Fig. 3.1
MicroRNA biogenesis is initiated via the activity of RNA polymerase II resulting in synthesis of a primary miRNA transcript (Pri-miRNA) that is both capped and polyadenylated. The appropriate spatial complementation of specific nucleotides within the transcript results in the formation of hairpin secondary structures that are recognized by the RNA processing complex consisting of DROSHA and DGCR8. The enzymatic activity of this protein complex results in cleavage and removal of the hairpin structure (now considered a pre-miRNA) from the primary transcript. Exportin 5 facilitates the transport of pre-miRNA from the nucleus into the cytoplasm of the cell where it is recognized by Dicer, and the loop is cleaved leaving a short duplex mature miRNA molecule. Upon dissociation, either strand from the duplex miRNA structure can be utilized by the RNA-induced silencing complex to contribute to posttranscriptional gene regulation though impacting mRNA stability and/or translation efficiency in addition to contributing to chromatin modifications to control gene expression
The endo-siRNA class is derived from long dsRNA, which can be either sense or antisense RNA pairs or long hairpins, and is also cleaved in the cytoplasm by Dicer (Ghildiyal et al. 2008; Okamura et al. 2008a, b; Tam et al. 2008). Synthesis of both endo-siRNA and miRNA occurs via Dicer activity and has a final mature product of an approximately 21 nucleotide single-stranded RNAs capable of association with members of the argonaute protein family (AGO 1–4) which enable the formation of the active RISC complex (Filipowicz 2005).
piRNA biogenesis is less well understood than miRNA and endo-siRNA, although they are produced from long single-stranded RNA precursors mediated through the action of Dicer (Houwing et al. 2007; Vagin et al. 2006). Mature piRNAs are approximately 26–31 nucleotides long and are expressed primarily in germ cells (Aravin et al. 2006; Houwing et al. 2007; Watanabe et al. 2006). Secondary processing of piRNA involves Miwi1, Miwi2, and Mili in mice (also known as Piwi1, Piwi2, and Pili) (Aravin et al. 2006, 2007; Klattenhoff and Theurkauf 2008). PiRNA action does not involve AGO protein association but is associated with transposon and repeat-associated siRNA inactivation (Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007; Vagin et al. 2006). The complex nature of piRNA sequences has been revealed via next-generation sequencing (Aravin et al. 2006; Girard et al. 2006). Class I piRNA are derived from clustered genomic loci in repeat sequences, indicating a role with respect to transposon defense (Houwing et al. 2008; Klattenhoff and Theurkauf 2008). Class II piRNAs are derived from transposon RNA cleavage, while the remainder of the piRNAs are thought to originate from diverse genomic regions perhaps including the 3′ untranslated region (UTR) of some mRNA as is the case in Drosophila, mouse, and Xenopus (Robine et al. 2009). The variation in piRNA biogenesis and sequence suggests their biological involvement in PTGR in addition to transposon repression.
The “ping-pong” model has been proposed as a regulatory model coupling piRNA generation and transposon repression (Brennecke et al. 2007). In Drosophila, Piwi and Aub proteins bind the genomic piRNA cluster loci-derived antisense primary piRNA. The proteins are then guided by the antisense primary piRNA, which binds and cleaves transposon mRNA to generate the 5′ ends of the sense secondary piRNA. AGO3 protein then binds the sense secondary piRNA resulting in transcript cleavage to produce the 5′ end of new primary piRNA (Brennecke et al. 2007). How 3′ ends of new piRNA are generated in the ping-pong model is not clear. The proportion of piRNAs generated via the ping-pong mechanism represent only a fraction of the piRNA population; therefore, how other piRNAs are generated from complex intergenic regions also remains uncharacterized (Cook and Blelloch 2013).
In mice, expression of piRNA is developmentally regulated (Aravin et al. 2007). During the prepachytene stage prior to meiosis in spermatogenesis, expression of one subtype of piRNA (26–28 nucleotides) is favored, whereas another piRNA subtype (29–31 nucleotides) is predominantly expressed during the pachytene meiotic stage. While piRNA expression is enriched in the mammalian and Drosophila germline, piRNAs are also expressed in somatic cells of Drosophila (Lin and Yin 2008), differentiated somatic cells of jellyfish (Seipel et al. 2004), and porcine cumulus cells (Yang et al. 2012a), suggesting additional piRNA roles in these cell types, which may be species dependent.
3.3 Small RNA Mechanism of Action
Mature miRNA and endo-siRNA interact with AGO proteins (AGO 1–4; also known as EIF2C1-4) to enable the association of RISC machinery (Filipowicz 2005). AGO proteins associated with either a miRNA or endo-siRNA interact with target mRNA via complementarity between the ncRNA and the 3′UTR of the mRNA target. If complete complementarity exists, cleavage of the target mRNA can occur. If there is incomplete complementarity, the primary mechanism of PTGR occurs by translation suppression (Fabian et al. 2010; Hutvagner and Zamore 2002; Martinez et al. 2002). There are, however, examples of weak complementation between the 3′ end of the ncRNA and the 3′UTR of the target gene which can still result in PTGR via mRNA degradation (Bagga et al. 2005).
