39
RNA Enveloped Viruses
CHAPTER CONTENTS
ORTHOMYXOVIRUSES
INFLUENZA VIRUSES
Influenza viruses are important human pathogens because they cause both outbreaks of influenza that sicken and kill thousands of people each year as well as infrequent but devastating worldwide epidemics (pandemics).
Influenza viruses are the only members of the orthomyxovirus family. The orthomyxoviruses differ from the paramyxoviruses primarily in that the former have a segmented RNA genome (usually eight pieces), whereas the RNA genome of the latter consists of a single piece.1 The term myxo refers to the observation that these viruses interact with mucins (glycoproteins on the surface of cells).
In addition, the orthomyxoviruses are smaller (110 nm in diameter) than the paramyxoviruses (150 nm in diameter). See Table 39–1 for additional differences.
Table 39–2 shows a comparison of influenza A virus with several other viruses that infect the respiratory tract. Table 39–3 describes some of the important clinical features of influenza virus and compares them with the clinical features of the other medically important viruses in this chapter.
In 1997, an outbreak of human influenza (avian influenza, bird flu) caused by an H5N1 strain of influenza A virus began. This outbreak and subsequent outbreaks are described on page 308. In 2009, there was an outbreak of human influenza caused by an H1N1 influenza A virus of swine origin (swine-origin influenza virus, S-OIV). This outbreak and the subsequent pandemic are described on page 309. In 2013, an outbreak of influenza caused by an H7N9 strain of influenza virus occurred.
1. Human Influenza Virus
Disease
Influenza A virus causes worldwide epidemics (pandemics) of influenza, influenza B virus causes major outbreaks of influenza, and influenza C virus causes mild respiratory tract infections but does not cause outbreaks of influenza. Pandemics occur when a variant of influenza A virus that contains a new hemagglutinin against which people do not have preexisting antibodies is introduced into the human population.
The pandemics caused by influenza A virus occur infrequently (the last one was in 1968), but major outbreaks caused by this virus occur virtually every year in many countries. Each year, influenza is the most common cause of respiratory tract infections that result in physician visits and hospitalizations in the United States.
In the 1918 influenza pandemic, more Americans died than in World War I, World War II, the Korean War, and the Vietnam War combined. Influenza B virus does not cause pandemics, and the major outbreaks caused by this virus do not occur as often as those caused by influenza A virus. It is estimated that approximately 36,000 people die of influenza each year in the United States.
Important Properties
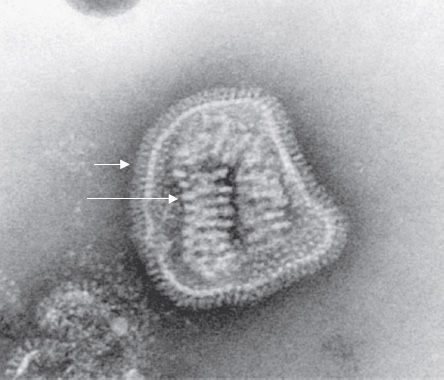
Influenza virus is composed of a segmented single-stranded RNA genome, a helical nucleocapsid, and an outer lipoprotein envelope (Figure 39–1). The virion contains an RNA-dependent RNA polymerase, which transcribes the negative-polarity genome into mRNA.
FIGURE 39–1 Influenza virus—electron micrograph. Long arrow points to the helical nucleocapsid of influenza virus. The nucleocapsid contains the segmented, negative-polarity genome RNA. Short arrow points to the spikes on the virion envelope. The spikes are the hemagglutinin and neuraminidase proteins. (Figure courtesy of Dr. Erskine Palmer and Dr. M. Martin, Public Health Image Library, Centers for Disease Control and Prevention.)
The envelope is covered with two different types of spikes, a hemagglutinin and a neuraminidase.2 Influenza A virus has 16 antigenically distinct types of hemagglutinin and 9 antigenically distinct types of neuraminidase. As discussed later, some of these types cause disease in humans, but most of the types typically cause disease in other animal species such as birds, horses, and pigs.
The function of the hemagglutinin is to bind to the cell surface receptor (neuraminic acid, sialic acid) to initiate infection of the cell. In the clinical laboratory, the hemagglutinin agglutinates red blood cells, which is the basis of a diagnostic test called the hemagglutination inhibition test. The hemagglutinin is also the target of neutralizing antibody (i.e., antibody against the hemagglutinin inhibits infection of the cell).
The neuraminidase cleaves neuraminic acid (sialic acid) to release progeny virus from the infected cell. The hemagglutinin functions at the beginning of infection, whereas the neuraminidase functions at the end. Neuraminidase also degrades the protective layer of mucus in the respiratory tract. This enhances the ability of the virus to gain access to the respiratory epithelial cells.
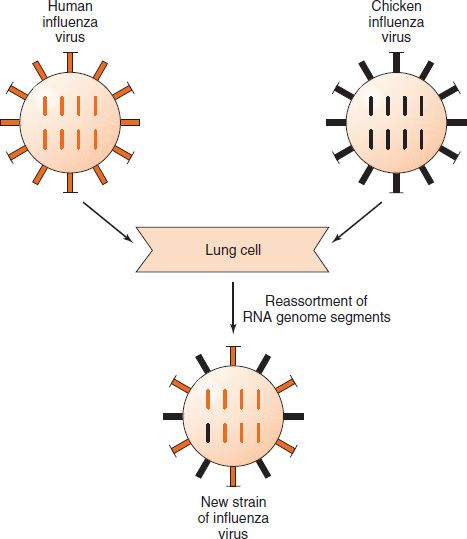
Influenza viruses, especially influenza A virus, show changes in the antigenicity of their hemagglutinin and neuraminidase proteins; this property contributes to their capacity to cause devastating worldwide epidemics (pandemics). There are two types of antigenic changes: (1) antigenic shift, which is a major change based on the reassortment of segments of the genome RNA; and (2) antigenic drift, which is a minor change based on mutations in the genome RNA. Note that in reassortment, entire segments of RNA are exchanged, each one of which codes for a single protein (e.g., the hemagglutinin) (Figure 39–2).
FIGURE 39–2 Antigenic shift in influenza virus. A human strain of influenza virus containing the gene encoding one antigenic type of hemagglutinin (colored orange) infects the same lung cell as a chicken strain of influenza virus containing the gene encoding a different antigenic type of hemagglutinin (colored black). Reassortment of the genome RNA segments that encode the hemagglutinin occurs, and a new strain of influenza virus is produced containing the chicken type of hemagglutinin (colored black).
Influenza A virus has two matrix proteins: The M1 matrix protein is located between the internal nucleoprotein and the envelope and provides structural integrity. The M2 matrix protein forms an ion channel between the interior of the virus and the external milieu. This ion channel plays an essential role in the uncoating of the virion after it enters the cell. It transports protons into the virion causing the disruption of the envelope, which frees the nucleocapsid containing the genome RNA, allowing it to migrate to the nucleus.
Influenza viruses have both group-specific and type-specific antigens.
(1) The internal ribonucleoprotein is the group-specific antigen that distinguishes influenza A, B, and C viruses.
(2) The hemagglutinin and the neuraminidase are the type-specific antigens located on the surface. Antibody against the hemagglutinin neutralizes the infectivity of the virus (and prevents disease), whereas antibody against the group-specific antigen (which is located internally) does not. Antibody against the neuraminidase does not neutralize infectivity but does reduce disease by decreasing the amount of virus released from the infected cell and thus reducing spread of the virus to adjacent cells.
An important determinant of the virulence of this virus is a nonstructural protein called NS-1 encoded by the genome RNA of influenza virus. NS-1 has several functions, but the one pertinent to virulence is its ability to inhibit the production of interferon mRNA. As a result, innate defenses are reduced and viral virulence is correspondingly enhanced.
Many species of animals (e.g., aquatic birds, chickens, swine, and horses) have their own influenza A viruses. These animal viruses are the source of the RNA segments that encode the antigenic shift variants that cause epidemics among humans. For example, if an avian and a human influenza A virus infect the same cell (e.g., in a farmer’s respiratory tract), reassortment could occur and a new variant of the human A virus, bearing the avian virus hemagglutinin, may appear (Figure 39–1).
There is evidence that aquatic birds (waterfowl) are a common source of these new genes and that the reassortment event leading to new human strains occurs in pigs. In other words, pigs may serve as the “mixing bowl” within which the human, avian, and swine viruses reassort. There are 16 types of hemagglutinin (H1 to H16) and 9 types of neuraminidase (N1 to N9) found in waterfowl. In humans, three types of hemagglutinin (H1, H2, and H3) and two types of neuraminidase (N1 and N2) predominate.
Because influenza B virus is only a human virus, there is no animal source of new RNA segments. Influenza B virus therefore does not undergo antigenic shifts. It does, however, undergo enough antigenic drift that the current strain must be included in the new version of the influenza vaccine produced each year. Influenza B virus has no antigens in common with influenza A virus.
A/Philippines/82 (H3N2) illustrates the nomenclature of influenza viruses. “A” refers to the group antigen. Next are the location and year the virus was isolated. H3N2 is the designation of the hemagglutinin (H) and neuraminidase (N) types. The H1N1 and H3N2 strains of influenza A virus are the most common at this time and are the strains included in the current vaccine. The H2N2 strain caused a pandemic in 1957.
Summary of Replicative Cycle
The virus adsorbs to the cell when the viral hemagglutinin interacts with sialic acid receptors on the cell surface. (The hemagglutinin on the virion surface is cleaved by extracellular proteases to generate a modified hemagglutinin that actually mediates attachment to the cell surface.) The virus then enters the cell in vesicles and uncoats within an endosome. Uncoating is facilitated by the low pH within the endosome. Protons pass through the ion channel formed by the M2 protein into the interior of the virion. This disrupts the virion envelope and frees the nucleocapsid to enter the cytoplasm and then migrate to the nucleus where the genome RNA is transcribed.
The virion RNA polymerase transcribes the eight genome segments into eight mRNAs in the nucleus. Synthesis of the eight mRNAs occurs in the nucleus because a methylated guanosine “cap” is required. The cap is obtained from cellular nuclear RNAs in a process called “cap snatching.” Most of the mRNAs move to the cytoplasm, where they are translated into viral proteins. Some of the viral mRNAs remain in the nucleus, where they serve as the template for the synthesis of the negative-strand RNA genomes for the progeny virions. Replication of the progeny genomes is performed by a different subunit of the viral RNA polymerase (acting as a replicase) from the subunit that functioned earlier as a transcriptase that synthesized the mRNAs. Two newly synthesized proteins, NP protein and matrix protein, bind to the progeny RNA genome in the nucleus, and that complex is transported to the cytoplasm.
The helical ribonucleoprotein assembles in the cytoplasm, matrix protein mediates the interaction of the nucleocapsid with the envelope, and the virion is released from the cell by budding from the outer cell membrane at the site where the hemagglutinin and neuraminidase are located. The neuraminidase releases the virus by cleaving neuraminic acid on the cell surface at the site of the budding progeny virions. Influenza virus, hepatitis delta virus, and retroviruses are the only RNA viruses that have an important stage of their replication take place in the nucleus.
Transmission & Epidemiology
The virus is transmitted by airborne respiratory droplets. The ability of influenza A virus to cause epidemics is dependent on antigenic changes in the hemagglutinin and neuraminidase. As mentioned previously, influenza A virus undergoes both major antigenic shifts as well as minor antigenic drifts. Antigenic shift variants appear infrequently, whereas drift variants appear virtually every year. The last major antigenic shift that caused a pandemic in humans was in 1968 when H3N2 emerged. Epidemics and pandemics (worldwide epidemics) occur when the antigenicity of the virus has changed sufficiently that the preexisting immunity of many people is no longer effective. The antigenicity of influenza B virus undergoes antigenic drift but not antigenic shift. The antigenic changes exhibited by influenza B virus are less dramatic and less frequent than those of influenza A virus.
Influenza occurs primarily in the winter months of December to February in the northern hemisphere, when influenza and bacterial pneumonia secondary to influenza cause a significant number of deaths, especially in older people. The morbidity of influenza in children younger than 2 years is also very high, second only to the morbidity in the elderly. In the southern hemisphere (e.g., in Australia and New Zealand), influenza occurs primarily in the winter months of June through August. In the tropics, influenza occurs year round with little seasonal variation.
Pathogenesis & Immunity
After the virus has been inhaled, the neuraminidase degrades the protective mucus layer, allowing the virus to gain access to the cells of the upper and lower respiratory tract. The infection is limited primarily to this area because the proteases that cleave the hemagglutinin are located in the respiratory tract. Despite systemic symptoms, viremia rarely occurs. The systemic symptoms, such as severe myalgias, are due to cytokines circulating in the blood. There is necrosis of the superficial layers of the respiratory epithelium. Influenza virus pneumonia, which can complicate influenza, is interstitial in location.
Immunity depends mainly on secretory IgA in the respiratory tract. IgG is also produced but is less protective. Cytotoxic T cells also play a protective role.
Clinical Findings
After an incubation period of 24 to 48 hours, fever, myalgias, headache, sore throat, and cough develop suddenly. Severe myalgias (muscle pains) coupled with respiratory tract symptoms are typical of influenza. Vomiting and diarrhea are rare. The symptoms usually resolve spontaneously in 4 to 7 days, but influenzal or bacterial pneumonia may complicate the course. One of the well-known complications of influenza is pneumonia caused by Staphylococcus aureus.
Reye’s syndrome, characterized by encephalopathy and liver degeneration, is a rare, life-threatening complication in children following some viral infections, particularly influenza B and chickenpox. Aspirin given to reduce fever in viral infections has been implicated in the pathogenesis of Reye’s syndrome.
Laboratory Diagnosis
Although most diagnoses of influenza are made on clinical grounds, laboratory tests are available. The test most commonly used is an enzyme-linked immunosorbent assay (ELISA) for viral antigen in respiratory secretions such as nasal or throat washings, nasal or throat swabs, or sputum. Several rapid ELISA tests suitable for a physician’s office laboratory are available. Two tests (FLU OIA and QuickVue Influenza Test) are based on detection of viral antigen using monoclonal antibodies, and a third test (ZSTATFLU) is based on detection of viral neuraminidase using a substrate of the enzyme that changes color when cleaved by neuraminidase. The rationale for using the rapid tests is that treatment with the neuraminidase inhibitors should be instituted within 48 hours of the onset of symptoms. Other tests such as direct fluorescent antibody and polymerase chain reaction (PCR) are also used.
Influenza can also be diagnosed by the detection of antibodies in the patient’s serum. A rise in antibody titer of at least four-fold in paired serum samples taken early in the illness and 10 days later is sufficient for diagnosis. Either the hemagglutination inhibition or complement fixation (CF) test can be used to assay the antibody titer. Because the second sample is taken 10 days later, this approach is used to make a retrospective diagnosis, often for epidemiologic purposes.
Treatment
Oseltamivir (Tamiflu) and zanamivir (Relenza) are used for both the treatment and prevention of influenza. They are members of a class of drugs called neuraminidase inhibitors, which act by inhibiting the release of virus from infected cells. This limits the extent of the infection by reducing the spread of virus from one cell to another. These drugs are effective against both influenza A and B viruses, in contrast to amantadine, which is effective only against influenza A viruses.
In 2009, most isolates of H1N1 influenza virus were resistant to Tamiflu. Most isolates of the novel H1N1 (swine) influenza virus (see later) are susceptible; however, Tamiflu-resistant mutants have emerged. H3N2 strains were still susceptible to Tamiflu. Both H1N1 and H3N2 strains remained susceptible to Relenza. In general, H5N1 strains of influenza virus that cause avian influenza (see later) are sensitive to Tamiflu and Relenza but resistant to amantadine and rimantadine.
Tamiflu is taken orally, whereas Relenza is delivered by inhalant directly into the respiratory tract. Clinical studies showed they reduce the duration of symptoms by 1 to 2 days. They also reduce the amount of virus produced and therefore reduce the chance of spread to others. To be effective as treatment, these drugs must be given within 48 hours of the onset of symptoms.
Amantadine (Symmetrel) is approved for both the treatment and prevention of influenza A. However, 90% of the H3N2 strains in the United States are resistant to amantadine (and rimantadine, see later), and so these drugs are no longer recommended. These drugs block the M2 ion channel, thereby inhibiting uncoating. Resistance is caused primarily by point mutations in the gene for the M2 protein.
Note that amantadine is effective only against influenza A, not against influenza B. Rimantadine (Flumadine), a derivative of amantadine, can also be used for treatment and prevention of influenza A and has fewer side effects than amantadine. It should be emphasized that the vaccine is preferred over these drugs in the prevention of influenza.
Prevention
The main mode of prevention is the vaccine, which contains both influenza A and B viruses. Prior to 2013, the vaccine was trivalent and contained recent isolates of two A strains (H1N1 and H3N2) and one B strain. In 2013, quadrivalent vaccines containing two A strains and two B strains became available. The vaccine is usually reformulated each year to contain the current antigenic strains.
There are two main types of influenza vaccines available in the United States, a killed vaccine and a live, attenuated vaccine. The vaccine that has been used for many years is a killed vaccine containing purified protein subunits of the virus (hemagglutinin and neuraminidase). The virus is inactivated with formaldehyde and then treated with a lipid solvent that disaggregates the virions. Note that the hemagglutinin is the most important antigen because it elicits neutralizing antibody. This vaccine is typically administered intramuscularly. In 2011, a killed influenza vaccine that can be administered intradermally became available.
The other vaccine is a live, attenuated vaccine containing temperature-sensitive mutants of influenza A and B viruses. These temperature-sensitive mutants can replicate in the cooler (33°C) nasal mucosa where they induce IgA, but not in the warmer (37°C) lower respiratory tract. The live virus in the vaccine therefore immunizes but does not cause disease. This vaccine is administered by spraying into the nose (“nasal mist”). The live vaccine should not be given to pregnant women or to immunocompromised individuals.
Most of the vaccines described above are made in chicken eggs, and anyone who has a significant allergy to egg proteins (e.g., anaphylaxis) should not receive these vaccines. However, in 2012, the U.S. Food and Drug Administration (FDA) approved a killed influenza vaccine (Flucelvax) made in calf kidney cell culture. This vaccine has two advantages: It can be given to those with egg allergy, and it has a short turnaround time, so the latest drift mutant can be used.
Also in 2012, the FDA approved a recombinant vaccine (Flublok) made by inserting the gene encoding the viral hemagglutinin into an insect virus (baculovirus) that is propagated in insect cell culture. This vaccine contains purified hemagglutinin as the immunogen. This vaccine can also be given to those with egg allergy.
Note that the killed vaccine is not a good immunogen, because little IgA is made and the titer of IgG is relatively low. Protection lasts only 6 months. Yearly boosters are recommended and should be given shortly before the flu season (e.g., in October). These boosters also provide an opportunity to immunize against the latest antigenic changes. The vaccine should be given to all persons 6 months and older who do not have a contraindication to receive the vaccine. It is especially important that people with chronic diseases, particularly respiratory and cardiovascular conditions, receive the vaccine. It should also be given to health care personnel who are likely to transmit the virus to those at high risk.
One side effect of the influenza vaccine used in the 1970s containing the swine influenza strain that caused influenza in humans was an increased risk of Guillain-Barré syndrome, which is characterized by an ascending paralysis. Analysis of the side effects of the influenza vaccines in use during the last 10 years has shown no increased risk of Guillain-Barré syndrome.
In addition to the vaccine, influenza can be prevented by using oseltamivir, which is described in the treatment section earlier. Oseltamivir is particularly useful in elderly people who have not been immunized and who may have been exposed. Note that this drug should not be thought of as a substitute for the vaccine. Immunization is the most reliable mode of prevention.
2. Avian Influenza Virus Infection in Humans
H5N1 Influenza Virus
In 1997, the H5N1 strain of influenza A virus that causes avian influenza, primarily in chickens, caused an aggressive form of human influenza with high mortality in Hong Kong. In the winter of 2003–2004, an outbreak of avian influenza caused by H5N1 strain killed thousands of chickens in several Asian countries. Millions of chickens were killed in an effort to stop the spread of the disease. Four hundred eight human cases of H5N1 influenza occurred between 2003 and February 2009, resulting in 254 deaths (a mortality rate of 62%). Note that these 408 people were infected directly from chickens. Both the respiratory secretions and the chicken guano contain infectious virus.
The spread of the H5N1 strain from person to person occurs rarely but remains a major concern because it could increase dramatically if reassortment with the human-adapted strains occurs. In 2005, the H5N1 virus spread from Asia to Siberia and into eastern Europe, where it killed thousands of birds but has not caused human disease. As of this writing (December 2009), there have been no cases of human influenza caused by an H5N1 virus in the United States. However, there have been two cases of human influenza caused by an H7N2 strain of avian influenza virus.
The ability of the H5N1 strain to infect chickens (and other birds) more effectively than humans is due to the presence of a certain type of viral receptor throughout the mucosa of the chicken respiratory tract. In contrast, humans have this type of receptor only in the alveoli, not in the upper respiratory tract. This explains why humans are rarely infected with the H5N1 strain. However, when the exposure is intense, the virus is able to reach the alveoli and causes severe pneumonia.
The virulence of the H5N1 strain is significantly greater than the H1N1 and H3N2 strains that have been causing disease in humans for many years. This is attributed to two features of the H5N1 strain, namely, relative resistance to interferon and increased induction of cytokines, especially tumor necrosis factor. The increase in cytokines is thought to mediate the pathogenesis of the pneumonia and acute respiratory distress syndrome (ARDS) seen in H5N1 infection.
The H5N1 strain is sensitive to the neuraminidase inhibitors, oseltamivir (Tamiflu) and zanamivir (Relenza), but not to amantadine and rimantadine. Tamiflu is the drug of choice for both treatment and prevention. There is no human vaccine available against the H5N1 strain, but there is one available for use in avian species. In 2008, the FDA approved an inactivated vaccine against H5N1 influenza virus, but as of this writing, it is not available to the public. The vaccine is being stockpiled in the National Emergency Reserve.
