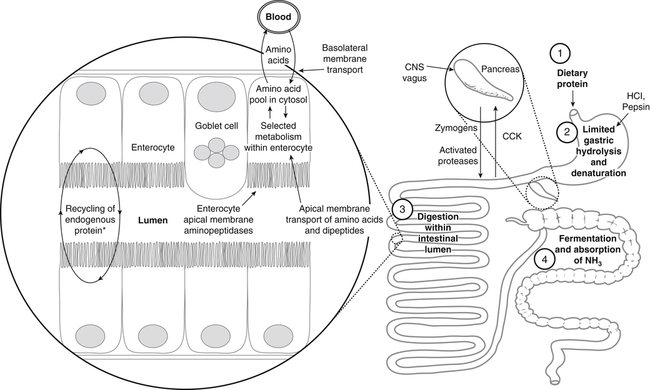
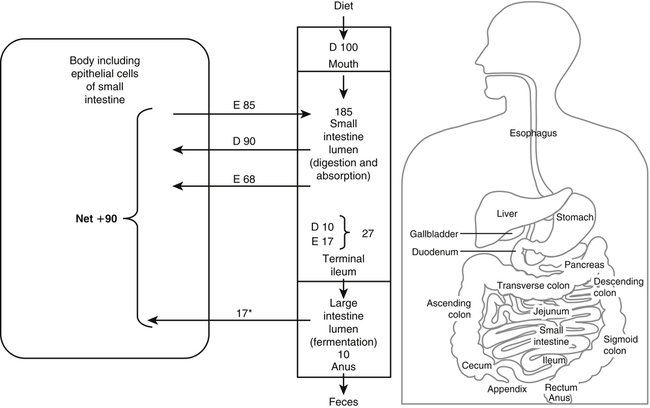
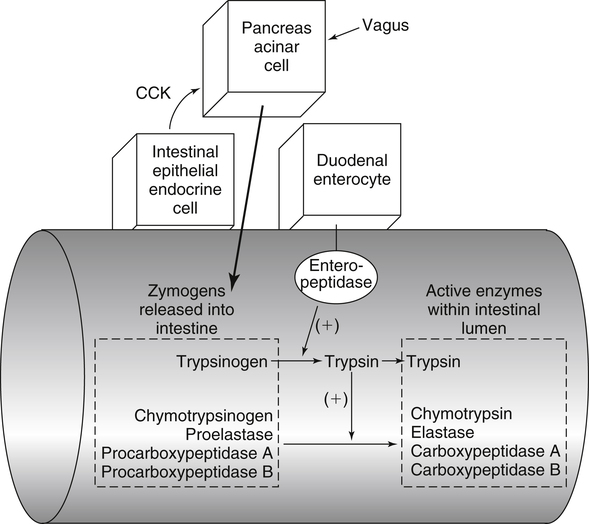
Paul J. Moughan, PhD, DSc, FRSC, FRSNZ and Bruce R. Stevens, PhD The Food and Agriculture Organization, World Health Organization, and United Nations University (FAO/WHO/UNU, 2007) define a “safe” daily intake of dietary protein as 0.83 g high- quality protein per kg body weight, or 58 g/day for the reference 70-kg man and 47 g/day for the reference 57-kg woman. The approximate median intake of protein for adults aged 31 to 50 years in the United States is 100 g/day for men and 65 g/day for women. In addition to food proteins, the body digests 50 to 100 g per day of endogenous protein that is secreted into or sloughed into the lumen of the gastrointestinal tract. These endogenous proteins include proteins in saliva, gastric juice, and other secretions; pancreatic enzymes; mucoproteins; sloughed intestinal cells; and proteins that leak into the intestinal lumen from the blood. Most of this mixture of exogenous and endogenous proteins (115 to 200 g/day) is efficiently digested and taken up by the absorptive enterocytes as free amino acids and dipeptides and tripeptides. Around 85% of the total protein is absorbed anterior to the end of the small intestine (terminal ileum), with around 10 to 20 g of protein entering the colon each day. Daily fecal nitrogen losses amount to the equivalent of about 10 g of protein. The nitrogen excreted in the feces represents primarily endogenous or dietary nitrogen that was not absorbed from the small intestine; this unabsorbed nitrogen was used in the large intestine by the microflora for growth and is therefore mainly present in the feces as part of the bacterial mass. An overall concept diagram of the major aspects of protein digestion and absorption is presented in Figure 9-1, and a typical flow of protein in the adult human is shown in Figure 9-2. The normal events of digestion and absorption are grouped into phases corresponding to physiological events. The six major phases covered in this chapter primarily involve the following: 1. Gastric hydrolysis of peptide linkages in the protein 2. Digestion of protein to smaller peptides by action of pancreatic proteases, which are secreted as zymogens and activated in the lumen of the small intestine where they then carry out digestion 3. Hydrolysis of peptide linkages in oligopeptides by apical (brush border) membrane peptidases and transport of amino acids, dipeptides, and tripeptides across the brush border membrane of the absorptive enterocytes 4. Further digestion of dipeptides and tripeptides by cytoplasmic peptidases in the enterocytes 5. Metabolism of some amino acids within the enterocytes 6. Transport of amino acids across the basolateral membrane of the enterocytes into the interstitial fluid from which the amino acids enter the venous capillaries and hence the portal blood Based on work that originated in the Russian laboratory of Ivan Pavlov in the late 1890s, research has established that protein digestion involves a multistep conversion of inactive zymogens to their active states within the lumen of the small intestine. The current understanding of the activation cascade for these pancreatic zymogens is summarized in Figure 9-3. the enterocytes of the proximal small intestine (duodenum/upper jejunum). Human enteropeptidase is a heavily glycosylated protein with an N-terminal transmembrane domain and a C-terminal extracellular serine protease domain. Enteropeptidase is classified as a serine protease (or serine endopeptidase) because it has a serine residue at its active site as part of a histidine/serine/aspartate catalytic triad. A benefit of synthesis of proteolytic enzymes as zymogens, with activation occurring after the proenzymes have been secreted into the intestinal lumen, is the prevention of proteolytic digestion and tissue damage within the pancreas and pancreatic duct. In addition to this protective mechanism, pancreatic juice normally contains a small peptide that inhibits trypsin to prevent any small amount of trypsin prematurely formed within the pancreatic cells or pancreatic ducts from catalyzing proteolysis. The absence of this protective mechanism leads to pancreatitis. Gain-of-function mutations and copy number variability of the trypsinogen gene, as well as loss-of-function variants for the pancreatic secretory trypsin inhibitor, have firmly established that prematurely activated trypsin causes chronic pancreatitis (Chen and Férec, 2009). The second group of proteolytic enzymes secreted by the pancreas, the carboxypeptidases, are exopeptidases that cleave off one amino acid at a time from the C-terminus of the substrate. These exopeptidases can attack the oligopeptides formed by the endopeptidases to sequentially cleave off free amino acids, leaving a mixture of free amino acids and small peptides of two to eight residues. Carboxypeptidases A and B are metalloenzymes that require Zn2+ at the active site, where the cation functions as a Lewis acid. (See Chapter 37 for a discussion of zinc metalloenzymes.) Carboxypeptidase B preferentially cleaves C-terminal lysine or arginine residues of peptides, and carboxypeptidase A selectively hydrolyzes most C-terminal amino acids, except proline, lysine, and arginine, with a preference for valine, leucine, isoleucine, and alanine. Neither carboxypeptidase A nor carboxypeptidase B readily cleaves C-terminal amino acids that are linked to a proline residue. These pancreatic enzymes act as a team within the small intestinal lumen to hydrolyze many of the peptide bonds in proteins and to efficiently digest protein to yield small peptides (two to eight residues) and free amino acids. It is also important to realize that the upper digestive tract of humans contains an active microflora, and bacteria undoubtedly have a role in the digestive breakdown of food and especially of some of the endogenous proteins. Although the size of the microflora in the colon is much greater than that in the upper digestive tract, there is considerable evidence for microbial activity in the upper tract (Moughan, 2003), and it seems likely, though more experimental evidence is needed, that bacterial enzymes complement to some extent the mammalian proteases. Free amino acids are initially taken up by transporters in the luminal-facing apical membrane of villous absorptive enterocytes and subsequently exit those cells via other basolateral membrane transporters. The amino acids can either pass through the enterocyte unmetabolized, be used for protein synthesis in the enterocyte, be partially (or completely) oxidized for energy, or undergo intermediary metabolic conversion into other amino acids or metabolites that, in turn, are subsequently transported out of the cell across the basolateral membrane. Following basolateral membrane transport to the interstitial fluid, the amino acids move into villus capillaries and on to the liver via the portal circulation, as summarized in Figure 9-1. The intestine is highly efficient in extracting the dietary essential and nonessential amino acids from the lumen as free amino acids. This occurs largely because of the activity of brush border and basolateral membrane transporter systems that handle specific amino acids. Some of the absorbed amino acids are used by the enterocytes themselves, most notably glutamine, which is used as the primary fuel source in enterocytes in place of glucose. Enterocyte basolateral membrane transporters also take up enterocyte-sustaining amino acids from the blood circulation, especially in the postprandial state. A transport “system” is defined as a physiological functional unit formed from one or more transporter protein subunits. Each transporter subunit type is encoded by a specific gene. A transport system activity may result from the action of a single transporter protein or the multimeric arrangement of transporter proteins within the membrane. Although it is technically correct to use the term transporter to mean only a single protein, scientists often informally also refer to multimeric functional units as transporters. Membrane amino acid transporter systems composed of a single protein (monomeric transport systems) are listed in Table 9-1, whereas heterodimeric transporter systems for amino acids are listed in Table 9-2. TABLE 9-1 Monomeric Amino Acid Transport Systems in Human Small Intestine or Colon
Digestion and Absorption of Protein
Digestion of Protein in the Gastrointestinal Tract
Small Intestinal Luminal Phase: Activation and Action of Pancreatic Proteolytic Enzymes
Pancreatic Zymogens and their Activation Cascade
Pancreatic Digestive Enzymes
Absorption of Free Amino Acids and Small Peptides
Amino Acid Transporters in the Apical and Basolateral Membranes
TRANSPORT “SYSTEM” FUNCTIONAL NAME
COMMON ALIAS
GENE (SLC=Solute carrier)
HUMAN GENE LOCUS
TYPICAL SUBSTRATES
ION DEPENDENCY
TISSUE
EPITHELIAL MEMBRANE
SLC1 FAMILY
XAG−
EAAT3
SLC1A1
9q24
L-Glutamate, D/L-aspartate, cystine (disulfide)
H+, Na+, K+
Small intestine
Apical
ASC
ASCT1
SLC1A4
2p13-p15
Alanine, serine, threonine, cysteine, glutamine
Na+
Small intestine
Apical
ASC
ASCT2 or ATB0
SLC1A5
19q13.3
Alanine, serine, threonine, cysteine, glutamine, branched neutrals
Na+
Small intestine, colon
Apical
SLC6 FAMILY
Creatine
CRTR
SLC6A8
Xq28
Creatine
Na+, Cl−
Small intestine
Apical
GLY
GLYT1
SLC6A9
1p33
Glycine
Na+, Cl−
Small intestine
Basolateral
B0,+
ATB0,+
SLC6A14
Xq23-q24
Neutrals and dibasics, arginine, D-serine
Na+, Cl−
Colon
Apical
B0 (or B)
B0AT1
SLC6A19
5p15.33
Neutrals, glutamine
Na+
Small intestine
Apical
IMINO
SIT1
SLC6A20
3p21.6
Proline, sarcosine, pipecolate
Na+
Small intestine, colon
Apical
SLC7 FAMILY
y+
CAT-1
SLC7A1
13q12-q14
Arginine, ornithine, lysine, histidine, dibasics
None
Small intestine, colon
Basolateral
SLC15 FAMILY
Pept1
PEPT1
SLC15A1
13q33-q34
Dipeptides & tripeptides, carnosine, β-lactam antibiotics, angiotensin-converting enzyme inhibitors
H+ with NHE3
Small intestine
Apical
SLC16 FAMILY
T
TAT1
SLC16A10
6q21-q22
Aromatics, L-DOPA
None
Small intestine
Basolateral
SLC22 FAMILY
OCTN2VT
OCTN2
SLC22A5
5q23.3
L-Carnitine, acetyl-L-carnitine
Na+
Small intestine
Apical
SLC36 FAMILY
Iminoacid
PAT1
SLC36A1
5q33.1
Proline, glycine, β-alanine, GABA, taurine, D-serine
H+ with NHE3
Small intestine, colon
Apical
SLC38 FAMILY
A
SNAT2
SLC38A2
12q
Alanine, asparagine, cysteine, glutamine, glycine, histidine, methionine, proline, serine
Na+
Small intestine
Basolateral
A
SNAT4
SLC38A4
12q13
Alanine, asparagine, cysteine, glycine, threonine
Na+
Small intestine
Basolateral
N
SNAT5
SLC38A5
Xp11.23
Glutamine, histidine, serine, asparagine, alanine
Na+, H+
Small intestine (crypt cells)
Apical
SLC43 FAMILY
LAT4
LAT4
SLC43A2
17p13.3
Branched-chain amino acids, phenylalaninine
None
Small intestine
Basolateral ![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Digestion and Absorption of Protein