13.1 INTRODUCTION TO REOVIRUSES
Icosahedral viruses with segmented genomes of dsRNA, isolated from the respiratory tracts and enteric tracts of humans and animals, and with which no disease could be associated (orphan), became known as reoviruses. Similar viruses have been found in mammals, birds, fish, invertebrates (including insects), plants, and fungi. Many of these viruses are causative agents of disease, but the original name has been preserved in the family name Reoviridae, and is found in the names of some genera (Table 13.1).
Table 13.1 Some of the genera in the family Reoviridae
| Genus | Hosts | Number of dsRNA segments |
| Cypovirus | Insects | 10 |
| Dinovernavirus | Insects | 9 |
| Orbivirus | Mammals, birds, invertebrates | 10 |
| Orthoreovirus | Mammals, birds | 10 |
| Phytoreovirus | Plants, insects | 12 |
| Rotavirus | Mammals, birds | 11 |
The original reoviruses are incorporated into the genus Orthoreovirus. The avian viruses are important disease agents, but most orthoreovirus infections of mammals are asymptomatic. The majority of humans become infected with orthoreoviruses early in life and have specific serum antibodies by early adulthood.
It is interesting to note that most of the plant-infecting reoviruses are transmitted between plants by insect vectors (Chapter 4). The viruses replicate in both the plant and the insect, generally causing disease in the plant, but little or no harm to the infected insect.
The main focus of this chapter is on the rotaviruses, which have been the subjects of intensive study because they are amongst the most important agents of gastroenteritis in humans and animals. Each year rotaviruses are responsible for the deaths of over half a million children under 5 years old.
13.2 ROTAVIRUS VIRION
Rotaviruses were first described in 1963, when they were observed during electron microscopy of fecal samples from monkeys and mice. Spherical virions about 75 nm in diameter, with structures resembling the spokes of a wheel, were described (Figure 13.1), so the viruses were named after the Latin word rota (wheel). Similar viruses were observed 10 years later during electron microscopy of fecal samples from children with diarrhea.
Figure 13.1 The rotavirus virion.
Source: (b) Chen et al. (2009) Proceedings of the National Academy of the USA, 106, 10644. Reproduced by permission of the National Academy of the USA and the authors. (c) by permission of Professor M. Stewart McNulty and The Agri-Food and Biosciences Institute.
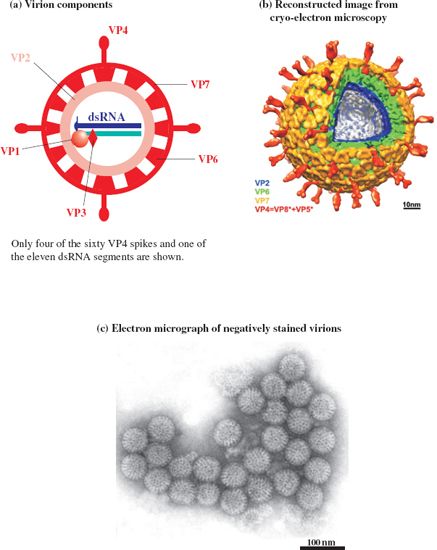
The virion, which has icosahedral symmetry, is also known as a triple-layered particle as the capsid has three layers, each constructed from a distinct virus protein (VP). The inner and middle layers, constructed from VP2 and VP6 respectively, are perforated by channels. The middle layer contains the “spokes” of the “wheel” and is the major component of the virion. The outer layer is constructed from VP7, which is glycosylated. It is unusual to find a glycoprotein in a naked virion, but VP7 is associated with a membrane within the cell before it is incorporated into the virion (Section 13.3.3). Three other protein species are found in the virion: VP1 and VP3 in the core, and VP4, which forms 60 spikes at the surface.
The proteins are numbered in order of their sizes. The three largest proteins are found towards the center of the virion; within the inner capsid layer (VP2), associated with the genome, are 12 copies each of VP1 and VP3. These two proteins are enzymes; VP1 is the RNA-dependent RNA polymerase and VP3 has guanylyl transferase and methyl transferase activities.
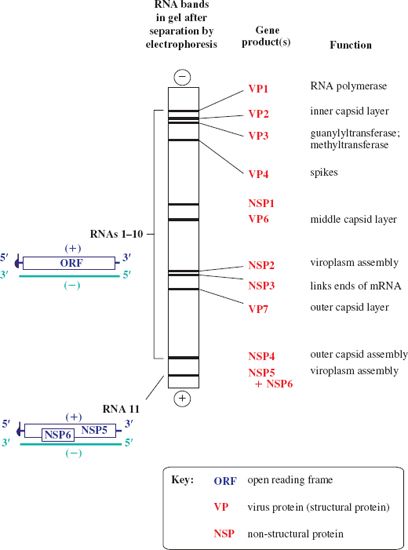
The 11 dsRNA segments that make up the rotavirus genome can be separated according to size by electrophoresis in a sodium dodecyl sulfate–polyacrylamide gel (Figure 13.2). Each RNA segment encodes one protein, with the exception of one segment, which encodes two proteins. Hence 12 proteins are encoded: six structural proteins (VP) and six non-structural proteins (NSP). In the diagrams the RNA strands are color-coded to distinguish the plus-strand (the coding strand) from the minus-strand.
Figure 13.2 The rotavirus genome and gene products. The plus-strand of each of the 11 RNA segments is capped. There is one ORF in each segment, except for the smallest (RNA 11), in which there are two ORFs in different reading frames. NSP5 and NSP6 therefore do not share a common sequence. The diagram shows a typical pattern of rotavirus RNA segments in a gel after electrophoretic separation, and the protein(s) encoded by each segment.
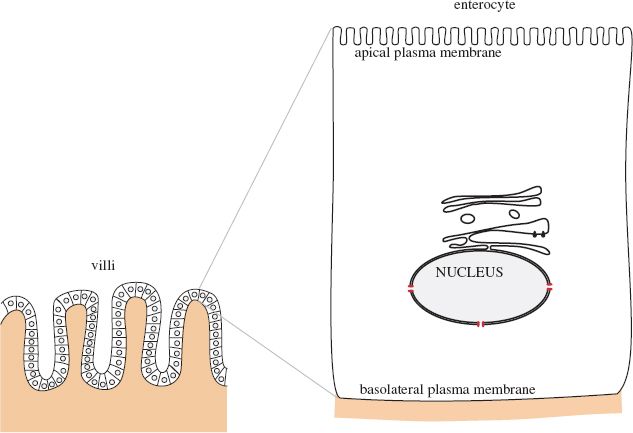
13.3 ROTAVIRUS REPLICATION
Rotaviruses infect cells called enterocytes at the ends of the villi (finger-like extensions) in the small intestine (Figure 13.3).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree