16.1 INTRODUCTION TO INFLUENZA VIRUSES
We are devoting a chapter to the influenza viruses because they are responsible for a huge disease burden and for hundreds of thousands of deaths each year. Furthermore, they are very interesting viruses because their replication cycle differs in a number of respects from that of most minus-strand RNA viruses. In particular, they replicate their RNA in the nucleus, whereas most other minus-strand viruses (Chapter 15) carry out all stages of their replication cycle in the cytoplasm.
Influenza viruses are classified in the family Orthomyxoviridae. There are three genera (Influenza-virus A, B, and C); the most important of these, and hence the most studied, are the influenza A viruses.
16.2 THE INFLUENZA VIRION
Influenza viruses are enveloped viruses with segmented genomes of ssRNA. The virions are spherical or elongated (Figure 16.1(a)), with diameters 80–120 nm. Isolates that have been passaged many times in the laboratory usually have spherical virions, whereas those that have been passaged only a few times tend to be filamentous.
Figure 16.1 Influenza A virus structure. (a) Cryo-electron micrograph of spherical and elongated virions. (b) The virion components. The envelope contains three integral membrane proteins: hemagglutinin, neuraminidase, and M2 (membrane 2). Underlying the envelope is a layer of M1 (membrane 1) protein, within which is a nuclear export protein and the genome (eight segments of ssRNA coated with nucleoprotein and associated with three polymerase proteins).
Source: (a) Moulès et al. (2011) Virology, 414, 51. Reproduced by permission of Elsevier Limited and the authors.
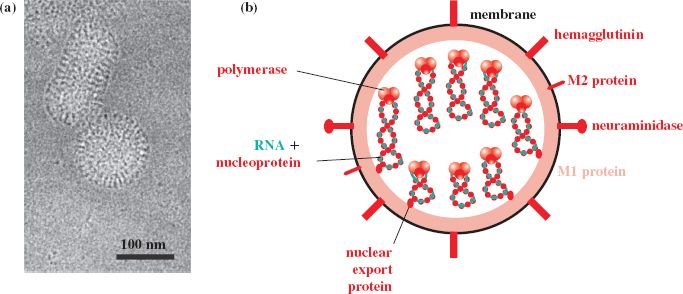
Influenza A and B virions each contain eight segments of ssRNA, while the influenza C virion contains seven segments. There are nine species of virus protein in the influenza A virion, the name of each protein describing a role or property of that protein (Figure 16.1(b)).
Three of the protein species are trans-membrane proteins. In decreasing order of abundance they are:
- hemagglutinin (can agglutinate red blood cells of various species);
- neuraminidase (an enzyme, the substrate for which is neuraminic acid);
- M2 (M for membrane; four M2 monomers form an ion channel).
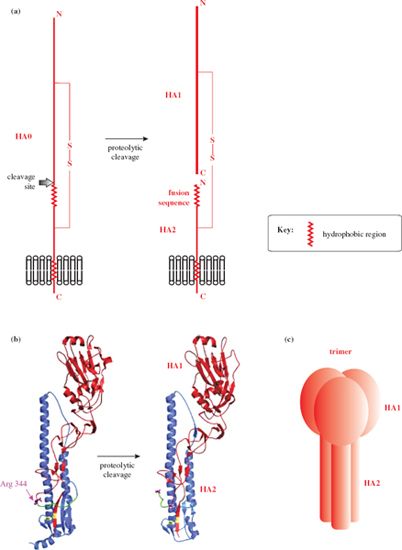
All three trans-membrane proteins have undergone post-translational modifications: for example, the hemagglutinin and neuraminidase are glycosylated, and the hemagglutinin has been cleaved into two subunits, HA1 and HA2, linked by a disulfide bond (Figure 16.2). HA1 forms a “head” which contains the attachment site for the host cell; HA2 forms a “stem” and is associated with the lipid membrane. Each hemagglutinin spike is a trimer (Figure 16.2 (c)), while each neuraminidase spike is a tetramer. Both the hemagglutinin and the neuraminidase occur as a variety of subtypes, and there is antigenic variation within each subtype. The result is that there are many strains of influenza A virus.
Figure 16.2 Influenza A virus hemagglutinin. (a) The main features of the hemagglutinin. The disulfide-linked subunits (HA1 and HA2) are formed by cleavage of a precursor protein (HA0). (b) Specific features of the 1918 influenza hemagglutinin. The colors in this diagram do not conform to the general color scheme of this book: HA1 (red), HA2 (blue), disulfide bond (yellow), fusion sequence (green), cleavage site of HA0 (Arg 344), C-terminus of HA1, and N-terminus of HA2 (purple). (c) Each hemagglutinin spike consists of a trimer of H1-H2.
Source: (b) Bertram et al. (2010) Reviews in Medical Virology, 20, 298. Reproduced by permission of John Wiley & Sons Ltd.
M1, another “membrane” protein, coats the interior surface of the envelope, and within this layer are the genome segments and a nuclear export protein. Each RNA segment is in a ribonucleoprotein (RNP) complex, with the RNA wrapped around nucleoprotein molecules. The 5′ and 3′ ends of each RNA segment are in close proximity and are associated with the RNA polymerase, which is a complex of three proteins. One of the polymerase proteins is acidic (PA: polymerase acidic), while the other two are basic (PB1 and PB2). The sizes of the RNP complexes vary with the lengths of the RNA molecules within them. Cell proteins, such as tubulin and cyclophilin A, have been reported as components of influenza virus particles.
16.3 INFLUENZA A VIRUS REPLICATION
Tissues commonly infected by influenza viruses are respiratory epithelium in humans and other primates, and intestinal epithelium in birds.
16.3.1 Attachment and entry
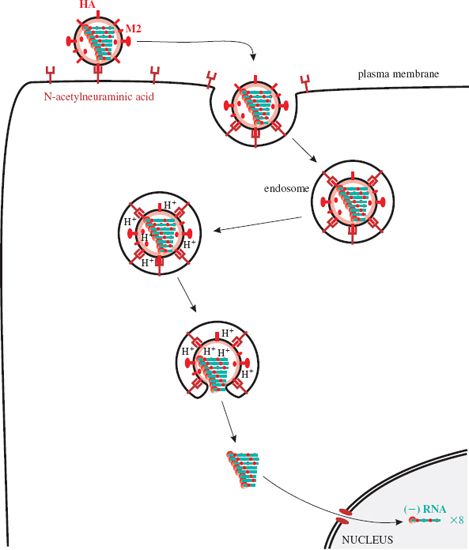
The virion attaches via a site on HA1 to the cell receptor, which is N-acetylneuraminic acid (neuraminic acid is a nine-carbon monosaccharide). The virion is endocytosed (Figure 16.3) and in the endosome it experiences a fall in pH, which causes a conformational change in the M2 protein ion channels. Hydrogen ions are pumped into the virion, and the drop in pH causes the RNP complexes to dissociate from M1 protein. As the pH in the endosome falls further the hemagglutinin molecules change shape, exposing fusion sequences in their HA2 subunits. The virion membrane fuses with the endosome membrane, releasing the RNP complexes, which are transported to the nucleus where virus RNA synthesis takes place.
Figure 16.3 Attachment and entry of influenza A virus. After endocytosis the endosome pH falls, changing the shape of M2 and HA molecules. M2 channels pump H+ ions into the virion, releasing the RNP complexes from M1 protein; HA fuses the membranes of the virion and the endosome. The RNP complexes are released and transported to the nucleus.
16.3.2 RNA replication
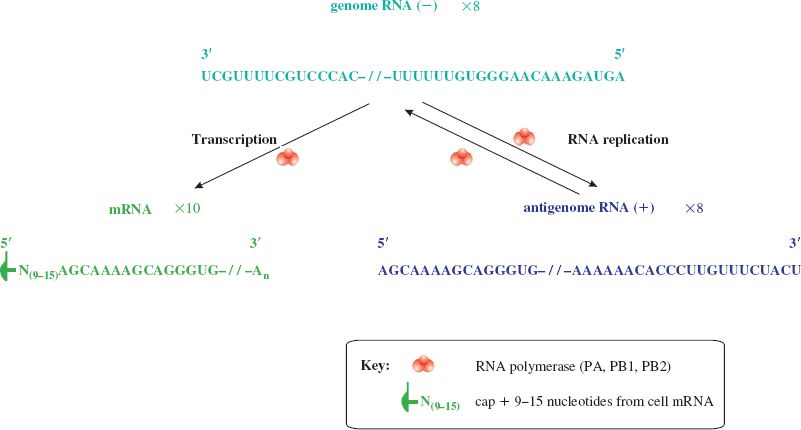
In the nucleus the polymerase (PA, PB1, PB2) associated with each RNP complex transcribes mRNA from the minus-strand genome RNA (Figure 16.4). RNA primers are required for this and these consist of short capped RNAs cut from the 5′ ends of cell mRNAs by an endonuclease activity in the polymerase complex. A virus non-structural protein (NS1) is synthesized and is transported to the nucleus where it inhibits the processing and export of cell mRNAs from the nucleus, making them available for cap-snatching.
Figure 16.4 Influenza virus transcription and genome replication. The nucleotide sequences shown are typical ends of influenza A virus RNAs. mRNAs (ten species) and antigenome RNAs are copied from the eight genome RNAs by the viral polymerase. The sequence UUUUUU towards the 5′ end of the (−) RNA is part of the polyadenylation signal; the sequence after this is not transcribed to mRNA. The antigenome RNAs act as templates for the synthesis of more (−) RNA.
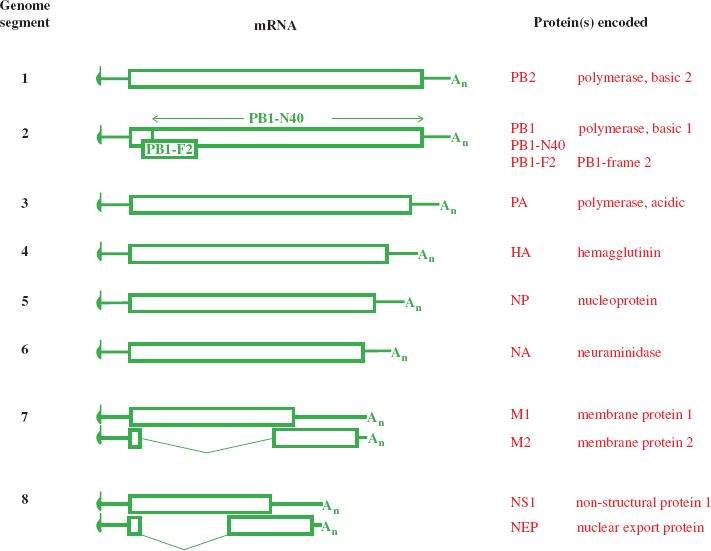
Ten mRNA species are derived from the eight genome segments, two mRNA species being produced from each of the two smallest segments (Figure 16.5). A proportion of the M1 transcripts are spliced to form the mRNAs for the M2 protein, and approximately 10% of the NS1 transcripts are spliced to form the mRNAs for the nuclear export protein (NEP).
Figure 16.5 Influenza A virus transcripts and the proteins they encode. Codon 40 of the PB1 ORF is the start codon for PB1-N40. The PB1-F2 ORF is within the PB1 ORF, in the +1 reading frame. M2 and NEP are encoded by spliced mRNAs.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


