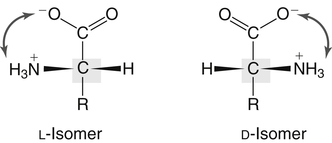
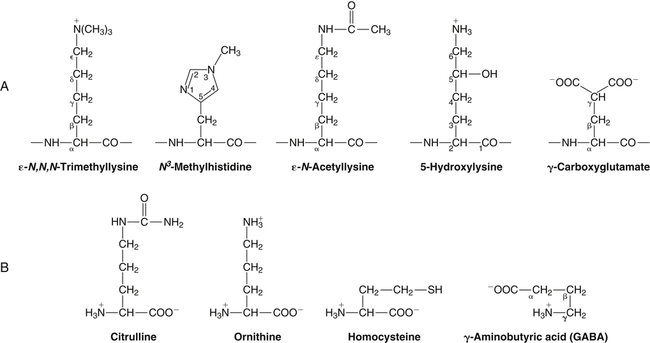
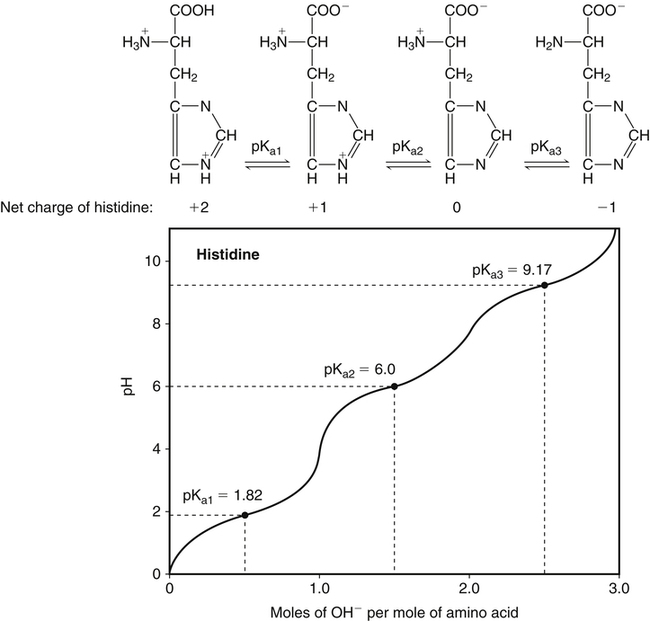
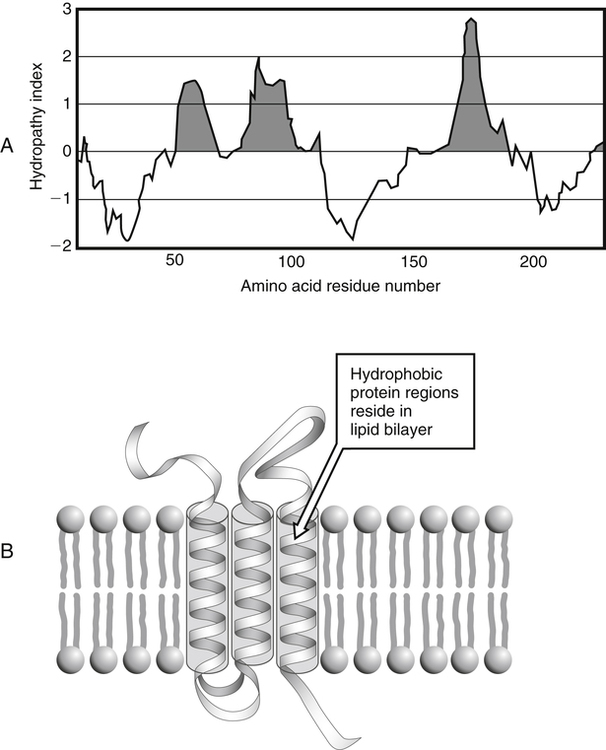
All peptides and proteins, regardless of their origin, are constructed from amino acids that are covalently linked together, usually in a linear sequence. Twenty-one amino acids are naturally incorporated into polypeptides in mammals. Twenty of these are directly encoded by the universal genetic code. The twenty-first of the amino acid precursors for protein synthesis, selenocysteine, is incorporated into a small number of proteins by a unique cotranslational mechanism requiring special secondary structure in the messenger RNA (mRNA) (i.e., a selenocysteine insertion sequence, SECIS) that causes the UGA stop codon to encode selenocysteine (see Chapter 39). Another unusual amino acid called pyrrolysine is considered the twenty-second proteinogenic amino acid, but it is found only in some methane-producing enzymes in methanogenic archaea. The structures of the 20 common amino acids and selenocysteine are shown in Figure 5-1. As shown in Figure 5-2, each amino acid contains an amino group and a carboxylic acid group. Both of these functional moieties are bonded directly to a central carbon atom designated as the α-carbon. Except for glycine, the α-carbon for each of the amino acids has four different functional groups bonded to it: an amino group, a carboxylic acid group, hydrogen, and its R group. The α-carbon of glycine does not have an R group or side chain, so two hydrogen atoms are attached to the α-carbon. In proteins and peptides, amino acids are found almost exclusively in the L form, although D-amino acids are found in some bacterial proteins and peptides (Petsko and Ringe, 2004). The almost exclusive presence of L-amino acids in proteins indicates that reactions that involve amino acid and protein synthesis must be highly stereospecific. The metabolic pathways for amino acid synthesis create predominantly amino acids in their L forms. Moreover, the biological machinery required for protein assembly recognizes L-amino acids almost exclusively. It should be noted that D-aspartate and D-serine are produced by the mammalian brain by enzymes that catalyze the racemization of L-aspartate and L-serine, respectively, and these D-amino acids are involved in activation of the N-methyl-D-aspartate type of excitatory amino acid receptors (Wolosker et al., 2008). In aqueous solutions, amino acids are easily ionized. The most abundant ionic species present when amino acids are dissolved in an aqueous medium at neutral pH are shown in Figure 5-1, and the pKas for all dissociable groups are shown in Table 5-1. The acid dissociation constant Ka is used to define characteristics of titratable groups in organic acids and amines. The negative log of the dissociation constant Ka is called the pKa of the titratable group. In a practical sense, this means that when the pH is equal to the pKa, the associated (AH, protonated) and dissociated (A–, unprotonated) species will be present in equal molar concentrations. TABLE 5-1 Properties of the Amino Acids That Serve as Common Building Blocks of Proteins The presence of a titratable group can be easily observed on a titration curve as a marked decrease in the change in pH per unit of base added; this will appear as a flattening of the curve when pH is plotted on the vertical axis and units of base are plotted on the horizontal axis. In essence, the titratable group acts as a buffer to resist changes in pH by donating protons to neutralize the base that is added. A curve obtained by the titration of histidine, which contains three titratable functional groups, is shown in Figure 5-3. On a titration curve, the pKa can be observed as the point of inflection near the center of the “plateau.” The inflection point is where the curvature changes from concave up to concave down. For histidine in Figure 5-3, three pKas can be detected: the carboxyl group has a pKa = 1.82 the imidazole group has a pKa = 6.0, and the α-amino group has a pKa = 9.17. The ionization state of these side chains affects the physical and chemical properties of proteins and is important for their interactions with other proteins, substrates or ligands, and other macromolecules as well as for their physiological functions. Within chromatin, the basic amino acid residues in histones form ionic bonds with the acidic sugar–phosphate backbone of DNA. Acidic amino acid residues are involved in chelation of calcium ions by calcium-binding proteins. The histidine side chain and the carboxylate of acidic amino acids often serve as coordinating ligands for metals in metalloproteins. Within the native protein structure, pKa values for ionizable groups can be substantially altered because of interactions with nearby residues or the hydrophobicity of the interior of the protein. Such alterations can be critical for the catalytic function of proteins such as enzymes (Harris and Turner, 2002). Jack Kyte and Russell Doolittle (1982) proposed a hydropathy index that is now widely used to predict aspects of protein structure; this scale assigns negative numbers to the most hydrophilic side chains and positive numbers to the most hydrophobic side chains (see Table 5-1). Other scales have been developed, some of which assign quite different values to some of the amino acids. Efforts to develop better methods of predicting protein structure continue. An example of the use of a hydropathy index to predict the transmembrane segments of a protein sequence is shown in Figure 5-4. Transmembrane segments of transmembrane proteins can be predicted from the average hydrophobicity scores for small regions of the polypeptide chain (e.g., segments of 9 to 19 amino acids). Transmembrane regions of proteins, which must pass through the lipid bilayers of cell membranes, tend to have high hydropathy scores (greater than 1.6 units). As was noted in the previous section, the properties of the common amino acids can vary markedly depending upon their innate characteristics (e.g., size, charge, polarity, and hydropathy). The characteristics of some amino acids can also be altered by additional enzymatic and nonenzymatic modifications of the R group. Such modifications can occur as cotranslational or posttranslational events following incorporation of the amino acid into proteins (see Chapter 13). These specific amino acid modifications are introduced to modulate or modify a given chemical property. Posttranslational modifications extend the structures and properties of amino acids in proteins well beyond those of the 20 (or 21, if selenocysteine is included) amino acids used for protein translation in mammals. Specific chemical properties can be altered subtly or dynamically by posttranslational modification. Some examples of posttranslational modifications of amino acid R groups in proteins include the methylation of lysine and histidine residues, the acetylation of lysine residues, the hydroxylation of proline and lysine residues, and the carboxylation of glutamate residues (Figure 5-5). These types of modifications of R groups are essential in defining the structural and functional properties of proteins. For example, the γ-carboxylation of glutamate residues in prothrombin and other blood clotting factors is critical for calcium binding and the proper function of these clotting factors in the blood clotting cascade.