H7N9 Influenza Virus
In 2013, an outbreak of influenza caused by an H7N9 strain of influenza virus occurred. Prior to this time, the H7N9 strain affected only birds, especially chickens. As of July 2013, 133 people have been diagnosed with influenza caused by this virus, 43 of whom have died (32% mortality rate). Cases have been limited to China and Taiwan. There has been no sustained person-to-person spread.
All of the genes of this virus are of avian origin. It acquired its H7 gene from ducks and its N9 gene from wild birds, and all the other genes are from an influenza strain that infects bramblings, a bird common in Asia and Europe. This H7N9 strain is susceptible to the neuraminidase inhibitors, oseltamivir and zanamivir. There is no vaccine.
3. Swine Influenza Virus Infection in Humans
In April 2009, a novel swine origin strain of influenza A (H1N1) virus (swine-origin influenza virus, S-OIV) caused an outbreak of human influenza, which appeared first in Mexico, then in the United States, followed by spread to 208 countries by December 2009. The Centers for Disease Control and Prevention (CDC) uses the name “novel influenza A (H1N1)” for this virus.
As of December 2009, millions of cases have occurred worldwide. There have been so many cases that most countries have stopped documenting the number of cases. Worldwide there have been 9596 deaths, of which 1445 have occurred in the United States. On June 11, 2009, the World Health Organization (WHO) declared a level 6 pandemic (the highest level alert). By August 2010, the number of cases had declined significantly and the pandemic warning was rescinded. As of this writing in November 2013, the number of cases in the United States and worldwide has significantly declined.
The disease affected primarily young people (60% of cases were 18 years old or younger). Symptoms were in general mild, with the few fatalities occurring in medically compromised patients. There was no outbreak of swine influenza in pigs prior to this human outbreak. Eating pork does not transmit the virus.
S-OIV is a quadruple reassortant: The hemagglutinin, nucleoprotein, and nonstructural protein genes are of North American swine origin, the neuraminidase and matrix protein genes are of Eurasian swine origin, the genes that encode two subunits of the polymerase are of North American avian origin, and the gene that encodes the third subunit of the polymerase is of human H3N2 origin.
A triple reassortant strain circulated in North American swine for several years prior to 2009 but caused human influenza only rarely. In the triple reassortant strain, all five of the genes that are not polymerase genes are of North American swine origin and the polymerase genes have the same origin as the quadruple reassortant. This strain does not have genes of Eurasian swine origin.
The key point is that most people worldwide do not have protective antibodies against the swine hemagglutinin of S-OIV even though they may have antibodies against the seasonal strain of H1N1 virus acquired either by immunization or by exposure to the virus itself. Note also that S-OIV spreads readily from human to human in contrast to the avian H5N1 strain that does not.
A PCR test for the diagnosis of S-OIV infection is available. S-OIV is sensitive to oseltamivir and zanamivir but resistant to amantadine and rimantadine. Both an inactivated and a live, attenuated vaccine against S-OIV became widely available in November 2009.
PARAMYXOVIRUSES
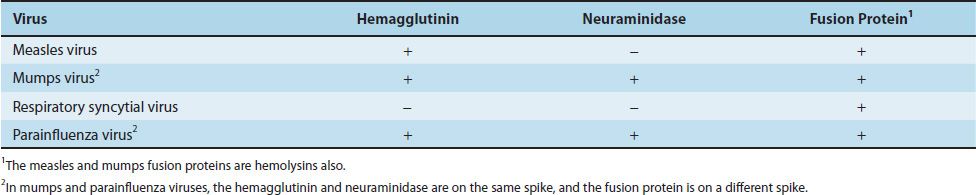
The paramyxovirus family contains four important human pathogens: measles virus, mumps virus, respiratory syncytial virus, and parainfluenza viruses. They differ from orthomyxoviruses in that their genomes are not segmented, they have a larger diameter, and their surface spikes are different (Table 39–1).
Paramyxoviruses are composed of one piece of single-stranded RNA, a helical nucleocapsid, and an outer lipoprotein envelope. The virion contains an RNA-dependent RNA polymerase, which transcribes the negative-polarity genome into mRNA. The genome is therefore not infectious. The envelope is covered with spikes, which contain hemagglutinin, neuraminidase, or a fusion protein that causes cell fusion and, in some cases, hemolysis (Table 39–4).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree