FIGURE 15–1. Viruses of diarrhea. All are photographed at the same magnification to illustrate the size and morphologic differences. A. Rotavirus, B. Calicivirus. C. Astrovirus. (Courtesy of Claire M. Payne.)
Viral diarrhea was a diagnosis of exclusion
Many viral particles seen in stool by electron microscopy
Confirmation by EIA or PCR is now available
Detection of a specific virus in the stools of symptomatic patients is not sufficient to establish the role of the virus in causing disease. Other criteria to be fulfilled include the following:
1. Establish that the virus is detected in patients who are ill significantly more frequently than in asymptomatic, appropriately matched controls, and that virus shedding temporally correlates with symptoms.
2. Demonstrate significant humoral or secretory antibody responses, or both, in patients shedding the virus.
3. Reproduce the disease by experimental inoculation of nonimmune human or animal hosts (usually the most difficult criterion to fulfill).
4. Exclude other known causes of diarrhea, such as bacteria, bacterial toxins, and protozoa.
Multiple criteria used for establishing etiologic relationship
Using these criteria, four groups of viruses have been clearly established as important causes of gastrointestinal disease: rotaviruses, caliciviruses, astroviruses, and some adenovirus serotypes (“enteric” adenoviruses). Other viruses have also been implicated, but many of the preceding criteria have not been fulfilled; therefore, they are currently regarded as “candidate” causes of gastrointestinal disease.
Rotaviruses, caliciviruses, astroviruses, and adenoviruses are established causes
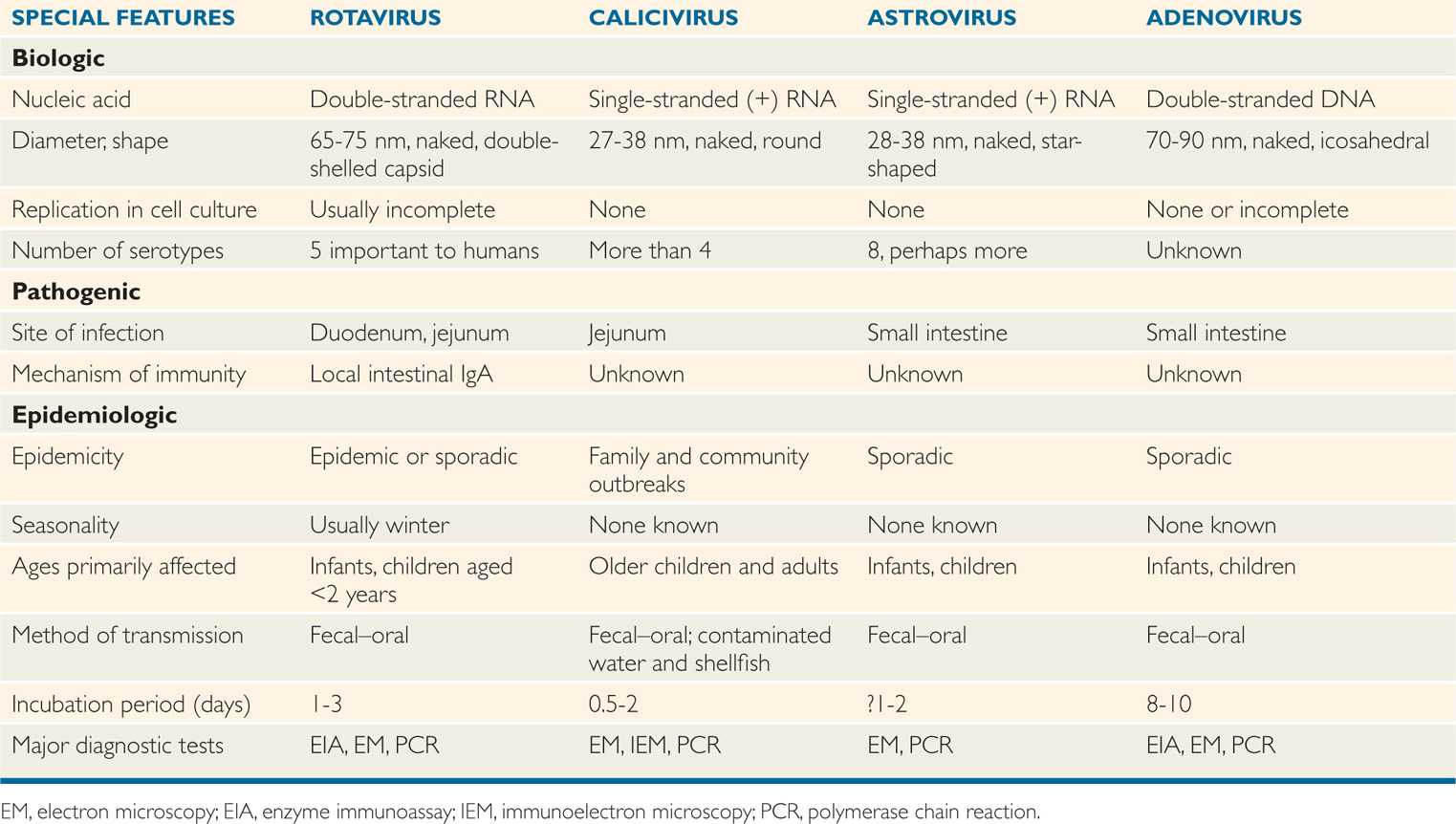
The currently established viruses are listed in Table 15–1, and all have several features in common, including a tendency toward brief incubation periods; fecal–oral spread by direct or indirect routes; and production of vomiting, which generally precedes or accompanies the diarrhea. The last feature has influenced physicians to use the term acute viral gastroenteritis to describe the syndrome associated with these agents.
TABLE 15–1 Biologic and Epidemiologic Characteristics of Viruses That Cause Diarrhea
“Candidate” viruses meet some criteria
Vomiting commonly follows short incubation period
ROTAVIRUSES
The human intestinal rotaviruses were first found in 1973 by electron microscopic examination of duodenal biopsy specimens from infants with diarrhea (Figure 15–2). Since then, they have been found worldwide and are believed to account for 40% to 60% of cases of acute gastroenteritis occurring during the cooler months in infants and in children less than 2 years of age. Worldwide, more than 500 000 deaths in children younger than 5 years of age annually are attributed to rotavirus infections; whereas such deaths in the United States are rather infrequent, the annual morbidity rate has been nonetheless considerable in recent years (Figure 15–3). Before the introduction of rotavirus vaccines in 2006, almost all children were infected in the United States before their fifth birthday. The routine use of rotavirus vaccine in infants has significantly reduced rotavirus infection in the United States. These viruses have been detected in intestinal contents and in tissues from the upper gastrointestinal tract.
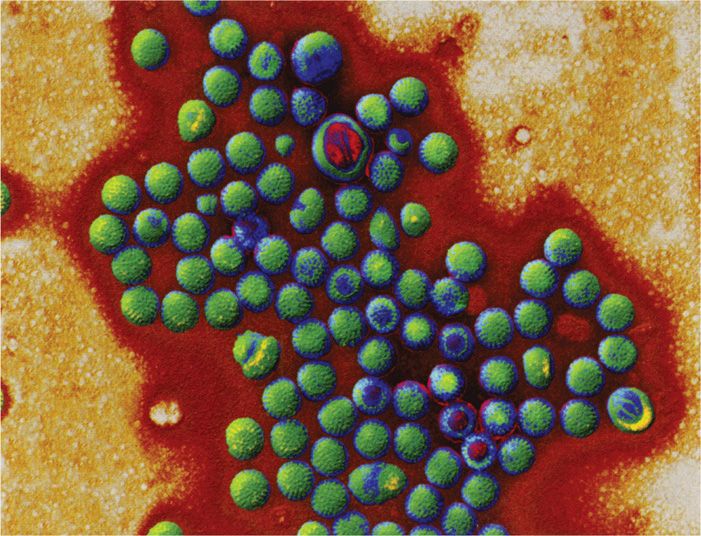
FIGURE 15–2. Rotavirus structure. (Reproduced with permission from Willey JM: Prescott, Harley, & Klein’s Microbiology, 7th edition. McGraw-Hill, 2008.)
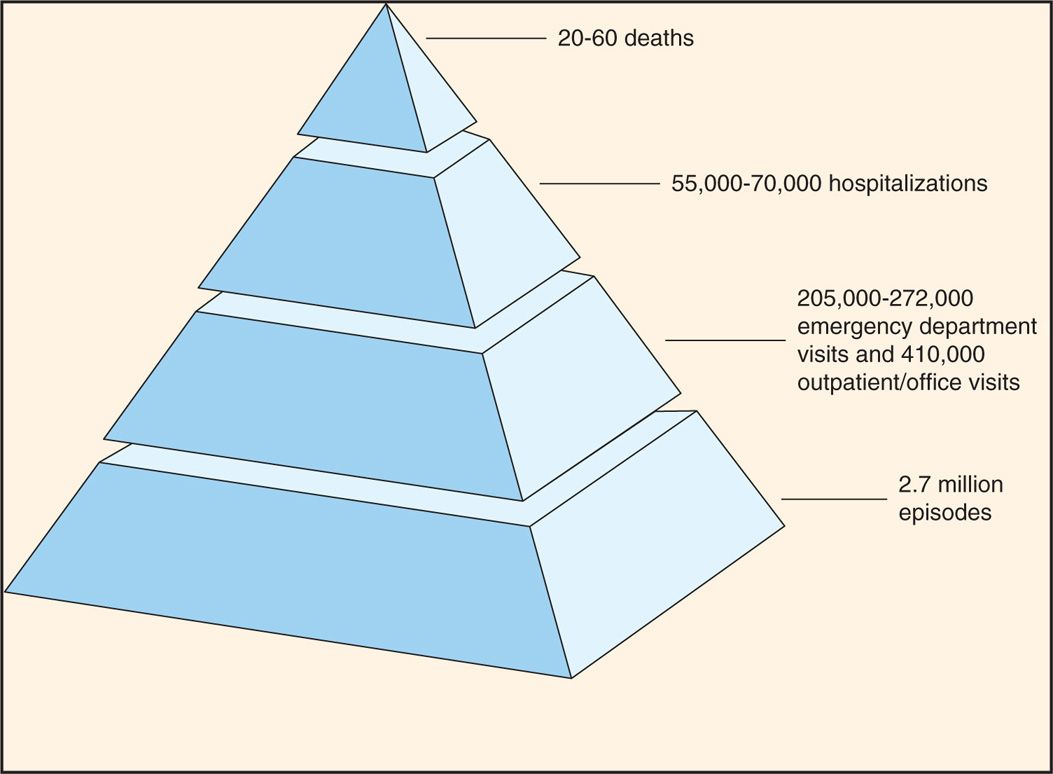
FIGURE 15–3. Estimated annual morbidity due to rotavirus infections in the United States. (Centers for Disease Control and Prevention.)
Most common cause of winter gastroenteritis in children less than 2 years of age
 VIROLOGY
VIROLOGY
The rotaviruses belong to the family Reoviridae. The genome of rotaviruses is unique in the sense that they have 11 segments of double-stranded RNA. The 11 segments of the genome encode six structural (VP1-VP4 and VP6-VP7) and six nonstructural (NSP1-NSP6) proteins (Figure 15–4A). There are three types of virus particles, including triple layered (previously called double shelled), double layered (previously called single shelled) and single layered (empty capsids, usually lacking genomes) (Figure 15–1A,15-2). The complete virus particle of rotavirus is a wheel-shaped virus and the name is derived from the Latin rota (“wheel”) because of the outer capsid, which resembles a wheel attached by short spokes to the inner capsid and core (Figures 15-1, 15-2, 15-4 A). Eleven segments of double-stranded RNA genome are packaged into an icosahedral capsid making the spherical particles of 65 to 75 nm in diameter in size (smaller forms have also been described) (Figure 15–4B–D). The virus particle has a virion-associated RNA-dependent RNA polymerase, and a double-shelled outer capsid; two segments encode proteins of the outer capsid (VP4 and VP7), which are targets for neutralizing antibodies. The major outer capsid proteins are VP4 and VP7. VP4 performs several functions, including viral attachment protein, whereas VP7 is a type-specific antigen and facilitates viral attachment and entry.
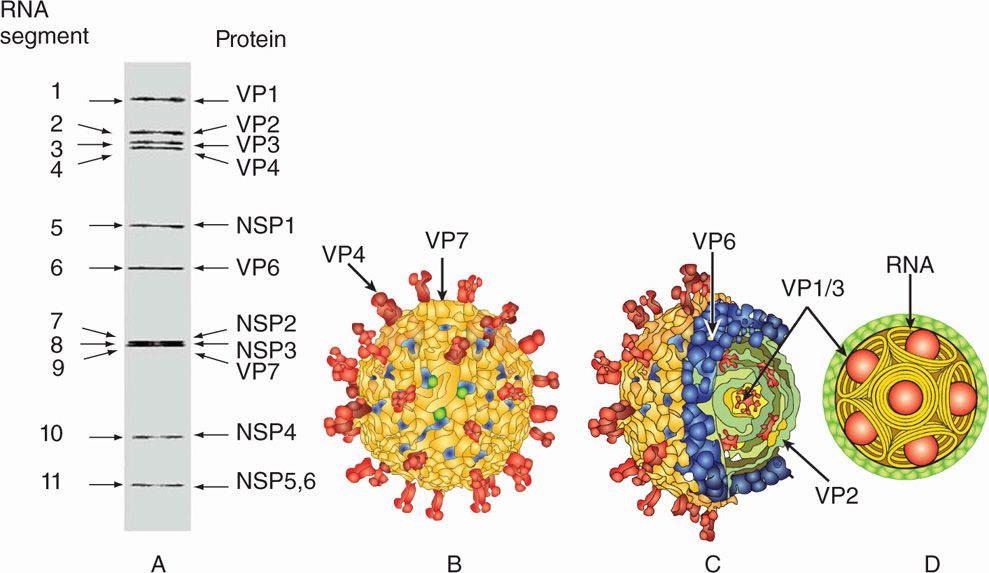
FIGURE 15–4A–D. Structure of Rotavirus (A). Eleven segments of rotavirus are shown on a gel, each segment encoding corresponding structural (VPI-VP7) or nonstructural (NSPI-NSP6) proteins are shown, (B) structure of rotavirus showing outer layer capsid proteins, including VP4 (spikes) and VP7 (outer capsid layer) (C). (Courtesy of BVV Prasad)
Wheel-shaped naked capsid spherical viruses
Double-stranded RNA genome replicates in the cytoplasm
Rotaviruses are classified into seven groups, A to G, based on the internal capsid protein, VP6. Human infections are predominantly caused by group A and less commonly by group B or C. Based on VP4 and VP7 type-specific antigens on the outer capsid, G (VP7 is a glycoprotein) and P (VP4 is protease-sensitive) serotypes have been designated. Five serotypes (G1, G2, G3, G4, and G9), are of major epidemiologic importance because they represent more than 90% of all serotypes detected worldwide. The outer capsid is proteolytically cleaved in the gastrointestinal tract to generate intermediate infectious subviral particle (ISVP), which activates the virus for infection. Rotaviruses can replicate in the cytoplasm of infected cell cultures in the laboratory but are difficult to propagate because the replicative cycle is usually incomplete, and mature, infectious virions are often not produced. However, successful propagation of human strains in vitro has been achieved in some instances.
Double-shelled (triple-layered) outer capsid
Group A rotaviruses predominantly infect humans
Five antigenic types (serotypes) based on capsid proteins VP4 and VP7 detected worldwide
Rotavirus replication is depicted in Figure 15–5. Rotavirus is transmitted by fecal–oral route, and the virus particle is partially digested in the gastrointestinal tract and activated by protease cleavage resulting in the loss of VP7 and cleavage of VP4 to generate ISVP. The VP4 binds to sialic acid containing glycoproteins on epithelial cells, and the ISVP penetrates the target cells. The generation of ISVP is necessary for rotavirus infection because the double-shelled virus particle, after entering the cells via receptor-mediated endocytosis, is unable to establish infection owing to a dead-end pathway. After entry of the ISVP, the core containing double-stranded RNA genomes and the RNA-dependent RNA polymerase is partially released into the cytoplasm. Rotaviruses use negative-sense RNA strategy for transcription and replication. RNA-dependent RNA polymerase directs the synthesis of early and late mRNAs followed by genome replication by using the negative-strand RNA of the double-stranded RNA genome. Early proteins are produced that are required for virus replication, whereas late proteins are mainly the structural proteins. Rotavirus assembles by associating its core with a nonstructural protein (NS28, a product of NSP4) and by acquiring VP7 and a membrane budding into the endoplasmic reticulum (ER). The virus eventually loses the membrane in the ER and is released upon cell lysis.
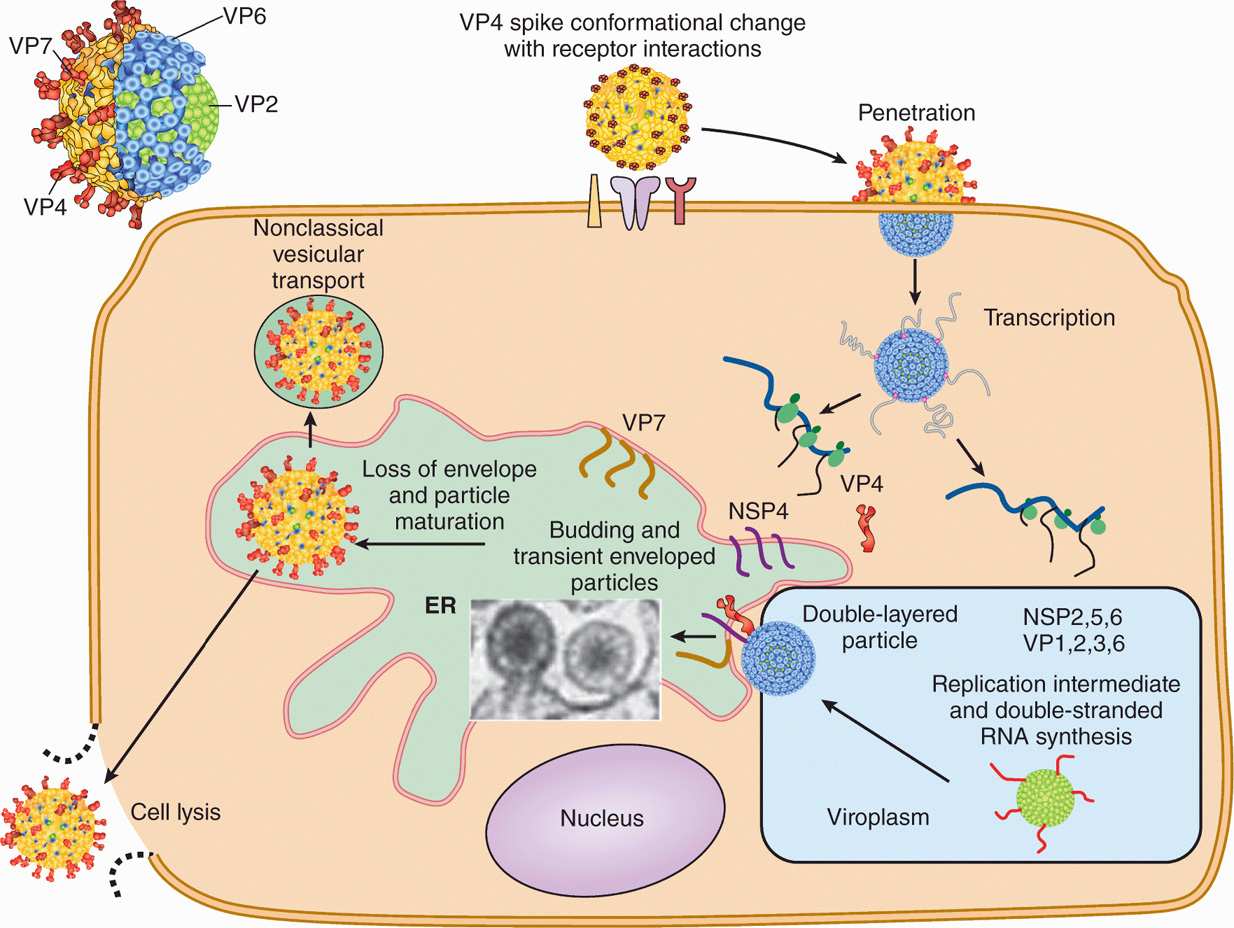
FIGURE 15–5. Schematic diagram of Rotavirus replication. Rotavirus outer capsid spike (VP4) binds to the receptor (sialic acid-containing glycoprotein) followed by a conformational change, removal of outer layer; and penetration of the virus in the target cells. Following partial uncoating, viral RNA-dependent RNA polymerase directs the transcription of viral mRNAs followed synthesis of viral proteins, by genome replication by using the negative-strand RNA of the double-stranded RNA genome. Rotavirus assembles by associating its core with a nonstructural protein (NSP4) and acquiring VP7 and a membrane budding from the ER. The virus eventually loses the membrane in the ER and is released upon cell lysis. (Courtesy of MK Estes)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