Structure, Nomenclature, and Properties of Proteins and Amino Acids
The Proteinogenic Amino Acids
Chirality and Optical Rotation
The Acid and Base Characteristics of Amino Acids
AMINO ACID
MOLECULAR MASS (g/mol)
pKa α-COOH
pKa α-NH3+
pKa R GROUP
HYDROPATHY INDEX (KYTE-DOOLITTLE SCALE)
Hydrophilic amino acids (charged and very polar)
Arginine
155
2.17
9.04
12.48
−4.5
Lysine
146
2.18
8.95
10.53
−3.9
Asparagine
132
2.04
9.82
−3.5
Aspartate
133
2.09
9.82
3.86
−3.5
Glutamine
146
2.17
9.13
−3.5
Glutamate
147
2.19
9.67
4.25
−3.5
Histidine
174
1.82
9.17
6.0
−3.2
Amino acids with intermediate hydrophobicity (Tyr and moderately/weakly polar amino acids)
Tyrosine
181
2.20
10.07
9.11
−1.3
Tryptophan
204
2.38
9.39
−0.9
Serine
105
2.21
9.15
−0.8
Threonine
119
2.63
10.43
−0.7
Glycine
75
2.34
9.60
−0.4
Proline
115
1.99
10.6 (NH2+)
1.6
Alanine
89
2.34
9.69
1.8
Methionine
149
2.28
9.31
1.9
Cysteine
121
1.71
10.78
8.33
2.5
Hydrophobic amino acids (uncharged and nonpolar)
Phenylalanine
165
1.83
9.13
2.8
Leucine
131
2.36
9.68
3.8
Valine
117
2.32
9.62
4.2
Isoleucine
131
2.36
9.68
4.5
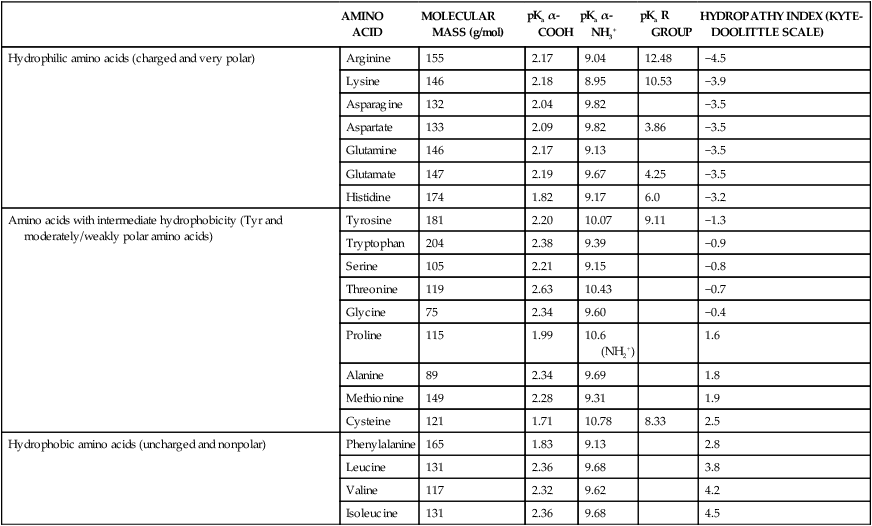
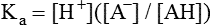
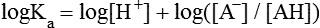
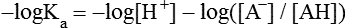

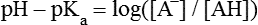
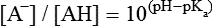
Hydrophobicity or Hydrophilicity of Amino Acid Residues
Modifications of Amino Acid Side Chains
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Basicmedical Key
Fastest Basicmedical Insight Engine