PIWI proteins are located in nuclear and/or perinuclear nuage (Brennecke et al. 2007; Carmell et al. 2007; Wang et al. 2009) where they repress genetic elements (Lim and Kai 2007). PIWI proteins have an N-terminal PAZ domain followed by an MID (middle) domain, which together recognize and bind the 3′ end of piRNAs (Jinek and Doudna 2009). The C-terminal domain of PIWI proteins has RNase H activity capable of recognizing the 5′ end of piRNA and facilitates the cleavage of the target sequence (Frank et al. 2010; Wang et al. 2009). In the germline, the PIWI-piRNA complex silences transposons both transcriptionally and posttranscriptionally, through chromatin silencing of transposable elements via histone modification and by altering the DNA epigenetic status (Klenov et al. 2007; Kuramochi-Miyagawa et al. 2008). Posttranscriptionally, transposable elements are repressed through their active RNA being cleaved by PIWI- and AUB-primary piRNA complexes, thereby producing secondary piRNA through the aforementioned “ping-pong” model (Brennecke et al. 2007). This mechanism is supported by the observation that transposon RNA is expressed at a higher level in the presence of PIWI protein mutations (Li et al. 2009). In Drosophila, piRNAs are involved in telomere function, including telomere protection complex assembly, thereby maintaining chromosome integrity (Khurana et al. 2010). Expression of PIWI proteins in human somatic stem cells (Sharma et al. 2001) and neoplastic cells (Lee et al. 2006, 2010; Liu et al. 2006b, 2010; Taubert et al. 2007) suggests that piRNA may also be involved in stem cell function regulation and carcinogenesis. However, the mechanism, major functions, and pathways regulated by the Piwi-piRNA complex remain poorly understood.
3.4 Small RNA Expression and Function in Reproductive Tissues
3.4.1 Germ Cell Development and Maintenance
PIWI proteins are expressed in germ cells, stem cells, and oocytes of multiple species. In the female germline of Xenopus laevis (Xiwi and Xili), Danio rerio (Ziwi and Zili), and Drosophila melanogaster (Aubergine, Piwi, and Ago2) (Houwing et al. 2007; Li et al. 2009; Wilczynska et al. 2009), several PIWI regulatory members have been identified and characterized. In mice, Miwi, Mili, and Miwi-2 exist (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2008), and in pigs three Piwi genes (Piwil1, Piwil2, and Piwil4) are expressed in the testes, ovaries, and oocytes (Kowalczykiewicz et al. 2012). Utilizing bioinformatic analysis, human Piwi proteins (Hiwi, Hili, and Hiwi-2) have been identified (Gu et al. 2010), with Hiwi being expressed in hematopoietic stem cells and germ cells (Qiao et al. 2002; Sharma et al. 2001). During germ cell specification in mice (Deng and Lin 2002) and gonad development of pigs (Kowalczykiewicz et al. 2012), expression patterns of individual PIWI proteins differ. Partial or complete loss of germ cells and transposon derepression in the germline have been demonstrated in Drosophila melanogaster and mice (Carmell et al. 2007; Cox et al. 1998; Li et al. 2009; Vagin et al. 2006) due to the loss of PIWI proteins, suggesting a conserved role for PIWI proteins in the maintenance of germ cell viability.
3.4.2 Oocyte Development and Maturation
Mammals are born with a finite oocyte number that originates from the primordial germ cell pool following migration to the genital ridge during embryonic development (Tingen et al. 2009). Following recruitment from the primordial follicle pool and selection for maturation, oocytes undergo germinal vesicle breakdown (GVBD) facilitating their progression through meiosis until subsequent arrest at metaphase II. The oocyte is transcriptionally quiescent following GVBD until after fertilization and subsequent activation of the embryonic genome, which occurs around the two- to eight-cell stage of development depending upon the species. The inability of the developing embryo to elicit a transcriptional response prior to genome activation suggests a potentially important role of ncRNA during this period of development. MiRNA, endo-siRNA, and piRNA are expressed in oocytes of multiple species at various stages of development (Abd El Naby et al. 2013; Golden et al. 2008; Tesfaye et al. 2009; Watanabe et al. 2006, 2008; Xu et al. 2011; Yang et al. 2012a).
Conditional knockout mice have utility for deciphering functions and contributions of endo-siRNA and miRNA in both the maturing oocyte and developing embryo. Mice with a ZP3-driven conditional loss of Dgcr8 (responsible for canonical miRNA production) are fertile (Ma et al. 2010; Suh et al. 2010), but litter size was substantially reduced, suggesting that some miRNA may contribute to developmental competency of the subsequently produced embryo even if not required in the maturing oocyte (Suh et al. 2010). The observation that Dgcr8 is not required for mouse oocyte maturation, since loss of Dgcr8 has no noticeable effect on mRNA regulation, coupled with the observation that similarly deleting Dicer or Ago2 does have a negative effect on maturing oocytes rendering them incompetent (Kaneda et al. 2009; Murchison et al. 2007; Suh et al. 2010; Tang et al. 2007), suggests that small ncRNAs are needed for the production of oocytes capable of producing viable embryos.
The small RNA population of both the oocyte and cumulus cells during in vitro maturation (IVM) have been sequenced and the portfolio of endo-siRNA, miRNA, and piRNA demonstrated in pigs (Yang et al. 2012a). During oocyte IVM and progression through meiosis, few alterations were evident, with the exception of miR-21 and miR-574-3p which were significantly upregulated and downregulated during IVM, respectively. In mice, granulosa cell miR-21 expression is luteinizing hormone dependent, and the in vivo use of oligonucleotide inhibitors against miR-21 suppressed ovulation rate but increased apoptosis, indicating a cell viability role for miR-21 (Carletti et al. 2010).
3.4.3 Fertilization and Early Embryo Development
Sperm possess a diverse portfolio of ncRNA (Amanai et al. 2006; Krawetz et al. 2011), and their abundance of miRNA may be related to the biogenesis and expression of small RNA contributing to spermatogenesis (Maatouk et al. 2008). Interestingly, miR-34c, a miRNA associated with the differentiation of male germ cells (Bouhallier et al. 2010), has been shown to influence embryonic development in mice following fertilization (Liu et al. 2012). Liu et al. (2012) identified six miRNAs, including miR-34c, as absent in mature oocytes but present in mouse sperm and zygotes. The impact on development occurs via the ability of miR-34c to interact with BCL2, thereby contributing to regulation of the first embryonic cleavage following oocyte activation (Liu et al. 2012).
Following fertilization and subsequent activation of the zygotic genome, the embryo begins to express RNA transcripts. Transcription of pri-miRNA has been observed in the mouse two-cell embryo, and mature transcripts are detectable at the four-cell stage (Tang et al. 2007; Zeng and Schultz 2005). Dicer and Dgcr8 activities are needed for epiblast formation (Kanellopoulou et al. 2005; Murchison et al. 2005; Wang et al. 2007, 2008), and in some organisms it is thought that miRNA contributes to maternal mRNA clearance before zygotic gene activation (ZGA) occurs (Giraldez et al. 2006; Hemberger et al. 2009; Sinkkonen et al. 2008; Svoboda and Flemr 2010).
Successful embryonic development requires broad transcriptional arrest and mRNA clearance to deplete maternally stored mRNA transcripts in coordination with both ZGA and subsequent mRNA and protein production. Some small RNAs, including miRNA, are abundantly expressed during oocyte maturation and early embryonic development in Xenopus laevis (Watanabe et al. 2005), Drosophila (Aboobaker et al. 2005; Biemar et al. 2005), zebra fish (Giraldez et al. 2006; Wienholds et al. 2005), mice (Tang et al. 2007), and pigs (Yang et al. 2012a). Maternal mRNA depletion is, in part, controlled via the 3′UTR of the expressed transcripts (Brevini et al. 2007; Tadros and Lipshitz 2005); thus, there is opportunity for small RNA to influence the posttranscriptional outcome of the resultant embryo.
3.4.4 Embryonic Stem Cells
Biogenesis of miRNA and the associated molecular machinery has been shown to be required for stem cell differentiation and the maintenance of pluripotency (Marson et al. 2008). The disruption of Dicer and Dgcr8 leads to a defective capacity of ES cells to be cultured in both mice and humans (Bernstein et al. 2003; Han et al. 2004; Kanellopoulou et al. 2005; Murchison et al. 2005). Embryonic lethality is the consequence of Dicer deletion in mice (Bernstein et al. 2003) while Dicer-null embryonic stem (ES) cells are deficient in proper differentiation (Kanellopoulou et al. 2005). Dgcr8 knockout mice ES cells show a phenotype similar to Dicer-deficient ES cells, with reduced proliferation and a defective capacity for differentiation (Wang et al. 2007). Taken together, these results support the requirement of canonical miRNA biogenesis for ES cell differentiation and proliferation.
There are a number of miRNA expression clusters that are likely to be involved in ES cell regulation including the let-7 family and the miR-290 and miR-17–92 cluster. Let-7 miRNA family members are highly expressed in differentiating cells (Gu et al. 2008; Lakshmipathy et al. 2007), and Let-7 g can be regulated by LIN28, which is highly expressed in pluripotent cells (Viswanathan et al. 2008), suggesting that let-7/LIN28 regulatory mechanism contributes to the maintenance of pluripotency. Pluripotent factors such as Oct4, Sox2, Nanog, and Tcf4 activate LIN28 gene expression, which in turn inhibits differentiation, and since the 3′UTR of LIN28 mRNA is targeted by Let-7, LIN28 inhibition of differentiation may be a result of let-7 g activity (Marson et al. 2008).
There are six miRNAs (miR-290 through miR-295) in the miR-290 cluster, all of which are transcribed in single polycistronic transcripts and regulated by a common promoter (Suh et al. 2004). All miR-290 members are expressed in undifferentiated mouse ES cells, but their abundance decreases after differentiation (Houbaviy et al. 2003). Embryonic lethality results in mice with a homozygous deletion of all six members of the miR-290 cluster (Ambros and Chen 2007), and exogenously delivered miR-290 cluster can partially rescue the self-renewal capacity of Dicer-null cells (Benetti et al. 2008; Sinkkonen et al. 2008).
The miR-17–92 cluster is strongly expressed in undifferentiated ES cells and forms a polycistronic transcript which generates miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1 (Gu et al. 2008; Houbaviy et al. 2003; Morin et al. 2008). The miR-17–92 cluster is activated by the oncogene, c-Myc (Chen and Daley 2008), which in combination with the pluripotency factors Oct4, Sox2, and Klf4 can induce pluripotency (iPS cells). Thus, it is suggested that the miR-17–92 cluster plays roles in pluripotency and stem cell renewal (Melton et al. 2010).
3.4.5 Embryo Implantation and Interaction with Maternal Endometrium
During the peri-implantation period of pregnancy, uterine epithelial cells and the conceptus trophectoderm develop adhesion competence in unison to initiate an adhesion cascade within the window of receptivity. Upregulation of 32 miRNA has been demonstrated in mouse endometrium during the window of implantation. Of the identified miRNA, miR-101, miR-144, and miR-199a* are predicted to interact with cyclooxygenase-2 (Cox2) mRNA (Chakrabarty et al. 2007). Proinflammatory immune signaling is associated with uterine receptivity in multiple species (Cha et al. 2012), and miRNA with the potential to influence the expression of inflammatory and immune response mediators includes let7, miR-17-5p, miR-20a, miR-106a, miR-125b, miR-146, and miR-155 (Meng et al. 2007, 2008; O’Connell et al. 2007; Rodriguez et al. 2007; Tili et al. 2007). For example, both miR-125b and miR-155 are involved in the development of T, B, and dendritic cells, key cells of the immune system. Transcription of these miRNAs is proposed to be activated by the nuclear factor of kappa light polypeptide gene enhancer in B cells (NF-κB) (Pan and Chegini 2008). The presence of numerous small RNAs has been identified in the porcine uterine endometrium during the implantation window; however, the mechanism by which specific small RNAs contribute to uterine function and ultimately facilitate embryo implantation remains largely unknown. Conditional deletion of Dicer in mice driven by the progesterone receptor or anti-Müllerian hormone receptor type 2 promoters resulted in sterility, abnormal development, and altered signaling pathways in the uterus suggesting important biological contributions of small RNA to the function of the uterine endometrium (Hawkins et al. 2012; Nagaraja et al. 2008).
