Polyomaviruses
James A. DeCaprio
Michael J. Imperiale
Eugene O. Major
History
Polyomaviruses are found ubiquitously in a broad range of avian and mammalian species. Polyomaviruses can cause severe illness and death on an epidemic scale in birds. In contrast, polyomaviruses typically cause lifelong and asymptomatic infections in healthy humans but severe illness in immunocompromised patients. Recognition of the ubiquitous presence of polyomaviruses in their natural hosts as well as their disease-causing ability has prompted a variety of research efforts.
Polyomaviruses are composed of nonenveloped capsids with a simple, double-stranded DNA (dsDNA) genome of approximately 5,000 base pairs containing a single origin of replication and a bidirectional promoter that drives expression of messenger RNA (mRNA) transcripts encoding five to nine proteins. The small size of the polyomavirus genome, comparable to a simple plasmid, and its limited number of genetic elements have enabled research that continues to be at the forefront of biology including DNA replication, gene expression, signal transduction, and oncogenesis.
The history of polyomavirus research began in the 1950s when Ludwig Gross103 noted that when he passaged a mouse leukemia virus in mice, the recipients occasionally developed salivary and parotid gland tumors rather than leukemia. He isolated this specific activity and demonstrated that the parotid agent differed from murine leukemia virus (MLV) in its sedimentation, filtration, and heat stability properties. Stewart et al247 observed the formation of multiple tumor types in newborn mice inoculated with this agent and coined the name polyomavirus, derived from the Greek word poly, meaning “many,” and oma, denoting “tumor.” The mouse polyomavirus is often called simply “polyomavirus” but will be referred to as MPyV in this chapter, consistent with the most recent taxonomic classification (Table 53.1).128 Initially discovered at the same time as MPyV, murine pneumotropic virus (MPtV), also known as Kilham virus or K virus, causes severe interstitial pneumonia in newborn mice.101,144
The next member of the family to be isolated was simian virus 40 (SV40) by Sweet and Hilleman256 in 1960. They were screening samples from poliovirus vaccine lots produced in rhesus monkey kidney cells for the presence of contaminating viruses. SV40 was the 40th virus isolated in this screen and caused cytopathic or vacuolating effects in African green monkey kidney cells but not in the rhesus monkey cells used for vaccine production. It soon became clear that the early production batches of poliovirus vaccine were contaminated with SV40. Although the Salk poliovirus vaccine was inactivated by formalin treatment, SV40 was relatively resistant to this treatment and survived. The presence of SV40 in the vaccine quickly became a public health concern when the oncogenic potential of purified virus was demonstrated in newborn hamsters.
Since that time, many polyomaviruses have been isolated from a variety of mammalian and bird species. The first two human polyomaviruses, JC (JCPyV) and BK (BKPyV), were isolated in 1971 from immunocompromised patients. JCPyV was isolated by the transfer of brain tissue from a patient with the demyelinating disease progressive multifocal leukoencephalopathy (PML) into cultures of human fetal brain tissue.199 BKPyV was isolated from the urine of a renal
transplant patient after inoculation into African green monkey kidney cells.89
transplant patient after inoculation into African green monkey kidney cells.89
Table 53.1 Taxonomic Classification of Polyomaviruses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The advent of advanced molecular biology techniques including polymerase chain reaction (PCR), rolling circle amplification, and deep DNA sequencing led to the identification of seven additional human polyomaviruses. WUPyV and KIPyV were cloned from respiratory secretions of young children by investigators at Washington University and the Karolinska Institute, respectively. The discovery of Merkel cell polyomavirus (MCPyV) reflected recognition that the incidence of Merkel cell carcinoma, a rare form of skin cancer, was more frequent in severely immunocompromised patients, suggesting an infectious cause.71 MCPyV was recovered from Merkel cell carcinomas by digital transcriptome subtraction, a method that used high-throughput sequencing of cellular transcripts to identify sequences that did not match the human genome but were distantly related to BKPyV.78 Two additional polyomaviruses, HPyV6 and HPyV7, were cloned from the skin or hair follicles of healthy adults using a technique called rolling circle amplification that takes advantage of the small circular nature of the polyomavirus dsDNA genome.226 Using the same technique, trichodysplasia spinulosa–associated polyomavirus (TSPyV) was found in a patient with a rare skin disease named trichodysplasia spinulosa.268 HPyV9 was identified in the serum of a renal transplant patient and on the skin of a patient with Merkel cell carcinoma.222,227 Undoubtedly, additional human polyomaviruses will be discovered. Although identification of novel polyomaviruses by DNA sequencing has become easier, isolation of virion particles remains a technical challenge.
Primate polyomaviruses from Old World monkeys include the simian agent 12 (SA12), isolated from a South African vervet monkey kidney culture in 1963, and B-lymphotropic polyomavirus (LPyV), isolated from an African green monkey lymphoblast cell line, as well as polyomaviruses directly isolated from animals including Bornean (OraPyV1) and Sumatran orangutan polyomavirus (OraPyV2), gorilla polyomavirus (GggPyV), and chimpanzee polyomavirus (CHPyV).160 The first New World monkey polyomavirus was isolated from squirrel monkey (SqPyV2).272
In addition to MPyV and MtPyV, a number of nonprimate mammalian polyomaviruses have been identified. Hamster polyomavirus (HaPyV) was discovered in a spontaneously occurring hair follicle epithelioma from a Syrian hamster. This is an interesting member of the polyomavirus family because the behavior of the virus in the tumors most closely resembles that of the papillomaviruses in that viral particles can be found in the highly differentiated layers of the tissue. However, analysis of the HaPyV DNA sequence and its genome organization revealed that it is indeed a polyomavirus and most closely related to MPyV.61 Rabbit kidney vacuolating virus (RKV) was originally isolated from a rabbit papilloma but was shown to be involved in subclinical infections in rabbits. Bovine polyomavirus (BPyV) contains the smallest of the polyomavirus genomes with fewer than 4,700 base pairs. Sea lion polyomavirus (SLPyV) was isolated from a sick animal with kidney swelling, interstitial nephritis, and an intestinal lymphoma.46
Polyomaviruses have been isolated from several bird species. The first of these, budgerigar fledgling disease virus (BFDV), was isolated from a parakeet in 1986.190 Unlike the strict host range restriction of mammalian polyomaviruses, BFDV can infect and cause disease in a wide variety of bird species and is now referred to as avian polyomavirus (APyV). Recently identified bird polyomaviruses include canary (CaPyV), crow (CPyV), finch (FPyV), and goose hemorrhagic polyomavirus (GHPyV).105,129,131 In general, the avian polyomaviruses cause a severe inflammatory illness that often results in death. For example, infection with GHPyV causes hemorrhagic nephritis and enteritis.130
For many years, the polyomaviruses were studied principally as model systems for understanding basic eukaryotic cell processes including DNA replication, RNA transcription, splicing and processing, and oncogenic transformation. The cloning and sequencing of the SV40 genome ushered in the era of recombinant DNA research. Indeed, the SV40 genome may be the most intensively studied DNA molecule per base pair. Several genetic elements from the SV40 genome are used in nearly every molecular biology laboratory in the world today.208
Interest in the polyomaviruses as human pathogens lagged behind these more basic biological studies because for many years the incidence of polyomavirus-associated diseases was rare and not well recognized. The onset of the human immunodeficiency virus type 1 (HIV-1)/acquired immunodeficiency syndrome (AIDS) epidemic, however, led to a dramatic rise in the incidence of PML, a JCPyV-induced disease. In addition, recent advances in immunosuppressive regimens for bone marrow and solid organ transplant recipients and biological therapies for autoimmune diseases led to increases in JCPyV-, BKPyV-, and TSPyV-associated diseases. More recently, the apparent transforming activity of the MCPyV and its contribution to Merkel cell carcinoma has generated widespread interest in the polyomaviruses. Studies have led to the emerging view that while many polyomavirus features are shared, there are many fundamental differences that distinguish each virus.
Classification
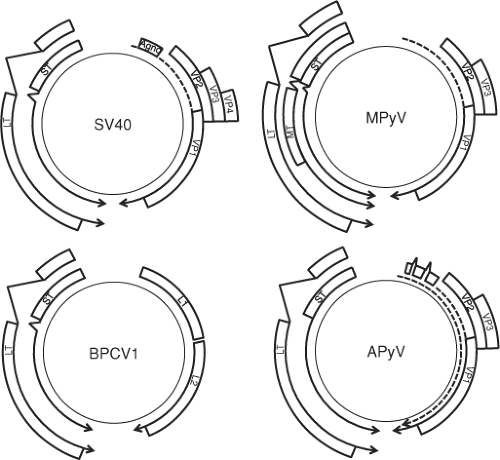
Polyomaviruses were originally classified within the Papovavirus family that included papillomavirus, polyomavirus, and vacuolating virus.181 The vacuolating or cytopathic effect of SV40 virus on host cells during lytic infection led to this distinction. Polyomaviruses were split from the papillomaviruses in 2000. Recently, a proposed classification organized polyomaviruses into three genera based on the DNA sequence of the viral genome and its normal host. The genera Orthopolyomavirus and Wukipolyomavirus contain polyomaviruses isolated from mammalian species, while Avipolyomavirus contains all avian species (Fig. 53.1).128 The Wukipolyomavirus genus contains WUPyV, KIPyV, HPyV6, and HPyV7, and the Orthopolyomavirus genus contains all other polyomaviruses including the bandicoot papillomatosis carcinomatosis virus type 1 (BPCV1) and type 2 (BPCV2).
Several polyomaviruses have not been officially classified within the family because the complete viral genomic sequence is not available. These include the baboon polyomavirus 2 (BPyV2), RKV, athymic rat polyomavirus (RatPyV), and cynomolgus polyomavirus (CyPV).
Virion Structure
Polyomavirus virions are nonenveloped, 45- to 50-nm particles consisting of three virally encoded capsid proteins,
VP1, VP2, and VP3, containing a circular dsDNA genome wrapped with cellular histones H2A, H2B, H3, and H4.262 The virion minichromosome exhibits the same nucleosome structure as cellular chromatin except for the absence of histone H1 that becomes associated with the viral genome only when in the infected cell. The particles have a T = 7 icosahedral symmetry and sediment at 240S in sucrose density gradients (Fig. 53.2). The density of mature virions is 1.34 g/mL and of empty capsids is 1.29 g/mL, as determined by cesium chloride equilibrium gradient centrifugation. The polyomaviruses are relatively resistant to heat and formalin inactivation, demonstrated by the isolation of viable SV40 from the Salk poliovirus vaccine.51 Because polyomaviruses are nonenveloped, they are resistant to lipid solvents. Similar to most viruses, preparations of polyomavirus virions contain many different types of particles. For example, in addition to mature virions, one can find empty capsids and capsids that contain cellular, rather than viral, DNA.
VP1, VP2, and VP3, containing a circular dsDNA genome wrapped with cellular histones H2A, H2B, H3, and H4.262 The virion minichromosome exhibits the same nucleosome structure as cellular chromatin except for the absence of histone H1 that becomes associated with the viral genome only when in the infected cell. The particles have a T = 7 icosahedral symmetry and sediment at 240S in sucrose density gradients (Fig. 53.2). The density of mature virions is 1.34 g/mL and of empty capsids is 1.29 g/mL, as determined by cesium chloride equilibrium gradient centrifugation. The polyomaviruses are relatively resistant to heat and formalin inactivation, demonstrated by the isolation of viable SV40 from the Salk poliovirus vaccine.51 Because polyomaviruses are nonenveloped, they are resistant to lipid solvents. Similar to most viruses, preparations of polyomavirus virions contain many different types of particles. For example, in addition to mature virions, one can find empty capsids and capsids that contain cellular, rather than viral, DNA.
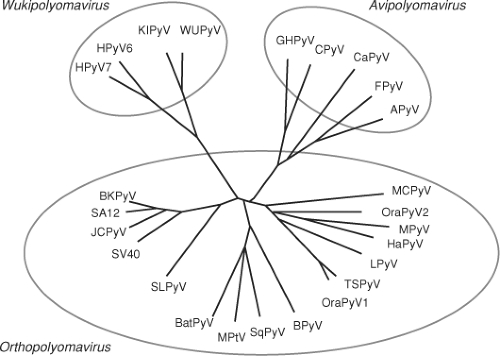 Figure 53.1. Phylogenetic relationships among the Polyomaviridae family based on whole genomic nucleotide sequences. The mammalian polyomaviruses are contained in the Orthopolyomavirus and Wukipolyomavirus genera, while the bird polyomaviruses are contained in the Avipolyomavirus genus. See Table 53.1 for full names and Genbank accession numbers. (Reproduced from Johne R, Buck CB, Allander T, et al. Taxonomical developments in the family Polyomaviridae. Arch Virol 2011;156(9):1627–1634, with kind permission from Springer Science + Business Media.) |
 Figure 53.2. Composite electron micrograph of a 40-nm polyomavirus icosahedral structure derived from assembly of VP1, negatively stained with 2% phosphotungstic acid (50,000¥). |
The polyomavirus capsid contains 360 molecules of VP1 arranged in 72 pentamers or capsomeres each containing 5 molecules of VP1 and 1 molecule of VP2 or VP3. Only the VP1 molecule is exposed on the surface of the capsid. The icosahedral capsid has both five- and sixfold axes of symmetry, with 12 pentamers surrounded by 5 other pentamers and 60 pentamers surrounded by 6 pentamers (Fig. 53.2). Capsomeres with fivefold symmetry are unusual but supported by the high-resolution structure of SV40.166,246 The C-terminus of each VP1 molecule extends out of the pentamer and contacts the neighboring capsomere. This structure is flexible and thereby provides the means to form an icosahedron. Capsomere contacts are stabilized by the presence of calcium ions, and mutations in residues that bind calcium result in premature disassembly.165 Treatment of virus with EGTA under reducing conditions results in the dissociation of the capsid into VP1 pentamers. In addition to VP1, VP2, and VP3, APyV expresses VP4 (agnoprotein 1a) that interacts with the C-terminus of VP1 and may be incorporated into viral capsids.234 The capsid contains posttranslational modifications including disulfide bridges that form between the pentameric capsomeres. In addition, VP2 undergoes myristoylation at its N-terminus. A recent report found a large number of posttranslational modifications on the BKPyV VP1 protein, although the role of these modifications during infection is not known.74
Genome Structure and Organization
The polyomavirus dsDNA circular genome contains approximately 5,000 base pairs and can be divided into three parts: the
early region encoding genes that are expressed prior to the onset of DNA replication; the late region encoding genes expressed after viral DNA replication commences; and the regulatory region, containing the origin of DNA replication as well as the promoters for early and late viral genes (Fig. 53.3). The early and late promoters give rise to primary transcripts from opposite strands of the DNA. The regulatory region including the origin is often referred to as the noncoding control region (NCCR). The numbering system for the polyomavirus genome differs from virus to virus with nucleotide position 1 defined in different ways.262 There has been precedent in recent years, however, to call the nucleotide preceding the A in the large T antigen ATG nucleotide 1, with numbering proceeding in the late direction, that is, away from the large T antigen open reading frame.
early region encoding genes that are expressed prior to the onset of DNA replication; the late region encoding genes expressed after viral DNA replication commences; and the regulatory region, containing the origin of DNA replication as well as the promoters for early and late viral genes (Fig. 53.3). The early and late promoters give rise to primary transcripts from opposite strands of the DNA. The regulatory region including the origin is often referred to as the noncoding control region (NCCR). The numbering system for the polyomavirus genome differs from virus to virus with nucleotide position 1 defined in different ways.262 There has been precedent in recent years, however, to call the nucleotide preceding the A in the large T antigen ATG nucleotide 1, with numbering proceeding in the late direction, that is, away from the large T antigen open reading frame.
The small size of the polyomavirus genome made it amenable to classical genetic approaches.262 Infection with temperature-sensitive mutants of SV40 led to the identification of five complementation groups, A, B, BC, C, and D. Mutations in group A mapped to the large T antigen gene; groups B, BC, and C to the VP1 gene; and group D to the VP3 gene. MPyV mutants have been classified into similar complementation groups, although no standard nomenclature was developed for this virus. Other mutants of MPyV known as host range (hr-t) mutants were selected for their inability to grow in established cell lines but retained the ability to replicate in primary cells or transformed cells. These mutations were later mapped to the middle T and small T antigen genes.240 The analysis of these early mutants set the stage for detailed and directed mutational studies enabled by recombinant DNA technology.
Another early genetic approach to the study of polyomaviruses was the selection for so-called evolutionary variants. In these experiments, viruses were passaged at high multiplicities of infection and variants were isolated. Many of these variants had alterations in the regulatory region that imparted a growth advantage to the virus. Passage of polyomavirus in culture often leads to duplications, deletions, and other rearrangements in the regulatory region.185 Although high-multiplicity passage of JCPyV in human glial cell cultures does not result in alterations in the regulatory region, the regulatory regions of BKPyV and JCPyV are frequently found to be rearranged when isolated from diseased tissues or patients’ blood.
The polyomavirus early and late promoters are contained within the regulatory region and overlap each other as well as the origin of replication. Early transcription progresses from the early promoter around the genome in one direction. Late transcription proceeds from the late promoter around the genome in the opposite direction. The early mRNAs are produced by posttranscriptional processing at a polyadenylic acid or poly(A) tail site that is located about halfway around the circular genome from the start site and by removal of introns by the cellular splicing machinery. SV40 was one of the first experimental systems where it was demonstrated that RNA polymerase II transcribes past the 3′ end of the mature mRNA molecule, implying that the 3′ end, including the poly(A) tail, was generated posttranscriptionally.82
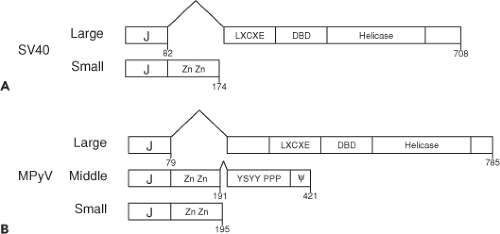
Each of the polyomaviruses encodes at least two early mRNAs by alternative splicing that are translated into the large T and small T antigens (Fig. 53.4). MPyV and HaPyV produce a third alternatively spliced mRNA that codes for middle
T antigen. The “T” in T antigen derives from the initial identification of these proteins as tumor antigens that were recognized by antisera from tumor-bearing animals inoculated with SV40.21,107 For many years, it was believed that large, middle, and small T antigens were the only early transcripts, but more recently it has been demonstrated that these viruses encode additional early mRNAs that differ in their splicing patterns, particularly for large T antigen. In SV40 and MPyV, an additional mRNA encodes a protein called 17KT and tiny T, respectively. Similarly, JCPyV produces a series of alternatively spliced mRNAs that encode proteins referred to as T′135, T′136, and T′165, and BKPyV encodes a molecule called truncated TAg. MCPyV encodes a full-length, 816-residue large T antigen as well as an alternatively spliced 57-kD T antigen that corresponds to the first 440 and last 100 residues of the full-length large T antigen.237
T antigen. The “T” in T antigen derives from the initial identification of these proteins as tumor antigens that were recognized by antisera from tumor-bearing animals inoculated with SV40.21,107 For many years, it was believed that large, middle, and small T antigens were the only early transcripts, but more recently it has been demonstrated that these viruses encode additional early mRNAs that differ in their splicing patterns, particularly for large T antigen. In SV40 and MPyV, an additional mRNA encodes a protein called 17KT and tiny T, respectively. Similarly, JCPyV produces a series of alternatively spliced mRNAs that encode proteins referred to as T′135, T′136, and T′165, and BKPyV encodes a molecule called truncated TAg. MCPyV encodes a full-length, 816-residue large T antigen as well as an alternatively spliced 57-kD T antigen that corresponds to the first 440 and last 100 residues of the full-length large T antigen.237
The late mRNA is transcribed in the opposite direction from the early mRNAs. As with the early transcripts, the late transcript has a single poly(A) site approximately halfway around the genome and is alternatively spliced (Fig. 53.3). The polyomavirus late transcript encodes three capsid proteins, VP1, VP2, and VP3. Notably, VP3 is translated in the same open frame as VP2 but uses an alternate AUG start codon downstream of the VP2 start codon and thereby shares all residues with VP2. In addition, SV40 encodes a VP4 protein that uses an internal AUG start codon even further downstream from VP3 and functions as a viroporin that promotes virus release from the infected cell.212 The late region transcript from SV40, JCPyV, and BKPyV as well as SA12, BatPyV, BPyV, SLPyV, and SqPyV encode an additional protein called agnoprotein.
Avian polyomaviruses express early transcripts encoding large and small T antigen, but the late transcripts have several distinct features (Fig. 53.3). There are two late transcription start sites, PL1 and PL2, that give rise to at least eight different transcripts due to alternative splicing.164 PL1 encodes two forms of VP4 (agnoprotein 1a) and VP4d (VPΔ4, agnoprotein 1b), while PL2 gives rise to two forms of agnoprotein 2a and 2b that use the same splice sites as VP4 and VP4d but are translated in a different reading frame. Avian agnoprotein 2a and 2b bears some similarity to the SV40 agnoprotein. All of the late avian transcripts also encode for VP1 or VP2 and VP3.
The late strand of several polyomaviruses encodes a microRNA (miRNA).39,253 The SV40 miRNA maps just 3′ of the late poly(A) site and appears to correspond to SAS (SV40-associated small RNA), a small RNA molecule identified 25 years earlier, albeit of then-unknown function. These miRNAs are complementary to the early mRNAs, target the early mRNAs for degradation, and may serve to limit the expression levels of the T antigens. MiRNA have been identified in SV40, BKPyV, JCPyV, MCPyV, SA12, MPyV, BPCV1, and BPCV2.39
Structure of T Antigens
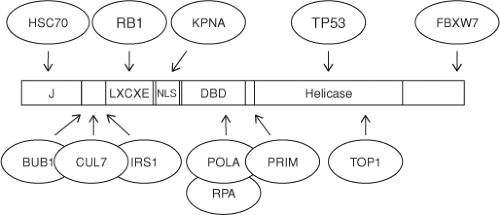
Much of what is known about the structure of polyomavirus T antigens comes from studies of SV40. All polyomavirus T antigens share an N-terminal region of approximately 80 residues that shows structural and sequence homology to the DnaJ or J domain found in host cell HSP40 homologs (Fig. 53.4). Full-length large T antigen is a nuclear phosphoprotein of approximately 700 residues. The molecule’s atomic coordinates have been assembled from crystallography of isolated domains, including the DnaJ and the retinoblastoma protein–binding domains; the DNA-binding domain, also known as the origin-binding domain; and a central domain consisting of residues 251 to 627 that forms a hexamer and contains the adenosine triphosphatase (ATPase) and helicase activities required for viral replication.163 Studies using scanning transmission electron microscopy, negative staining with atomic force microscopy, and single-particle reconstruction of cryoelectron microscopy (cryo-EM) images revealed that large T antigen forms a double hexamer in a head-to-head arrangement when bound to the origin of DNA.50
A number of functional domains contained within large T antigen are required for viral replication. Functions intrinsic to SV40 large T antigen include the ATPase/helicase domain and
the DNA-binding domain that mediates direct interactions with specific DNA sequences at the origin of replication. In addition to these intrinsic functions, the SV40 large T antigen domains serve to recruit host factors important for viral replication (Fig. 53.5). For example, the N-terminal J domain, containing the canonical residues HPDK, binds and activates the ATPase activity of host cell HSC70.31 The DNA-binding domain binds to replication protein A, while the helicase domain binds to the DNA polymerase α/primase complex.118 In addition, the helicase domains of many but not all polyoma large T antigens bind to p53. The outer surface of each SV40 large T antigen hexamer subunit can bind directly to the DNA-binding domain of p53.167 The large T antigen from MPyV is a notable exception among polyomaviruses because it does not bind to p53, although the ability of all the other polyomavirus large T antigens to bind to p53 has not yet been reported.
the DNA-binding domain that mediates direct interactions with specific DNA sequences at the origin of replication. In addition to these intrinsic functions, the SV40 large T antigen domains serve to recruit host factors important for viral replication (Fig. 53.5). For example, the N-terminal J domain, containing the canonical residues HPDK, binds and activates the ATPase activity of host cell HSC70.31 The DNA-binding domain binds to replication protein A, while the helicase domain binds to the DNA polymerase α/primase complex.118 In addition, the helicase domains of many but not all polyoma large T antigens bind to p53. The outer surface of each SV40 large T antigen hexamer subunit can bind directly to the DNA-binding domain of p53.167 The large T antigen from MPyV is a notable exception among polyomaviruses because it does not bind to p53, although the ability of all the other polyomavirus large T antigens to bind to p53 has not yet been reported.
Smaller functional motifs within large T antigen include the nuclear localization signal (NLS).132,154 Mutations that disrupt the NLS result in the cytoplasmic localization of SV40 large T antigen and inability to support the viral lytic life cycle. All mammalian polyomavirus large T antigens contain the conserved residues LXCXE (where X is any residue) that bind directly to the retinoblastoma family of tumor suppressor proteins including pRb (RB1), p107 (RBL1), and p130 (RBL2).60,68 The large and small T antigens from GHPyV, CPyV, and FPyV each contain the LXCXE motif, while those from APyV contain a related sequence, LXAXE. It is not known if any of the bird polyomavirus T antigens can bind to pRb or p53.
SV40 large T antigen contains a series of posttranslational modifications, including phosphorylation, O-glycosylation, acylation, poly(ADP)-ribosylation, and acetylation. In addition, phosphorylation of C-terminally located threonine residues in SV40 large T antigen creates a phospho-degron motif that binds directly to FBXW7, an F-box substrate adapter. Large T antigen binding to FBXW7 blocks binding to cyclin E and prevents its degradation by the CUL1FBXW7 RING ubiquitin ligase (Fig. 53.5).277 Phosphorylation regulates some of the functions of the molecule, including its subcellular localization and its ability to participate in the initiation of viral DNA synthesis. These regulatory events will be discussed later in the context of the life cycle.
Polyomavirus small T antigen is found in both the nucleus and the cytoplasm. Small T antigen is a cysteine-rich protein ranging in size from 124 to 198 residues and shares its N-terminus with large T antigen (i.e., those residues encoded up to the 5′ large T antigen splice site) but contains a unique C-terminal region. Small T antigen contains the same N-terminal J domain as large T antigen and a unique C-terminal domain (Fig. 53.4). The unique domain of the mammalian polyomavirus small T antigens contains a highly conserved set of cysteine and histidine residues that bind to two zinc molecules.43,44 These zinc-binding domains serve an important role in binding to the cellular protein phosphatase 2A (PP2A).200 PP2A is a trimeric complex consisting of two regulatory subunits A and B that bind to the catalytic C subunit. Small T antigen binds directly to the PP2A A subunit where the B subunit normally binds and thereby displaces or replaces the B subunit. SV40 small T antigen binds specifically to the Aα (PPP2R1A) subunit, while MPyV small T antigen binds to both the Aα and Aβ (PPP2R1B) subunits.4 The small T antigen–PP2A complex contains also the Cα (PPP2CA) or Cβ (PPP2CB) catalytic subunit. There are at least 18 different B PP2A subunits identified in mammalian cells. At the very least, SV40 small T antigen can displace the B56α (PPP2R5A), B56γ (PPP2R5C), and PR72/PR130 (PPP2R3A).219 It is likely that the polyomavirus small T antigen–PP2A complex not only serves to disrupt the cellular PP2A complexes but also is likely to retain specific phosphatase activity directed toward substrates. Notably, the bird polyomavirus small T antigens do not contain the conserved cysteine/histidine residues that serve to bind zinc, and it is not known if they are capable of PP2A binding.
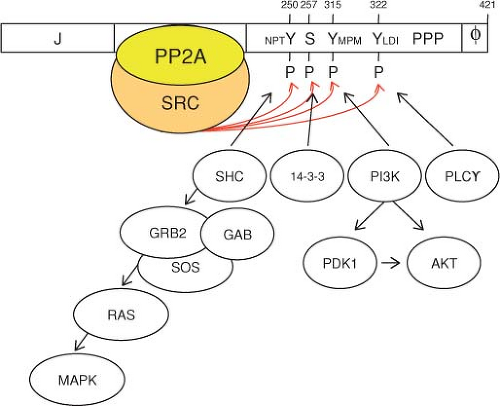
The middle T antigen of MPyV shares its N-terminal J domain and PP2A-binding domain with small T antigen (Figs. 53.4 and 53.6). The MPyV middle and small T antigens are identical for the N-terminal 191 residues until a splice junction that removes four nucleotides resulting in an additional 230 residues at the C-terminus of middle T antigen. In contrast, small T antigen contains only four unique amino acids after this intron. The unique middle T antigen C-terminus is encoded by an alternate reading frame used for coding the second exon of large T antigen. Notably, middle T antigen contains an N-terminal J domain in common with both large and small T antigen and, like small T antigen, binds to the A and C subunits of PP2A. The additional MPyV middle T residues mediate binding to several proteins involved in signal transduction, including the SRC tyrosine kinase, the SHC1 phosphotyrosine docking protein, phospholipase C (PLCG1), and phosphatidylinositol 3-kinase (PIK3CA and PIK3R1).223 MPyV middle T antigen can also bind to the SRC-related tyrosine kinases YES1 and FYN. The C-terminus of middle T antigen contains
a 22-residue hydrophobic domain that moves the newly translated middle T antigen from the cytoplasm through the endoplasmic reticulum (ER) to the inner plasma membrane.285 The combination of membrane localization with recruitment and activation of several enzymes enables middle T antigen to function as a constitutively activated tyrosine kinase that triggers downstream signaling in the RAS and MEK pathways.
a 22-residue hydrophobic domain that moves the newly translated middle T antigen from the cytoplasm through the endoplasmic reticulum (ER) to the inner plasma membrane.285 The combination of membrane localization with recruitment and activation of several enzymes enables middle T antigen to function as a constitutively activated tyrosine kinase that triggers downstream signaling in the RAS and MEK pathways.
Stages of Replication
Mechanism of Attachment
Prior to the 21st century, the identity of the cell surface receptors for polyomaviruses was poorly understood. Early studies on SV40 indicated that it used the major histocompatibility complex (MHC) class I antigens to bind to cells.29 Supporting evidence for this model included observations that antibodies against MHC class I blocked virus binding to rhesus monkey kidney cells and the inability of virus to bind well to human cells that do not express MHC class I antigens. Experimentally induced expression of MHC class I in nonexpressing cells restored binding. Although the MHC class I antigens were implicated as the SV40 receptor, they were not sufficient to account for all binding. For example, virus binding occurred only on the apical surface of polarized monkey epithelial cells, while MHC class I antigens were expressed on both the apical and basolateral surfaces. In addition, expression of MHC class I on human kidney epithelial cells was not sufficient for SV40 infection.
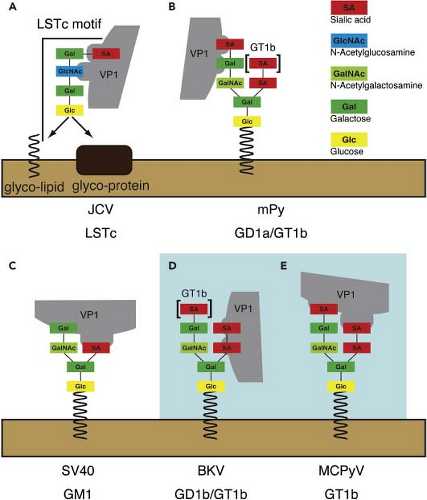
Subsequent results challenged the notion that SV40 uses a protein molecule as its receptor, indicating instead that it uses the branched ganglioside, GM1 (Fig. 53.7).265 This finding is more consistent with the route of entry of the virus through endosomes. In these studies, a rat cell line that did not express gangliosides and was unable to be infected by SV40 was rendered susceptible by preincubation with GM1. Gangliosides are glycosphingolipids that combine a sialylated oligosaccharide with ceramide consisting of sphingosine and a fatty acid. The sialic acid is critical for viral binding to the cell. In addition to providing a binding site for the virus, the gangliosides direct the virus to the correct endocytic pathway.209 The efficiency of SV40 infection is dependent on the relative concentration of GM1 on the cell surface as well as its ability to activate focal adhesion kinase (PTK2).244
Other polyomaviruses use different forms of gangliosides as receptors (Fig. 53.7). For example, MPyV uses GT1b or GD1b.92,265 After initial binding to the ganglioside, MPyV interacts with α4β1 integrin that may serve as a secondary or cell type–specific receptor for viral entry.35 BKPyV also uses gangliosides to enter the cell, as judged by restoration of infectivity to otherwise resistant cells upon preincubation with GT1a and GD1a.171 These branched gangliosides are found on the renal tubular epithelial cells that BKPyV normally infects. Another report indicated that an N-linked glycoprotein containing sialylated oligosaccharides can also mediate BKPyV binding.66 Both the glycolipid and glycoprotein contain sialic acid, consistent with early reports that BKPyV can hemagglutinate human red blood cells and that this activity was neuraminidase sensitive. There are conflicting reports regarding the receptor for MCPyV. Although it has been shown that MCPyV VP1 capsomeres can bind the ganglioside GT1b,72 pseudovirions can bind heparin moieties.225
JCPyV binds to lactoseries tetrasaccharide c(LTSc), a linear sialylated oligosaccharide that differs from the branched forms reported for other polyomaviruses (Fig. 53.7). LTSc is a pentasaccharide with the terminal sialic acid linked by an α2,6 bond to the penultimate galactose.194,266 In addition, JCPyV uses the 5HT2A serotonin receptor, perhaps as a cell type– specific receptor, for viral entry.69 This receptor is expressed on
glial cells, the major target cell for JCPyV. Antibodies to the 5HT2A receptor and receptor antagonists block JCPyV infection, while expressing the receptor in otherwise noninfectible cells renders them susceptible to infection.
glial cells, the major target cell for JCPyV. Antibodies to the 5HT2A receptor and receptor antagonists block JCPyV infection, while expressing the receptor in otherwise noninfectible cells renders them susceptible to infection.
Entry, Intracellular Trafficking, and Uncoating
The polyomaviruses use several pathways to enter into the cell and pass through the endosomes to the ER (Fig. 53.8). The pathway from the endosomal compartment to the ER and from the ER to the nucleus is not well understood. After binding to gangliosides on the cell surface, SV40 and BKPyV enter the cell using a pathway involving caveolin. The virus is delivered to a neutral pH organelle called the caveosome by endocytosis and then to the ER, where it could be detected by electron microscopy. Recent work has indicated that SV40 may also traffic through a more traditional endocytic pathway to the ER that does not involve a caveosome.70 Evidence also indicates that entry of SV40 into the cell requires engagement of a signal transduction cascade through interactions at the cell surface.70
Studies with various inhibitors of intracellular structures and processes have shed some light on how the virus travels within the cytosol. Molecules that interfere with microtubules and prevent movement of vesicles from the endosome to the ER interfere with SV40 infection.235 Infection can be blocked by the drug, brefeldin A, which inhibits trafficking between the ER and the Golgi apparatus,197 and a number of other drugs that interfere with endosome maturation and trafficking to the ER.70
The polyomavirus capsid begins to disassemble in the ER. Evidence for this includes the appearance of epitopes on VP2 and VP3 that become accessible for immunostaining.197 In addition, structural changes to the capsid, including disulfide bond reduction and isomerization mediated by ER-resident protein disulfide isomerases, can be detected biochemically at the point when the virus enters the ER.93,172,224,274 Furthermore, transmission EM experiments have detected changes in the morphology of virions that were isolated from the ER.121 It is thought that this change leads to exposure of hydrophobic surfaces of the VP1 molecule on the capsid that facilitates transport across the ER membrane. This step in trafficking also involves ER chaperone proteins. Similar to SV40, it has been shown that the conformation of the MPyV capsid begins to change in the ER due to the action of ERp29, a member of
the protein disulfide isomerase family.172,265 Capsid disassembly in the ER or the cytosol may be required because the viral particle is bigger than the functional capacity of the nuclear pore. Evidence for a multistep disassembly process for SV40 includes the exposure of certain VP2 and VP3 epitopes in the ER, while further disassembly occurs in the cytoplasm at a later time point as evidenced by immunoassay detection of the viral genome.150 The ER-associated degradation pathway, which functions to target misfolded cellular ER proteins for proteasome-mediated degradation, has also been implicated in polyomavirus disassembly.91,96,125
the protein disulfide isomerase family.172,265 Capsid disassembly in the ER or the cytosol may be required because the viral particle is bigger than the functional capacity of the nuclear pore. Evidence for a multistep disassembly process for SV40 includes the exposure of certain VP2 and VP3 epitopes in the ER, while further disassembly occurs in the cytoplasm at a later time point as evidenced by immunoassay detection of the viral genome.150 The ER-associated degradation pathway, which functions to target misfolded cellular ER proteins for proteasome-mediated degradation, has also been implicated in polyomavirus disassembly.91,96,125
The question of how the genome gets carried to the nucleus remains to be answered. For SV40, it has been speculated that release from the vesicular compartment might involve VP1, as noted earlier, or VP2, by virtue of its myristoylated N-terminus inserting into the lipid bilayer.197 An NLS in VP3 may mediate entry through the nuclear pore complex, because mutations in this NLS inhibit entry of the virion into the nucleus but do not affect capsid assembly or the production of new virions.191
JCPyV enters the cell through clathrin-coated pits, as indicated by use of inhibitors of this pathway as well as the demonstration that JCPyV co-localizes with transferrin, which is known to use clathrin-mediated entry into the cell.206 Both microtubules and microfilaments play a role in trafficking of JCPyV to the nucleus.7 As with SV40, binding of JCPyV to its receptor induces a signal transduction cascade required for efficient entry that can be inhibited by genistein, a tyrosine kinase inhibitor.211 JCPyV appears to signal through the extracellular signal-regulated kinase (ERK) or mitogen-activated protein kinase (MAPK) pathway. Using fluorescently labeled virus-like particles (VLPs), it has been shown that JCPyV particles do not disassemble before they reach the nuclear pore and that the nuclear localization signal of VP1 is required for entry into the nucleus.210
Transcription
After the polyomavirus genome enters the nucleus, it serves as a template for transcription by the cellular RNA polymerase II. Once inside the nucleus, the viral genome becomes wrapped in nucleosomes containing histone H1 in addition to the four core histones that are present in the virus particle.270 Within the cell, the SV40 viral genome contains 24 nucleosomes with a nucleosome-free region of 400 base pairs encompassing the NCCR of early and late promoters and origin of replication. This is in contrast to the virion, where all regions of the SV40 genome are covered with nucleosomes devoid of H1. Chromatin immunoprecipitation can detect transcriptionally active chromatin, defined by the presence of RNA polymerase II, as early as 30 minutes postinfection with SV40.11 The chromatin at this time also contains hyperacetylated histone H3 and H4,10,182 which have been associated with chromatin undergoing transcription initiation and elongation.
Transcription of polyomavirus genes is governed by cis-acting sequences in the regulatory region. The SV40 regulatory region has been the most intensively studied and serves as a paradigm for the other polyomaviruses. Seminal studies involving SV40 include the demonstration that AT-rich sequences designated TATA boxes act to direct RNA polymerase II to the proper initiation site for transcription.15 The SV40 early promoter also contains a series of GC-rich sequences within the 21–base-pair repeat region.73,84 The protein SP1, one of the first eukaryotic transcription factors to be cloned, binds to these sequences.67 The SV40 early promoter also contains a duplicated element called the 72–base-pair repeat, which was the first cis-acting DNA sequence to be deemed a transcriptional enhancer because it could activate transcription when placed several thousand base pairs distant from the transcription start site.104,187 Interestingly, most clinical isolates of SV40 carry only a single copy of the 72–base-pair repeat, and it appears that duplication can be selected during passage in culture.157
In MPyV, the enhancer element consists of two neighboring enhancers called A and B, alternatively α and β, that can act independently. The activity of the MPyV enhancers is dependent on the cell environment, as viral variants that are selected for growth on differentiated or undifferentiated embryonal carcinoma cells have mutations that map to the enhancers.54,180 The MPyV early promoter is regulated by the cellular factors characterized as polyomavirus enhancer A binding proteins (PEA) or RUNX1, CBFB, and ETV4 that are expressed when cells are growth stimulated with serum.122,176
The JCPyV promoter shows a distinct tissue-specific activity that correlates with the sites of acute infection. While normal JCPyV virions can attach and enter many types of cells, its host range is restricted to those cells that have the appropriate DNA-binding proteins.267 The JCPyV early promoter
contains sequences that act as transcriptional promoters. These sequences include the TATA box as well as binding sites for the transcription factors SP1, YB1 (YBX1), Pur α (PURA), AP-1, a heterodimer of JUN and FOS, nuclear factor 1 (NFAT), and NF1X. A nuclear factor-κB (NF-κB) binding site that includes the NFAT site has been shown to be active for late transcription and possible positioning of other DNA-binding proteins to initiate efficient transcription.86 NFAT consists of a family of proteins that are expressed in many cells but have multiple classes with tissue-specific expression. The NF-1 class X (NFIX) is highly expressed in human glial cells, stromal cells, B lymphocytes, and CD34+ stem cells that have all been reported to support JCPyV transcription and replication.79 Interactions between YB1 and PURA may also provide cell-specific regulation.86
contains sequences that act as transcriptional promoters. These sequences include the TATA box as well as binding sites for the transcription factors SP1, YB1 (YBX1), Pur α (PURA), AP-1, a heterodimer of JUN and FOS, nuclear factor 1 (NFAT), and NF1X. A nuclear factor-κB (NF-κB) binding site that includes the NFAT site has been shown to be active for late transcription and possible positioning of other DNA-binding proteins to initiate efficient transcription.86 NFAT consists of a family of proteins that are expressed in many cells but have multiple classes with tissue-specific expression. The NF-1 class X (NFIX) is highly expressed in human glial cells, stromal cells, B lymphocytes, and CD34+ stem cells that have all been reported to support JCPyV transcription and replication.79 Interactions between YB1 and PURA may also provide cell-specific regulation.86
The transcription factors that govern kidney tissue-specific activity of the BKPyV early promoter are not known. While numerous candidate transcription factor binding sites have been identified,185 it remains unclear which of those factors bind the BKPyV regulatory region in an infected kidney cell.
Rearrangements in the NCCR have also been found in BKPyV and JCPyV.30,124,214 Viral regulatory regions referred to as archetype are thought to be associated with naturally circulating polyomaviruses, while rearranged regulatory regions arise when high levels of viral replication occur in culture or during disease. For example, JCPyV early promoter undergoes duplication in virus isolated from patients with PML. These rearrangements may serve to allow higher levels of large T antigen expression and viral replication that are tolerated by the patient’s immunocompromised state.98 Notably, the δSV40/JCV hybrid virus contains JCPyV T antigen and VP1, VP2, and VP3 coding sequences but a hybrid regulatory region with elements of the SV40 enhancer. This laboratory-generated hybrid virus grows with faster kinetics and to higher titers than wild-type JCPyV and has an expanded host range to human and monkey kidney cells as well as monkey glial cells.267
The SV40 early promoter undergoes negative feedback regulation by large T antigen.259 When cells are infected with a temperature-sensitive large T antigen mutant of SV40 at the nonpermissive or restrictive temperature, early mRNAs are overproduced. The ability to repress early transcription is dependent on the binding of large T antigen to the promoter. Mutations that disrupt the DNA-binding domain of large T antigen also typically lead to higher levels of large T antigen.133 Three binding sites, referred to as sites I, II, and III, in the regulatory region are all involved in repression by large T antigen. Because site III overlaps the early promoter elements, it is possible that large T antigen binding acts to prevent binding or displace other transcription factors to that region. Early transcription of MPyV is also regulated by large T antigen, but to a lesser degree and in a DNA binding site–independent manner.
Early gene expression in SV40 is down-regulated by miRNAs encoded by the late transcript.253 The other primate polyomaviruses have similar sequences that are predicted to be able to form the characteristic hairpin structure of miRNA.228 Somewhat surprisingly, an SV40 mutant virus lacking the miRNA does not produce more virus than wild type in cultured monkey kidney cells. However, cells infected with the mutant virus are more sensitive to killing by cytotoxic T lymphocytes in vitro. This observation led to the proposal that the miRNA serves to limit production of antigens recognized by the immune system and thereby protecting the infected cell.253 A mutant MPyV that cannot express its miRNA shows no difference in pathogenesis in animals.254 Therefore, the role of the miRNAs during infection remains to be fully elucidated.
Similar to the early promoter, the late promoter elements span the regulatory region and have been defined in a variety of in vivo and in vitro systems. In SV40, maximal late transcription requires sequences in the 21–base-pair GC repeats and the 72–base-pair enhancers. Late gene transcription occurs concomitantly with the onset of DNA replication, although replication is not required for activation of late transcription. Large T antigen can promote viral replication as well as late gene expression. Large T antigen can stimulate late transcription, although large T antigen binding the origin of replication is not required for this activity. Rather, transcription activation is accomplished through large T antigen interactions with components of the basal transcription machinery such as TATA-binding protein, a component of TFIID and TBP-associated factor 1 (TAF1), as well as transcription factors late SV40 factor (LSF, TFCP2), TEF-1 (TEAD1), and SP1. These interactions increase the binding of TBP and another basal transcription factor, TFIIA, to the TATA element.53
The control of late gene expression in MPyV is more complicated. Host range or hr-t mutants that do not express functional small or middle T antigen produce equivalent amounts of late proteins, as does wild-type virus.87 However, infection with a virus containing similar mutations in cells with a different genetic background demonstrated a stimulatory role of the two T antigens during the late phase. This does not appear to be solely due to an indirect effect because of amplification of the genome, but also involves a direct stimulation of transcription because the RNA/DNA ratio increases.40 These same studies also indicated a role for middle and small T antigens in the induction of early gene expression. As these two T antigens induce signaling pathways that lead to activation of transcription factors known to bind the polyomavirus enhancer, this effect is not surprising. The murine virus also differs from its primate counterparts in how its late primary RNA transcript is processed. The MPyV late poly(A) site is a relatively weak site, resulting in the RNA polymerase circling the viral genome multiple times.1,161 The first exon of the late transcript can therefore be spliced to itself multiple times, although the protein-coding sequence is only present once on each mature mRNA.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree