods that do not depend on visual changes in the growth medium or colony formation are also used to detect bacterial growth in culture. These techniques include optical, chemical, and electrical changes in the medium, produced by the growing numbers of bacterial cells or their metabolic products. Many of these methods are more sensitive than classic techniques and, thus, can detect growth hours, or even days, earlier than traditional methods. Some have also been engineered for instrumentation and automation. For example, one fully automated system that detects bacterial metabolism fluorometrically can complete a bacterial identification and antimicrobial susceptibility test in 2 to 4 hours.
Optical, chemical, and electrical methods can detect growth
Culture Media
Over the last 100 years, countless media have been developed by microbiologists to aid in the isolation and identification of medically important bacteria and fungi. Only a few have found their way into routine use in clinical laboratories. These may be classified as nutrient, selective, or indicator media.
Nutrient Media The nutrient component of a medium is designed to satisfy the growth requirements of the organism to permit isolation and propagation. For medical purposes, the ideal medium would allow rapid growth of all agents. No such medium exists; however, several suffice for good growth of most medically important bacteria and fungi. These media are prepared with enzymatic or acid digests of animal or plant products such as muscle, milk, or soybeans. The digest reduces the native protein to a mixture of polypeptides and amino acids that also includes trace metals, coenzymes, and various undefined growth factors. For example, one common broth contains a pancreatic digest of casein (milk curd) and a papaic digest of soybean meal. To this nutrient base, salts, vitamins, or body fluids such as serum may be added to provide pathogens with the conditions needed for optimum growth.
Media are prepared from animal or plant products
Selective Media Selective media are used when specific pathogenic organisms are sought in sites with an extensive microbiota (eg, Campylobacter species in fecal specimens). In these cases, other bacteria may overgrow the suspected etiologic species in simple nutrient media, either because the pathogen grows more slowly or because it is present in much smaller numbers. Selective media usually contain dyes, other chemical additives, or antimicrobial agents at concentrations designed to inhibit contaminating flora but not the suspected pathogen.
Unwanted organisms are inhibited with chemicals or antimicrobials
Indicator Media Indicator media contain substances designed to demonstrate biochemical or other features characteristic of specific pathogens or organism groups. The addition to the medium of one or more carbohydrates and a pH indicator is frequently used. A color change in a colony indicates the presence of acid products and thus of fermentation or oxidation of the carbohydrate by the organism. The addition of red blood cells (RBCs) to plates allows the hemolysis produced by some organisms to be used as a differential feature. In practice, nutrient, selective, and indicator properties are often combined to various degrees in the same medium. It is possible to include an indicator system in a highly nutrient medium and also make it selective by adding appropriate antimicrobials. Some examples of culture media commonly used in diagnostic microbiology are listed in Appendix 4–1, and more details of their constitution and application are provided in Appendix 4–2.
Metabolic properties of bacteria are demonstrated by substrate and indicator systems
Atmospheric Conditions
Aerobic After inoculation, cultures of most aerobic bacteria are placed in an incubator with temperature maintained at 35°C to 37°C. Slightly higher or lower temperatures are used occasionally to selectively favor a certain organism or organism group. Most bacteria that are not obligate anaerobes grow in air; however, CO2 is required by some and enhances the growth of others. Incubators that maintain a 2% to 5% concentration of CO2 in air are frequently used for primary isolation, because this level is not harmful to any bacteria and improves isolation of some. A simpler method is the candle jar, in which a lighted candle is allowed to burn to extinction in a sealed jar containing plates. This method adds 1% to 2% CO2 to the atmosphere. Some bacteria (eg, Campylobacter) require a microaerophilic atmosphere with reduced oxygen (5%) and increased CO2 (10%) levels to grow. This can be achieved by using a commercially available packet that is placed in a jar which is then sealed similar to the anaerobic system described further.
Incubation temperature and atmosphere vary with organism
Anaerobic Strictly anaerobic bacteria do not grow under the conditions just described, and many die when exposed to atmospheric oxygen or high oxidation–reduction potentials. Most medically important anaerobes grow in the depths of liquid or semisolid media containing any of a variety of reducing agents, such as cysteine, thioglycollate, ascorbic acid, or even iron filings. An anaerobic environment for incubation of plates can be achieved by replacing air with a gas mixture containing hydrogen, CO2, and nitrogen and allowing the hydrogen to react with residual oxygen on a catalyst to form water. A convenient commercial system accomplishes this chemically in a packet that is added before the jar is sealed. Specimens suspected to contain significant anaerobes should be processed under conditions designed to minimize exposure to atmospheric oxygen at all stages.
Anaerobes require reducing conditions and protection from oxygen
Clinical Microbiology Systems
Routine laboratory systems for processing specimens from various sites are needed because no single medium or atmosphere is ideal for all bacteria. Combinations of broth and solid-plated media and aerobic, CO2, and anaerobic incubation must be matched to the organisms expected at any particular site or clinical circumstance. Examples of such routines are shown in Table 4–1. In general, it is not practical to routinely include specialized media for isolation of rare organisms such as Corynebacterium diphtheriae or Legionella pneumophila. For detection of these and other uncommon organisms, the laboratory must be specifically informed of their possible presence by the physician. Appropriate media and special procedures can then be included.
TABLE 4–1 Routine Use of Gram Smear and Isolation Systems for Selected Clinical Specimensa
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