Postimplantation survival of a growing fetus is entirely dependent on the coordination of gas exchange, nutrient supply, and waste product removal via the placenta, while maintaining immunological protection of the embryo. Human placenta is abundant in miRNA, with distinctive expression profile patterns (Barad et al. 2004; Landgraf et al. 2007; Liang et al. 2007). In addition, miRNA biogenesis protein machinery also seems to be essential for placental development and function (Cheloufi et al. 2010).
Expression of chromosome 19 miRNA cluster (C19MC) and miR-371-3 cluster changes throughout pregnancy (Morales Prieto and Markert 2011) and differs in placental tissue from females who undergo preterm labor compared to normal term pregnancy (Mayor-Lynn et al. 2011). These clusters are located within imprinted genes that are known to be involved with embryonic development and cellular differentiation (Tsai et al. 2009). Interestingly, C19MC is one of the largest miRNA gene clusters in the human genome (Bentwich et al. 2005; Lin et al. 2010) and is expressed from the paternally inherited chromosome and controlled by upstream promoter region methylation (Noguer-Dance et al. 2010). Regulation of C19MC is not well characterized; however, the expression appears to be restricted to the reproductive system and the placenta (Liang et al. 2007; Lin et al. 2010) though expression of miR-498, a component of the C19MC cluster, has been shown to be expressed in the fetal brain (Flor and Bullerdiek 2012). Interestingly, no homologues of this cluster have been found in rat, mouse, or dog (Zhang et al. 2008). The miR-371-3 cluster consists mainly of three miRNAs (miR-371a-3p, miR-372, and miR-373-3p) sharing the same seed sequence, AAGUGC (Griffiths-Jones 2006), and is predominantly placentally expressed (Bentwich et al. 2005).
3.5 miRNA Regulation During Spermatogenesis
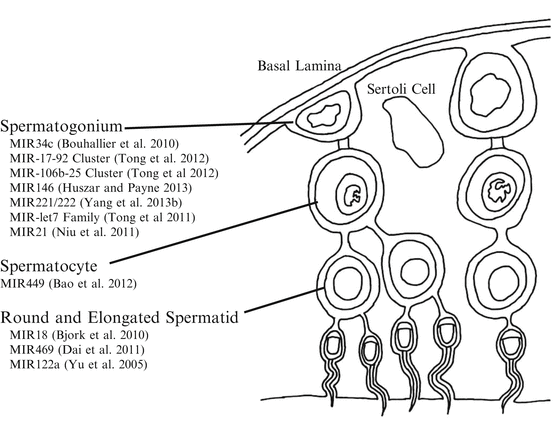
Spermatogenesis is the process by which diploid spermatogonium differentiates into motile spermatozoa. The process begins after birth when spermatogonial stem cells enter the differentiation pathway. The stages of spermatogenesis vary both in location within the seminiferous tubule and by morphological changes to the germ cell as stem cell-like spermatogonia near the basal lamina are recruited and undergo several rounds of mitotic divisions, followed by progression through meiosis, morphogenesis, and eventual release of mature spermatids into the lumen of the seminiferous tubule (Kotaja et al. 2004). MiRNA expression and function throughout numerous stages of spermatogenesis has been observed by numerous investigators (Fig. 3.2).
3.5.1 miRNA Biogenesis During Spermatogenesis
The miRNA biogenesis and RISC-associated proteins Dicer, Drosha, AGO1, AGO2, AGO3, and AGO4 have all been detected in pachytene stage spermatocytes, round and elongated spermatids, and Sertoli cells (Gonzalez-Gonzalez et al. 2008). The importance of small RNA regulation within the maturing sperm cell has been exhibited by extensive rodent knockout models. Primordial germ cells as well as spermatogonia from Dicer-deficient mice exhibit poor proliferation (Hayashi et al. 2008). Furthermore selective knockout of Dicer driven by promoters for DEAD box polypeptide 4 (Ddx4) or tissue-nonspecific alkaline phosphatase (Tnap) at the early onset of spermatogenesis also leads to infertility implying the importance of small RNA production and function to enable spermatogenesis. There is an observable loss of sperm differentiation, abnormal morphology, and loss of motility with the absence of Dicer (Maatouk et al. 2008; Romero et al. 2011). Neurogenin 3 (Ngn3) promoter-driven conditional knockout of Dicer results in reduced testis size and disruption of spermatogenesis within the seminiferous tubules as well as the epididymis (Korhonen et al. 2011). Furthermore, selective knockout of Dicer in mice Sertoli cells driven by the Mis (anti- Müllerian hormone) promoter also leads to infertility due to complete absence of spermatozoa and progressive testicular degradation (Papaioannou et al. 2009). Other evidence that small RNA regulation is important in the maturing male gamete is the presence of chromatoid bodies. Chromatoid bodies are a unique cloud-like structure found within spermatids that are thought to contain miRNA, RISC-associated proteins, and potentially the mRNA targets of miRNA (Kotaja et al. 2006; Kotaja and Sassone-Corsi 2007; Meikar et al. 2011).
3.5.2 miRNA Expression Patterns During Spermatogenesis
MiRNAs are differentially expressed during testicular development (Yu et al. 2005). High-throughput sequencing experiments have detected 141 miRNAs expressing in the mouse testis and 29 novel miRNAs in the human testis (Ro et al. 2007), and another study detected 770 known and five novel miRNAs in the human testis (Yang et al. 2013a). Immunohistochemistry detected miRNAs in the Sertoli cell nucleus and in the dense body of the pachytene spermatocytes (Marcon et al. 2008). Microarray experiments have shown dynamic changes in miRNA profiles when comparing immature and mature testis in mice, rhesus monkeys, and pigs (Luo et al. 2010; Yan et al. 2007, 2009).
When investigating specific miRNA or miRNA clusters, investigators have found changes in abundance of miRNA comparing stage of spermatogenesis (Fig. 3.2). There is high abundance of miR-34c in the spermatocyte and spermatid. There is also high abundance of miR-34c in the testis compared to the spleen, brain, or liver tissue (Bouhallier et al. 2010). When mouse embryonic stem cells (mESCs) are transfected with pri-miR-34c-GFP and combined with retinoic acid induction, miR-34c promotes mESCs to differentiate into spermatogenic-like cells (Zhang et al. 2012). However, in caprine germline stem cells, overexpression of miR-34c leads to apoptosis and suppressed proliferation (Li et al. 2013). The potential mechanisms of miR-34c during spermatogonial differentiation in mice are thought to be through targeting Atf1 and inducing apoptosis (Liang et al. 2012) or by targeting Nanos2 mRNA (Yu et al. 2014).
Both the miR-17–92 and miR-106b-25 clusters are downregulated during retinoic acid-induced spermatogonial differentiation both in vivo and in vitro (Tong et al. 2012). A mouse loss-of-function model for miR-17–92 cluster in male germ cells causes smaller testis, decreased number of epididymal sperm, and mild defects in spermatogenesis (Tong et al. 2012). The abundance of miR-146 is highly increased in mice undifferentiated spermatogonia (Huszar and Payne 2013), suggesting that miR-146 plays a role in differentiation. Impaired function of the X chromosome-clustered miR-221 and miR-222 in mouse undifferentiated spermatogonia induces loss of stem cell capacity to regenerate spermatogenesis and induces the transition from a KIT− to a KIT+ state (or expressing the CD117 receptor), a hallmark of the transition into differentiation. In accordance with this, growth factors that promote maintenance of undifferentiated spermatogonia increase miR-221/miR-222 abundance (Yang et al. 2013b). When comparing abundance of miR-135a in rat descended testis or undescended testis, abundance is greatest in descended testis. A specific example to PTGR occurring in the testis involves miR-135a and forkhead box protein O1 (FOXO1), both of which have been localized in spermatogonial stem cells. Transfection of miR-135a into spermatogonia in vitro resulted in decreased FOXO1 abundance (Moritoki et al. 2014). Another example of miRNA regulation during spermatogenesis is the significant increase in the expression of the miR-let7 family of miRNAs during mouse spermatogonial differentiation through suppression of Lin28 (Tong et al. 2011).
3.5.3 miRNA Associated with Male Fertility
Infertility is estimated to affect 15 % of the couples worldwide, and male infertility is expected to be responsible for 50 % of this (Dohle et al. 2005; Hellani et al. 2006). Idiopathic, or spontaneously occurring, male infertility is accompanied by qualitative and quantitative abnormalities (Ferlin et al. 2007; Hargreave 2000). This has led research groups to be able to diagnose the molecular components and morphology of infertile men or species and compare miRNA abundance between the two. Abu-Halima et al. (Abu-Halima et al. 2013) compared the miRNA profiles in 27 diagnosed male patients: nine normozoospermia, nine asthenozoospermia, and nine oligoasthenozoospermia. When comparing the miRNA profiles of asthenozoospermia to normozoospermia patients through miRNA microarray, 50 miRNAs were upregulated and 27 were downregulated. In comparing oligoasthenozoospermia to normozoospermia, 42 miRNAs were upregulated and 44 were downregulated. Interestingly, miR-34c was downregulated in oligoasthenozoospermia compared to normozoospermia.
More recently Abu-Halima et al. (Abu-Halima et al. 2014) have suggested the use of five miRNAs, miR-34b*, miR-34b, miR-34c, miR-429, and miR-122, as potential biomarkers for the diagnosis and assessment of male infertility. In this study 80 semen samples from patients showing abnormal semen parameters were compared with 90 semen samples showing normal parameters. The abundance of the five miRNAs was compared using qRT-PCR, and miR-449 was increased and the other four were decreased in the abnormal semen groups compared to normal control subjects. Using support vector machine classification combined with these five miRNAs, the study was able to discriminate individuals with subfertility from control subjects with accuracy of 98.65 %, specificity of 98.83 %, and sensitivity of 98.44 % (Abu-Halima et al. 2014).
3.6 Small RNA and Female Reproductive Disorders
3.6.1 Relationship Between Obesity and Infertility Mediated by miRNA
Obesity has detrimental effects on female reproductive function, including increasing the likelihood for polycystic ovarian syndrome (PCOS), ovulation defects, reduced fecundity, and poor quality oocytes (Brewer and Balen 2010; Maheshwari et al. 2007; Rachon and Teede 2010). There is association between obesity and increased risk of birth defects, prematurity and stillbirths, and gestational diabetes (Bellver et al. 2007; Maheshwari et al. 2007). Obesity contributes to the development of type 2 diabetes, characterized by hyperglycemia and impaired insulin signaling (Akamine et al. 2010). In contrast to many peripheral tissues that display insulin resistance (Akamine et al. 2010; Kalra et al. 2006; Kashyap and Defronzo 2007), the ovary remains insulin sensitive during obesity-induced type 2 diabetes. Ovaries from female mice fed with a diet containing 60 % kcal of fat for 12 weeks maintained insulin sensitivity, despite that other classical tissues like the muscle and liver became insulin resistant (Wu et al. 2012).
The exact mechanism(s) by which obesity affects ovarian function remains poorly understood; however, systemic low-grade inflammation has been implicated in the development of infertility and other obesity-associated adverse reproductive health outcomes (Blencowe et al. 2012; Carmichael et al. 2010; Nestler 2000; Pasquali and Gambineri 2006; Pasquali et al. 2003; Pettigrew and Hamilton-Fairley 1997; Rubens et al. 2010). Chronic inflammation alters miRNA levels in immune cells (Fernandez-Valverde et al. 2011; Williams and Mitchell 2012; Xie et al. 2009), and miRNAs have been shown to regulate the activity of key cellular processes, including insulin release in pancreatic β cells, adipocyte differentiation (Williams and Mitchell 2012; Xie et al. 2009), and insulin sensitivity (Trajkovski et al. 2011), and are therefore potentially involved in obesity-induced infertility in females. We have demonstrated elevated tumor necrosis factor alpha (Tnfa) mRNA expression in the ovaries from obese mice (Nteeba et al. 2013a). miR-125b has been shown to negatively regulate Tnfa mRNA (Huang et al. 2012), while increased Tnfa itself downregulates miR-143 (Xie et al. 2009). Both miR-125b and miR-143 were decreased in ovarian tissue during obesity supporting that miRNAs may mediate physiological alterations involving an inflammatory mechanism (Nteeba et al. 2013a). Further, decreased circulating miR-125b has also been associated with increased fat mass in morbidly obese men (Ortega et al. 2013), while miR-143 is reported to be critical for the formation of the primordial follicle pool in utero (Zhang et al. 2013), raising concerns about the impact of obesity on the neonatal ovary.
Another mechanism by which miRNA can impact fertility may be through the phosphatidylinositol-3 kinases (PI3K) pathway. PI3K are lipid kinases that phosphorylate the 3′-OH group on the inositol ring of inositol phospholipids. Activation of PI3K results in conversion of the plasma membrane lipid phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 can recruit proteins containing lipid-binding domains from the cytoplasm (Pawson and Nash 2000) such as the serine/threonine kinases 3′-phosphoinositide-dependent kinase-1 (PDK1) and AKT (Cantley 2002) to the plasma membrane where their proximity results in their phosphorylation. Once phosphorylated, AKT can translocate to the nucleus, where it regulates a number of cellular responses such as growth, survival, and cell cycle entry (Datta et al. 1999). PI3K signaling has many ovarian roles, being involved in steroidogenesis, oocyte viability (Brown et al. 2010), and primordial follicle activation (Jagarlamudi et al. 2009; Liu et al. 2006a), and can be regulated through PTGR via miRNA activity (Näär 2011; Trajkovski et al. 2011; Xu and Mo 2012; Yu et al. 2008). In order to evaluate if obesity has any effect on miRNA that could at least partially explain alterations observed with respect to PI3K signaling, levels of miR-103, miR-21, miR-184, and miR-205 in ovaries from obese female mice were investigated. Obese mice had decreased ovarian miR-21 and miR-103. MiR-21 expression is important for the regulation of apoptosis (Donadeu et al. 2012; McBride et al. 2012), and decreased miR-21 abundance has been reported to increase cell apoptosis in a variety of cell culture systems including the granulosa cells from mouse preovulatory follicles both in vivo and in vitro (Carletti et al. 2010). In addition, miR-21 has been identified as promoting follicular cell survival during ovulation, and miR-21 inhibition also has been reported to reduce ovulatory rates (Carletti et al. 2010). Although many different cell types undergo apoptosis in response to inhibition of miR-21 action, the miR-21 targets implicated vary widely for different cells, and the mechanism by which miR-21 suppresses apoptosis in granulosa cells remains to be identified (Kim et al. 2012; Sirotkin et al. 2010; Yang et al. 2008).
MiR-184 has been shown to act as a physiological suppressor of general secretory activity of progesterone and estradiol (Sirotkin et al. 2009, 2010). Obese females have increased levels of miR-184 (Nteeba et al. 2013b), and miR-184 is believed to play a critical role in development in addition to being a mediator of apoptosis. Upregulation of miR-184 has been reported to interfere with the ability of miR-205 to repress PI3K signaling (Yu et al. 2008). Several studies from human and rodent models have reported obesity-induced miR-103 upregulation (Perri et al. 2012; Rottiers and Näär 2012; Trajkovski et al. 2011). We observed a trend for decreased ovarian miR-103 due to obesity (Nteeba et al. 2013b). Importantly, silencing miR-103 has been proposed to improve insulin sensitivity in adipocytes mainly through increased caveolin-1 expression, which in turn leads to stabilization of the insulin receptor thus enhancing insulin signaling (Trajkovski et al. 2011). In contrast, miR-103 expression has been found to be downregulated in the mouse model of genetic insulin resistance and obesity (ob/ob mice) (Xie et al. 2009), consistent with our observations using a diet-induced obesity model.
3.6.2 Endometriosis
Endometriosis is a condition affecting approximately 10–15 % of females whereby cells of the endometrial lining of the uterus relocate via retrograde menstruation and localize to other regions outside of the endometrium. Recent studies investigating the miRNA profiles of endometriosis and associated tissues have shown that some miRNAs are associated with the induction and persistence of the disease. Transcriptional profiling has previously been used to identify numerous candidate genes that may contribute to the etiology of the disease (Kao et al. 2003). The large number of endometrial mRNA being altered in endometriosis may be due to miRNA function influencing the transcriptome.
Among the multitude of miRNAs associated with endometriosis, several appear to be promising markers in both diagnosis and progression of the disease. Zhao et al. demonstrated that miR-20a is overexpressed in patients with ovarian endometriosis, being greatest in patients in advanced stages of the disease (Zhao et al. 2014). Using small RNA sequencing of both endometriotic lesions and adjacent tissues, Saare et al. (2014) demonstrated that miR-499a, miR-200a, miR-200b, miR-141, and miR-34c were in significantly greater abundance in endometriotic lesions compared to neighboring healthy tissues (Saare et al. 2014). Using microarray analysis, Ohlsson Teague et al. (2009) were able to identify differential miRNA expression differences between ectopic and eutopic endometrial samples of women with endometriosis (Ohlsson Teague et al. 2009). The authors identified 14 miRNAs with elevated abundance and eight with reduced abundance in ectopic endometrium in comparison to eutopic endometrial samples. The miRNAs observed to be dysregulated in ectopic endometrium were predicted to interact with mRNA known to impact critical biological processes such as cell migration, invasion, and cell proliferation which are all putative mechanisms contributing to the endometriotic tissue formation (Ohlsson Teague et al. 2009).
3.6.3 Polycystic Ovarian Syndrome
Polycystic ovarian syndrome is an anovulatory disorder that is characterized by the pathological development of ovarian cysts. The underlying mechanisms resulting in PCOS have to this point been elusive; thus, multiple groups have worked towards the identification of miRNA associated with PCOS (Sorensen et al. 2014). Using microarray and quantitative PCR, five (let-7i-3 pm, miR-5706, miR-4463, miR-3665, miR-638) miRNAs were found in greater abundance in circulation of women with PCOS compared to healthy controls, while four (miR-124-3p, miR-128, miR-29a-3p, let-7c) were less abundant in women with PCOS (Ding et al. 2014). Others have taken similar approaches in the comparison of global miRNA expression profiling in women with PCOS compared to healthy controls. Sang et al. (2013) utilized deep sequencing to characterize miRNA content in microvesicles isolated from the follicular fluid of women with PCOS and identified miR-132 and miR-320 as being suppressed in the follicular fluid compared to control patients (Sang et al. 2013). In addition, the authors demonstrate the ability of miR-132 and miR-320 stimulation to increase estradiol production in a human granulosa-like tumor cell line, while inhibition of these miRNA had a suppressive effect on estradiol production.
Not only do aberrations in serum and follicular miRNA populations appear to be associated with women afflicted with PCOS, in some cases, differences are also observed in the developing embryo. Blastocysts derived from women afflicted with PCOS appear to have suppressed abundance of let-7a, miR-19a, miR-19b, miR-24, miR-92, and miR-93 (McCallie et al. 2010), suggesting that compromised embryos resulting from abnormalities during oocyte development may be a mechanism by which PCOS compromises fertility.
3.6.4 Chemically Induced Infertility Mediated Through miRNA
Exposure to environmental or occupational chemicals can disrupt female reproductive function (Mattison and Schulman 1980). Additionally, the reproductive age and status of an individual alter the susceptibility and the outcome following exposure to a reproductive toxicant (Fig. 3.3). A number of studies have shown that exposure to ovarian toxicants can lead to oocyte depletion [reviewed in Bhattacharya and Keating (2012a) and Hoyer and Keating (2014)]. The stage of development at which the follicle is lost determines the reproductive impact; if large or antral follicles are depleted, temporary interruptions to reproductive function are observed since these follicles can be replaced by recruitment from the pool of primordial follicles (Hoyer and Keating 2014). Due to the irreplaceable nature of the ovarian reserve, chemicals that destroy oocytes contained in primordial follicles can lead to permanent infertility and premature ovarian failure (POF). Also, the level and duration of exposure to an environmental toxicant can influence the reproductive impact. Chronic, low-dose exposures, likely to be environmental in nature, are difficult to identify because their ovarian impact may go unrecognized for years. Ongoing selective damage of small preantral follicles may not initially raise concern until the onset of POF that will eventually result. Further, the age at which exposure occurs can impact the outcome. Prepubertal exposure may not cause the same extent of follicle loss as that postpubertal, due to the higher number of follicles present during childhood. However, damage to oocytes by chemical exposures in utero and/or during childhood presents a concern, which would not be detected until the reproductive years.
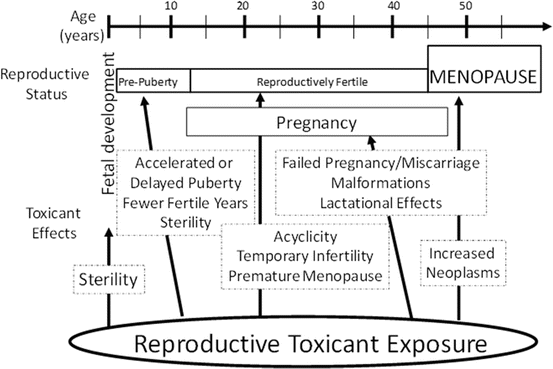

Fig. 3.3
Potential age-related effects of reproductive toxicants in females. The impact(s) of reproductive toxicants is partially dependent upon the reproductive status of the exposed individual. In most cases, direct ovarian toxicity can lead to premature ovarian failure and infertility (menopause). From Keating and Hoyer, 2009 with copyright permission
Ovotoxic chemicals can accelerate activation of primordial follicles from the ovarian reserve (Fernandez et al. 2008; Keating et al. 2009), leading to POF. Since this process is, at least partially, regulated by PI3K signaling, the involvement of miRNAs is plausible. It is known that miR-21 inhibits phosphatase and tensin homologue (PTEN), an antagonist of PI3K (Carletti et al. 2010; Ling et al. 2012). Also, increased abundance of miR-184 can interfere with AKT action, repressing PI3K action (Baley and Li 2012; Yu et al. 2008). Furthermore, miR-451 (Tian et al. 2011) and miR-7 (Fang et al. 2012) inhibit PI3K, while the miR-17–92 cluster (Ji et al. 2011) and miR-21 (Darido et al. 2011) activate PI3K.
It is important to note that the ovary has the capability to biotransform chemicals to more or less toxic metabolites, and these metabolic processes are highly active in ovarian tissues (Bhattacharya and Keating 2011, 2012b; Bhattacharya et al. 2012, 2013; Igawa et al. 2009; Keating et al. 2008a, b, 2010; Madden and Keating 2014) and contribute to the extent of ovotoxicity observed. We have demonstrated that many genes encoding ovarian chemical biotransformation enzymes are regulated by PI3K signaling (Bhattacharya and Keating 2012b; Bhattacharya et al. 2012), including aryl hydrocarbon receptor (AhR) and nuclear erythroid-related factor (Nrf2) (Bhattacharya and Keating 2012b), transcription factors that regulate xenobiotic metabolism. MiRNA-mediated regulation of Nrf2 (Eades et al. 2011; Yang et al. 2011) and Ahr (Huang et al. 2011) has recently been demonstrated. Thus, taken together, the potential for miRNA to have major roles in mediating chemical-induced ovotoxicity is being realized and is likely to be deciphered in greater detail in the coming years.
3.7 RNA-Binding Proteins Impact miRNA Function and Fertility
Despite their presence and activity in cells contributing to reproduction and development, the ability of ncRNAs to impact the cellular phenotype can be tempered by a variety of molecular regulators, and RNA-binding proteins, such as dead end homologue 1 (DND1), may be involved. The 3′ UTR of mRNA in close proximity to potential miRNA-binding sites is frequently the location of AU-rich elements (ARE) which contribute to mRNA stability (Chen and Shyu 1995; Fan et al. 1997). Since DND1 is capable of binding to regions where miRNA-mediated PTGR is conferred, it is thought that DND1 interaction with specific mRNA near miRNA-binding sites could potentially alter miRNA-induced PTGR (Kedde and Agami 2008; Kedde et al. 2007).
Germ cell viability for both male and female and early embryonic development requires DND1 function (Bhattacharya et al. 2007; Saga 2008). We have also demonstrated that DND1 abundance in the maturing pig oocyte and early embryo is greatest during the period of transcriptional quiescence following GVBD and prior ZGA (Yang et al. 2012b). The ability of DND1 to bind specific mRNA transcripts associated with pluripotency (POU5F1 (aka OCT4), SOX2, and LIN28) was demonstrated in the germinal vesicle stage oocyte (Yang et al. 2012b). Considering the numbers of miRNA present in the developing oocyte and the period of oocyte transcriptional quiescence following GVBD, it is possible that DND1 influences embryonic developmental competency by contributing to the mRNA and protein repertoire maintenance in the oocyte and early embryo. The biological roles of DND1 in addition to other RNA-binding proteins, such as fragile X mental retardation syndrome-related protein 1 (FXR1) which is also capable of mediating miRNA PTGR (Vasudevan and Steitz 2007; Vasudevan et al. 2007), will likely be continued areas of investigation in determining the biological ability of small RNA to influence reproductive function and fertility.
3.8 Conclusion
Computational and biochemical approaches have certainly improved the understanding of ncRNA regulation, and it is clear that ncRNA, in particular small RNA, impacts reproductive function. Advances in high-throughput sequencing technologies have rapidly amplified the discovery and characterization of multiple RNA classes in relation to specific biological functions of specific reproductive tissues and in relation to reproductive dysfunction and infertility. Together with knockout models deciphering the specific contributions of RNA classes in reproductive tissues, a more comprehensive understanding of small RNAs and their contribution to cellular function in reproductive tissues and fertility is being developed. Small RNAs, in particular endogenously produced miRNA, have distinct and critical biological roles contributing to fertility and reproductive success for both males and females. Additionally, RNA-binding proteins add biological complexity through their potential to also influence small RNA function in both a tissue-specific and target transcript-specific manner. As a result of the intricacies associated with small RNA biogenesis, the complexities of the interactions with specific transcripts, and to what degree those interactions result in PTGR based on what other contributing factors are present, bioinformatic predictions alone to characterize the role of small RNA in reproduction function are insufficient. Thus, there remains a dearth of information on how small RNA directly and indirectly influences fertility, and future studies are required to focus on determining the specific mechanistic function of particular small RNA in reproductive tissues.
References
Abd El Naby WS, Hagos TH, Hossain MM, Salilew-Wondim D, Gad AY, Rings F, Cinar MU, Tholen E, Looft C, Schellander K, Hoelker M, Tesfaye D (2013) Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 21:31–51PubMed
Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC (2005) Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA 102(50):18017–18022PubMedCentralPubMed
Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, Backes C (2013) Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril 99(5):1249–1255PubMed
Abu-Halima M, Hammadeh M, Backes C, Fischer U, Leidinger P, Lubbad AM, Keller A, Meese E (2014) Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril 102(4):989–997PubMed
Akamine EH, Marcal AC, Camporez JP, Hoshida MS, Caperuto LC, Bevilacqua E, Carvalho CR (2010) Obesity induced by high-fat diet promotes insulin resistance in the ovary. J Endocrinol 206(1):65–74PubMed
Amanai M, Brahmajosyula M, Perry AC (2006) A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod 75(6):877–884PubMed
Ambros V, Chen X (2007) The regulation of genes and genomes by small RNAs. Development 134:1635–1641PubMed
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207PubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree