Pharmacogenetics of Cytochrome P450
LEARNING OBJECTIVES
Cytochrome P450s (CYPs) are a heme-containing superfamily of enzymes responsible for the metabolism of a wide range of structurally diverse substrates. In addition to the metabolism of human drugs, they also have biologically important roles, such as metabolism of several hormones, synthesis and elimination of cholesterol, regulation of blood homeostasis, metabolism of arachidonic acid, and detoxification of foreign pollutants. Genetic polymorphisms in genes encoding drug-metabolizing enzymes can result in variability in drug metabolism and drug elimination, which often affect treatment outcome at various degrees, depending on the severity of mutation and the extent of penetrance of that gene. There are large environmental factors, drug-drug interactions, and clinical factors that influence clinical outcomes, providing further complexity in understanding individual differences in drug responses. Because CYPs play a critical role in the metabolism of human drugs, accounting for approximately 80% of all phase I drug metabolism, CYP genes have been important targets for pharmacogenetics and pharmacogenomics research. Tremendous efforts on pharmacogenetics and pharmacogenomics during the last several decades have led to the discovery of many clinically relevant genetic polymorphisms in CYPs. This chapter presents an overview of functional CYP polymorphisms with regard to biochemical aspects as well as clinical consequences.
PHARMACOGENOMICS OF CYP2C9
Introduction
Among the four CYP2C genes in humans, CYP2C9 is the most abundantly expressed enzyme in the human liver,1,2 accounting for the metabolism of approximately 15-20% of prescribed and over-the-counter drugs.3 CYP2C9 metabolizes a number of clinically important drugs, including the antidiabetic drugs tolbutamide and glipizide,4,5 the anticonvulsant phenytoin,6 the anticoagulant warfarin,7 the antihypertensive drug losartan,8 the diuretic torasemide,9 and nonsteroidal anti-inflammatory drugs (NSAIDs) such as ibuprofen, diclofenac, piroxicam, tenoxicam, and mefenamic acid.10 CYP2C9 also metabolizes endogenous substrates, such as arachidonic acid and linoleic acid.11 Since CYP2C9 is polymorphic, it is involved in interindividual variation in the metabolism and disposition of the drugs described above. In particular, drugs with a narrow therapeutic index, such as S-warfarin and phenytoin, can present serious problems in dose adjustments while achieving appropriate drug concentrations without causing high dose-induced drug toxicity.
Functional Polymorphism of CYP2C9
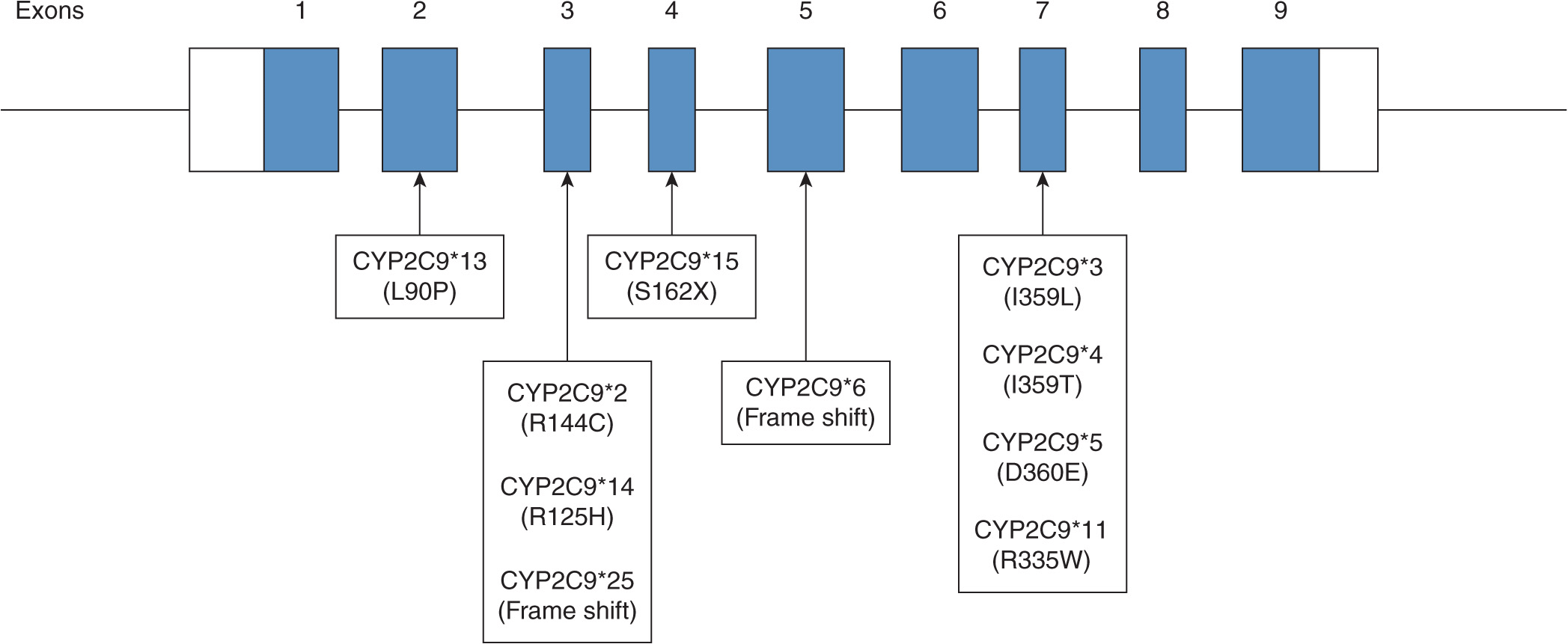
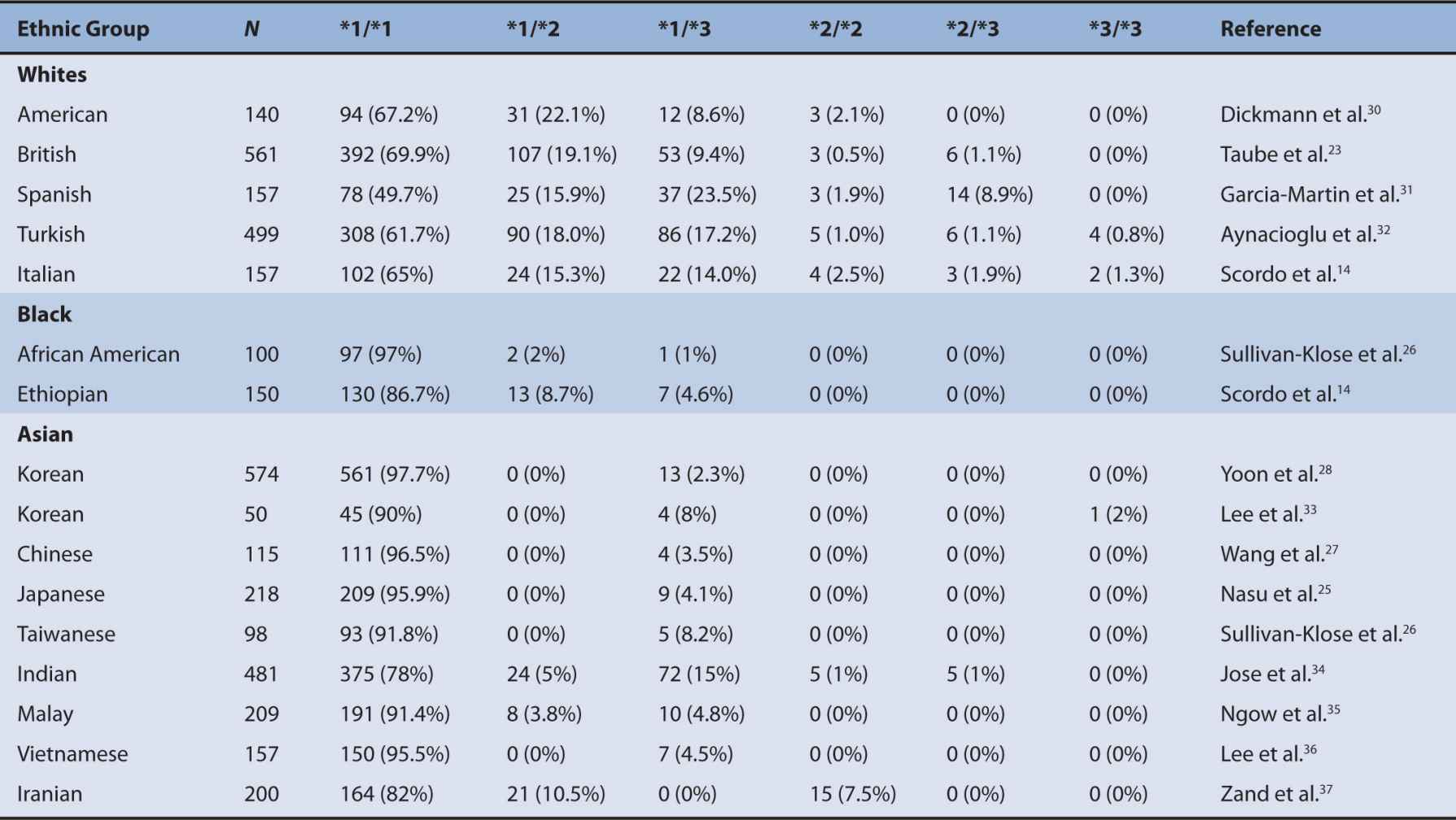
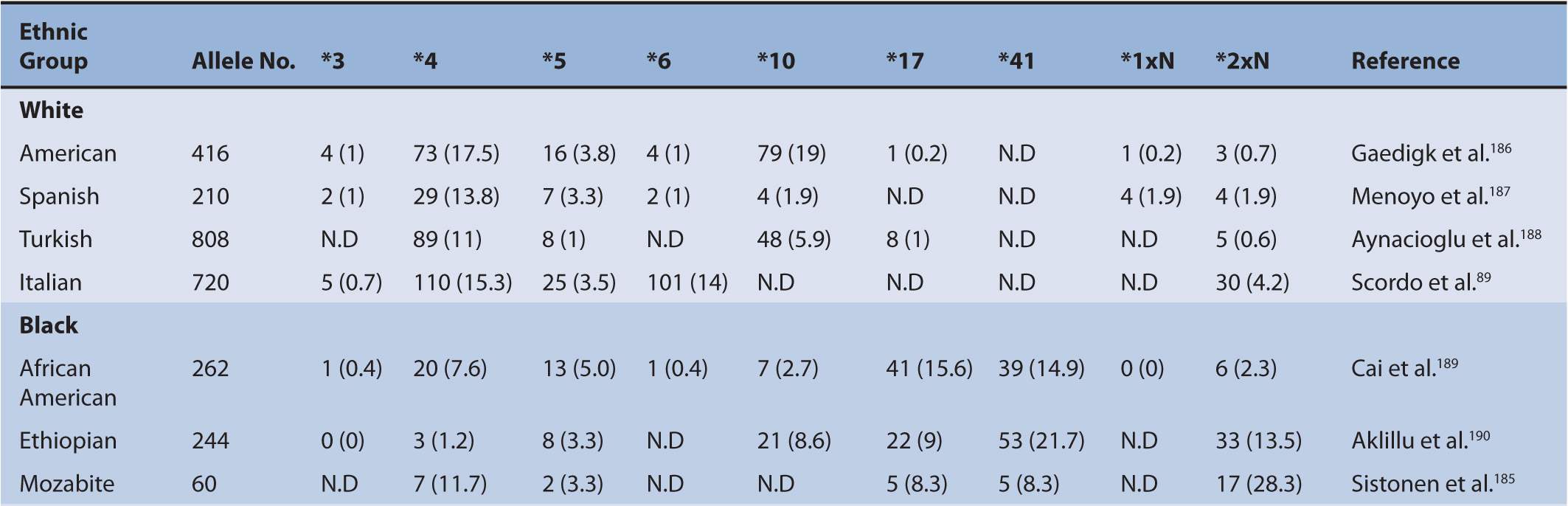
A number of polymorphisms of CYP2C9 have been shown to contribute to altered CYP2C9 activity. The two most prevalent and clinically relevant defective variants in white populations are CYP2C9*2 and *3.12–15 Null variant alleles include CYP2C9*6, *15, and *25.16–18 CYP2C9 variants identified from phenotyped individuals include CYP2C9*2, *3, *4, *5, *6, *11, *13, and *1419–22 (Figure 6A-1). The frequency of CYP2C9*2 is 11% in whites and about 1% in blacks.23,24 However, the frequency of CYP2C9*2 in Asians is very rare or not detected in certain populations, such as Korean, Japanese, Taiwanese, and Chinese.25–28 The frequency of CYP2C9*3 is 7% in whites, 1% in blacks, and 3% in Asians.29 Ethnic differences in the frequency of these CYP2C9 genotypes are summarized in Table 6A–1. CYP2C9*2 carries an Arg144Cys mutation and CYP2C9*3 encodes an Ile359Leu change. The investigation of CYP2C9*2 and *3 alleles showed that these variants are associated with significantly reduced S-warfarin 7′-hydroxylation in the various recombinant enzyme assay systems as well as the genotyped human liver microsomes.13,15,30,38 However, no single in vitro system has been adopted as the standard method for exact evaluation of the functional change of the CYP2C9 polymorphisms compared with the wild-type CYP2C9*1. Therefore, quantitative comparisons of CYP2C9 variants with the wild-type CYP2C9 should be interpreted with great caution. Many functional studies evaluating the activity of CYP2C9*2 and *3 have consistently demonstrated a significantly reduced CYP2C9 activity compared with CYP2C9*1.13,38–41 In general, the CYP2C9*3 variant has consistently exhibited a greater reduction, a 10-30% activity of the wild-type CLint, in the metabolism of all CYP2C9 substrate drugs than the CYP2C9*2 allele that exhibits a 70-90% activity of the wild-type CLint in various in vitro studies.21 In most CYP2C9 substrates, individuals with the CYP2C9*3 heterozygous mutation exhibited approximately 50% of the wild-type total oral clearance and individuals carrying the CYP2C9*3 homozygous mutation showed a 5- to 10-fold reduction.42–44 Therefore, it is believed that the CYP2C9*3 allele is a more defective allele affecting drug pharmacokinetics and pharmacodynamics. Substrate-specific differences in the metabolic activity of CYP2C9 alleles were reported,42 suggesting that the particular CYP2C9 alleles can affect drug metabolism differently depending on the substrate. Although in vitro systems provide characterization of CYP2C9 alleles in regard to many substrates, the corresponding response of the drug by these alleles in vivo is still a difficult issue. However, for detecting changes in the metabolic activity of the CYP2C9 allele for certain drugs such as warfarin and phenytoin, in vitro systems are a useful predictor of the drug response in vivo.
FIGURE 6A–1 Functional CYP2C9 alleles that were identified and evidenced in phenotyped human subjects. White boxes are 5′- and 3′-untranslated regions. Black boxes are exons in open reading frame. Arrow heads indicate mutated locations that result in amino acid change, stop codon (X), or frameshift.
TABLE 6A–1 Distribution of major CYP2C9 genotypes in different ethnic populations.
Clinical Relevance of CYP2C9 Genetic Polymorphism
The clinical relevance of CYP2C9 polymorphisms was initiated and identified by two main coding variants, CYP2C9*2 and *3. A number of individual case reports with CYP2C9 substrate drugs have described the clinical significance of CYP2C9 polymorphisms with the new discovery of CYP2C9 defective alleles (official CYP2C9 allele nomenclature: http://www.cypalleles.ki.se/cyp2c9.htm). Among these case reports, most clinical problems with toxicity and dose adjustment have resulted from warfarin and phenytoin administration in CYP2C9 “poor metabolizer” (PM) genotypes.45,46 Warfarin consists of equal amount of R-and S-warfarin. S-Warfarin is more potent and metabolized principally by CYP2C9, whereas R-warfarin is metabolized by CYP1A1, CYP2C19, and CYP3A4.47 Patients having a CYP2C9*2 or *3 allele are significantly prone to experience a bleeding event and prolonged hospitalization due to unstable anticoagulation when compared with the wild-type. Therefore, it is suggested that lower warfarin doses need to be employed in patients having defective alleles. For example, in a similar demographic condition, stable warfarin daily dose requirements were differently observed as an average value of 7.9 mg per day for CYP2C9*1/*1 patients ![]() and 2.2 mg per day for CYP2C9*1/*3 patients
and 2.2 mg per day for CYP2C9*1/*3 patients ![]() Genotyping for CYP2C9 is a significant factor during the unstable period of warfarin dose initiation.23,49,50 However, it appears that once antithrombotic stability is attained, experienced clinicians are likely to maintain a patient’s INR and minimize his or her bleeding risk with less dependence on CYP2C9 genotype.23 Studies from meta-analysis indicated that the CYP2C9 genotype accounted for 12% of the variability of warfarin dosing and VKORC1 polymorphisms accounted for 25%.51 CYP2C9 genotyping prior to warfarin initiation improves the safety profile. The FDA has approved warfarin pharmacogenetic tests, which can be used for initial dose determination. Although cost-effectiveness with the CYP2C9 genotype testing remains to be determined, warfarin dosing models incorporating clinical factors and genetic profiles are under strong investigation.
Genotyping for CYP2C9 is a significant factor during the unstable period of warfarin dose initiation.23,49,50 However, it appears that once antithrombotic stability is attained, experienced clinicians are likely to maintain a patient’s INR and minimize his or her bleeding risk with less dependence on CYP2C9 genotype.23 Studies from meta-analysis indicated that the CYP2C9 genotype accounted for 12% of the variability of warfarin dosing and VKORC1 polymorphisms accounted for 25%.51 CYP2C9 genotyping prior to warfarin initiation improves the safety profile. The FDA has approved warfarin pharmacogenetic tests, which can be used for initial dose determination. Although cost-effectiveness with the CYP2C9 genotype testing remains to be determined, warfarin dosing models incorporating clinical factors and genetic profiles are under strong investigation.
Although both CYP2C9 and CYP2C19 metabolize phenytoin, CYP2C9 is responsible for the major pathway of phenytoin metabolism, accounting for approximately 80-90% of its elimination.52,53 Since phenytoin exhibits a narrow therapeutic range with a concentration-related toxicity, small changes of CYP2C9 activity can lead to nervous system intoxication.4,45 Multiple case reports and clinical observations suggested that the CYP2C9 genotype is an important determinant for the prediction of phenytoin disposition in humans.16,32,54 For example, patients carrying at least one variant of defective CYP2C9 allele required a 30% lower phenytoin maintenance dose than the patients carrying CYP2C9*1/*1 genotype.55 Similarly, individuals having CYP2C9*3/*3 exhibited a 3-fold increase in half-life and a 4-fold increase in AUC (area under the plasma concentration vs. time curves) compared with individuals having CYP2C9*1/*1.4 The importance of CYP2C9 in the metabolism of phenytoin was evidenced by a case report of an individual who had a homozygous mutation for CYP2C9*6 null allele, indicating that this individual has no active CYP2C9 protein.16 This individual was taken to the emergency department with severe phenytoin toxicity. Clearance of phenytoin in this individual was estimated to be 17% of the general population with 13 days of elimination half-life.16 Clinical implications of CYP2C9 genotype are evident in phenytoin therapy. However, since there are many different CYP2C9 variant alleles, the impact of each variant on the clinical outcome of phenytoin remains to be determined.
Tolbutamide has been used as a phenotypic probe for CYP2C9 activity in vivo and as a prototypic substrate for the assessment of hepatic CYP2C9 activity in vitro.5,56 Therefore, tolbutamide “PM” phenotype could be attributed to the homozygous mutation for CYP2C9 defective alleles. Investigations using CYP2C9 variants demonstrate that functional CYP2C9 polymorphisms are associated with significant changes in tolbutamide pharmacokinetics.26 Similarly, pharmacokinetics of the antidiabetic drug glibenclamide and glimepiride has been significantly affected by CYP2C9 genotypes, indicating that individuals having CYP2C9*3 exhibited higher AUC and reduced oral clearance.57,58 However, clinical consequences, such as blood glucose response, of CYP2C9 polymorphisms in the treatment of type 2 diabetic patients using oral hypoglycemic agents remain unclear.57,58 Further prospective studies for the relationship between pharmacokinetics and pharmacodynamics would be necessary to approach the dose adjustment strategy on the basis of CYP2C9 genotype in diabetic patients.
Perspective of Clinical Application
A number of investigations have evaluated the clinical significance of CYP2C9*2 and *3 on the various CYP2C9 substrate drugs. In particular, individuals homozygous for CYP2C9*2 or *3 were associated with significant alterations in pharmacokinetics and pharmacodynamics of the drugs such as warfarin and phenytoin. Contribution of CYP2C9 genotype to drug metabolism is dependent on the substrate specificity, because most CYP2C9 substrates are also metabolized to certain degrees by other enzymes. For example, phenytoin is predominantly metabolized by CYP2C9, but minor metabolic pathway includes CYP2C19.52 The metabolism of tolbutamide includes CYP2C19 as a minor metabolic pathway and the same is true of losartan with CYP3A4.8,59 Since CYP2C9 metabolizes a wide range of clinically used drugs with different degrees of specificity,42 comprehensive pharmacogenomics information together with the extent of CYP2C9’s contribution to the total drug metabolism would be necessary to understand interindividual variations when taking CYP2C9-dependent drugs.
PHARMACOGENOMICS OF CYP2C19
Introduction
The cytochrome P450 CYP2C19 is a clinically important enzyme that metabolizes a number of drugs such as the antiulcer drug omeprazole,60 the anticonvulsant mephenytoin,60,61 the antimalarial proguanil,62,63 the anxiolytic drug diazepam,61,64,65 and certain antidepressants such as citalopram,66 imipramine,67 amitriptyline,68 and clomipramine.69 The metabolism of these drugs has been reported as polymorphic in humans.12 The metabolic ratio (MR) between the plasma concentration of omeprazole and that of its 5-hydroxy metabolite is used as a measure of CYP2C19 activity in vivo.70 Because mephenytoin, a racemic mixture of R– and S-enantiomers, is metabolized differently and S-mephenytoin is much more rapidly hydroxylated by the CYP2C19, the S-/ R-ratio has been used as a measure of CYP2C19 activity in humans.1,70,71 Individuals can be categorized as “extensive metabolizers” (EMs) or “PMs” of drugs metabolized by CYP2C19 in population studies. Genetic polymorphisms of CYP2C19 have been shown to cause clinical consequences resulting in undesirable side effects, such as prolonged sedation and unconsciousness after administration of diazepam in CYP2C19 PMs.61,64,65 In contrast, the proton pump inhibitor (PPI) drugs such as omeprazole and lansoprazole exhibit a greater cure rate for gastric ulcers with Helicobacter pylori infections in PMs than in EMs due to higher plasma concentration of the parent drugs in PMs.72,73
Functional Polymorphism of CYP2C19
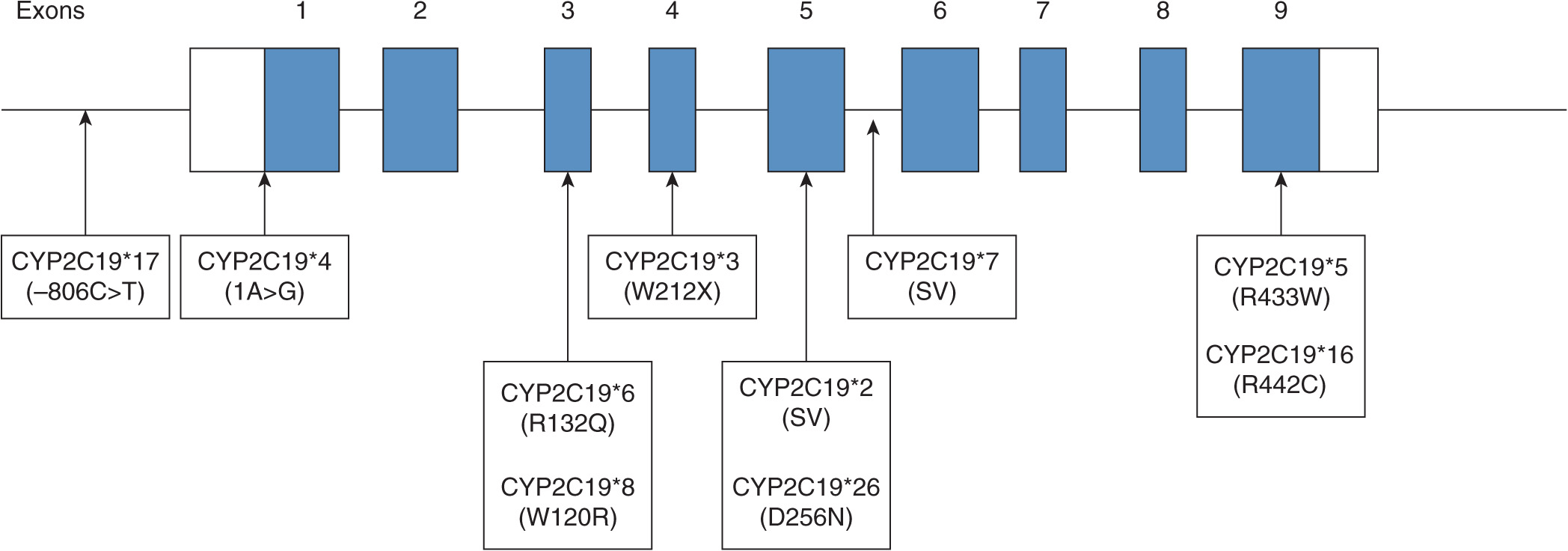
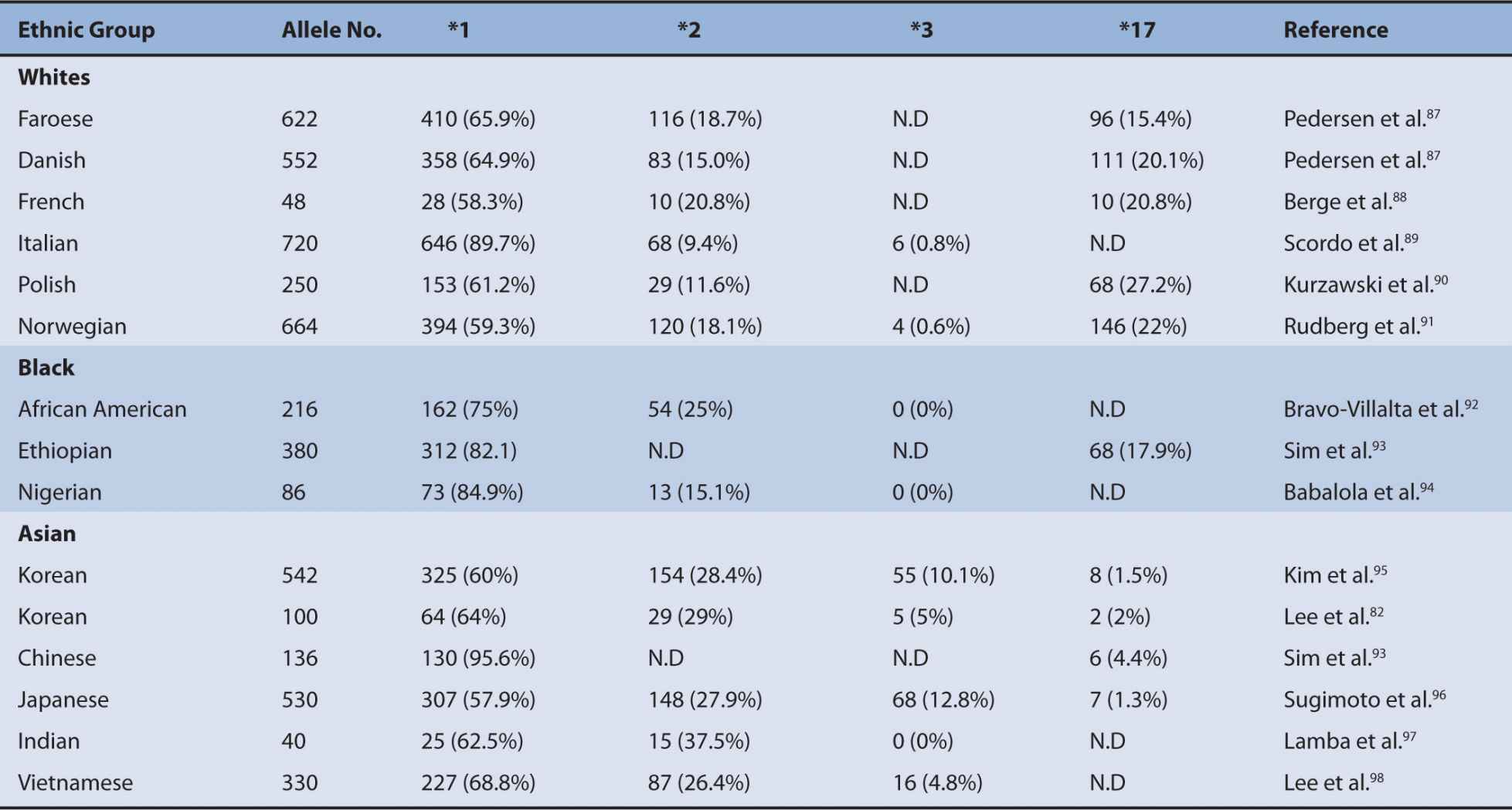
The CYP2C subfamily in humans consists of four CYP2C genes that include CYP2C8, CYP2C9, CYP2C18, and CYP2C19. Among the four CYP2C genes, CYP2C19 is a major polymorphic P450.74 A number of polymorphisms of CYP2C19 have been shown to contribute to the altered CYP2C19 activity. The most common defective variants are CYP2C19*2 and *3.74,75 CYP2C19*2 is a splice mutation in exon 5 that produces a truncated nonfunctional protein and CYP2C19*3 produces a stop codon in exon 4.76,77 Other null variant alleles include CYP2C19*4 (a mutation in the initiation codon) and CYP2C19*7 (a splice mutation in intron 5).78,79 PMs can be defined as individuals carrying two defective CYP2C19 alleles. Several CYP2C19 variants were discovered by DNA sequencing of CYP2C19 gene in individuals who had been characterized by clinicians as PMs by phenotyping with mephenytoin or omeprazole.70,71 These functional variants identified from phenotype studies include CYP2C19*5, *6, *8, *16, and *2679–84 (Figure 6A–2). Among the defective CYP2C19 alleles, CYP2C19*2 and *3 are responsible for the majority of PM genotypes in the metabolism of CYP2C19 substrate drugs.75,85 The PM trait is inherited as an autosomal recessive mutation and these polymorphisms are apparently different among ethnic groups. PMs represent 3-6% of whites and blacks, 13-23% of Asians, and 38-79% of Polynesians and Micronesians.75,86 Ethnic differences in the frequency of functional CYP2C19 alleles are summarized in Table 6A–2. High interindividual variations in subjects homozygous for CYP2C19*1 are also observed and part of these variations can be explained by the genotyping for CYP2C19*17.93 CYP2C19*17 confers an increased CYP2C19 activity due to an increase in CYP2C19 transcription and therefore individuals with homozygous carriers of CYP2C19*17 exhibit the rapid clearance of CYP2C19 substrate drugs.91,93,99 Individuals having CYP2C19*17 are likely to exhibit a lack of response to certain PPIs and antidepressants when compared with the EMs (CYP2C19*1/*1).
FIGURE 6A–2 Summary of CYP2C19 alleles that exhibited altered functions compared with the wild-type in phenotyped human subjects. White boxes are 5′- and 3′-untranslated regions. Black boxes are exons in open reading frame. Arrow heads indicate mutated locations that result in amino acid change, stop codon (X), or splicing variant (SV).
TABLE 6A–2 Frequencies of CYP2C19 *2, *3, and *17 alleles in different ethnic populations.
Clinical Relevance of CYP2C19 Genetic Polymorphism
Lack or low activity of CYP2C19 enzyme can result in the prolonged accumulation of the CYP2C19 substrate drugs and this would lead to the drug toxicities or therapeutic failures. The altered CYP2C19 activity can be, at least in part, predicted by CYP2C19 genotype. The most extensively studied polymorphic variants of CYP2C19 are CYP2C19*2, *3, and *17 in clinical settings.12,75,85 For example, the low extent of biotransformation from the antimalarial drug proguanil to its active metabolite cycloguanil can lead to the insufficient efficacy of proguanil response.100,101 However, clinical implications of CYP2C19 genotype on the efficacy of proguanil are uncertain.12,102,103 This stems from the fact that other P450s such as CYP3A4 and CYP1A2 contribute to the formation of the active metabolite.104,105
Diazepam is demethylated by CYP2C19.106 The plasma half-life of diazepam is about 4-fold longer in individuals for PM genotypes than in individuals homozygous for the wild-type CYP2C19*1.64,65 Many Hong Kong physicians routinely prescribe a lower dose of diazepam for Chinese patients than for Caucasisans,107 which is now attributed to the high frequency of CYP2C19*2 and *3 in Asians.61 Toxic doses of diazepam may occur in PMs and therefore more care would be required for the decision of diazepam dose, particularly in Asians.
Individual variations in plasma concentrations of various PPIs can be accounted for the degree of CYP2C19 activity determined by S-mephenytoin 4′-hydroxylation assay.108 After oral administration of omeprazole, the AUC of S-mephenytoin is much higher in PMs than in the EMs.109 The extent of the contribution of CYP2C19 to the metabolism of various PPIs is different. For example, the ratios of the AUC values in PMs versus EMs are different in the decreasing order of omeprazole, pantoprazole, lansoprazole, and rabeprazole.101,110 Although all of these PPIs are affected by CYP2C19 genotypes, rabeprazole is less dependent on the CYP2C19 genotype compared with the other PPIs, since rabeprazole can be nonenzymatically converted to its metabolite. CYP2C19 genotype affects cure rates for H. pylori infection in peptic ulcer patients. For example, the cure rate of peptic or duodenal ulcers for Japanese patients who received dual therapy with omeprazole (20 mg per day for 2 weeks) and amoxicillin (2,000 mg per day for 2 weeks) was 100% in CYP2C19 PMs, 60% in individuals heterozygous for one mutant allele, and 29% in individuals homozygous for CYP2C19*1 allele.72 All of the past and current studies suggest that CYP2C19 PM genotype is beneficial in the treatment of gastric ulcer and gastroesophageal reflux disease due to the impaired metabolism of PPIs and therefore higher plasma concentrations of the PPIs in individuals having the CYP2C19 PM genotype.110
Increased enzyme activity of CYP2C19 can cause drug toxicity or increased drug efficacy. For instance, enhanced response to clopidogrel with the increased risk of bleeding was reported from CYP2C19*17 carriers due to the higher rate of biotransformation into the active metabolite.111 However, another study reported an improved protective effect of clopidogrel after myocardial infarction in patients carrying the CYP2C19*17 allele.112 The major contributors of clopidogrel activation have been identified by not only CYP2C19 but also CYP3A4 and paraoxonase-1.68,113 Furthermore, clopidogrel itself is a potent inhibitor of CYP2C19 and CYP3A4.114 Clinical relevance of CYP2C19 genotype on the efficacy of clopidogrel is under strong investigation for the genotype-guided therapy.115 In general, patients who carry one or two CYP2C19 loss-of-function alleles exhibit a higher adverse event rate and diminished platelet inhibition than those with the EM genotype in the use of clopidogrel.115–118 CYP2C19 PMs may not benefit from clopidogrel and therefore it is recommended to consider alternative treatments. However, no definitive recommendations have been established regarding dose adjustment of clopidogrel with CYP2C19 genotype testing. Further investigation on a large scale using genotype-guided clopidogrel therapy compared with other therapy options would reveal the cost-effectiveness of CYP2C19 genotype testing for clopidogrel therapy.
Perspective of Clinical Application
Translation of genetic information into the clinical practice requires two important issues, clinical significance and cost-effectiveness. CYP2C19 polymorphisms have been reported to affect the pharmacokinetics of the CYP2C19 substrate drugs such as diazepam, various PPIs, clopidogrel, and certain antidepressants (moclobemide, amitriptyline, clomipramine, sertraline, and citalopram). However, a significant relationship between CYP2C19 genotype and the clinical outcome has been limited to certain cases. CYP2C19 PMs have a markedly longer half-life of the common anxiolytic drug diazepam and the antiulcer drug omeprazole. Individuals with CYP2C19 PMs would require a much lower dose of diazepam for clinical relevance. Patients with CYP2C19 PMs exhibited an increased efficacy of omeprazole. However, CYP2C19 PMs have less benefit from the use of clopidogrel. The varied frequencies of CYP2C19 polymorphisms in different ethnic groups are evident, which results in variations in risk and efficacy of CYP2C19 substrate drugs in different ethnic populations. Many rare defective alleles have been reported in the CYP2C19 gene. Although there is a lack of statistically significant data, such as in vivo pharmacokinetics and pharmacodynamics, due to their low incidences in populations, these rare polymorphisms are important since CYP2C19 metabolizes many important clinical drugs with great penetrance. Functional studies of CYP2C19 polymorphisms and their genotype/phenotype relationships need to be established with respect to their contributions to the toxicity or efficacy of the CYP2C19 drugs, which would be useful in the development of a dose prediction model for personalized medicine in the future.
PHARMACOGENOMICS OF CYP2D6
Introduction
CYP2D6 is one of the most polymorphic P450s and metabolizes approximately 20% of currently prescribed drugs.99,119,120 However, hepatic content of CYP2D6 compared with major metabolic P450s is relatively small, about 2-4% of all P450 content.121–123 CYP2D6 is located in a highly polymorphic gene cluster together with two inactive pseudogenes, CYP2D7P and CYP2D8P.124–126 Although an active form of CYP2D7, a 138delT in exon 1 of CYP2D7P, was reported,127 it is generally accepted that humans carry only one active CYP2D6 gene.123,128 Representative substrates of CYP2D6 include antipsychotic drugs (haloperidol, clozapine, and risperidone), antiarrhythmic agents (flecainide and perphenazine), tricyclic antidepressants (imipramine, clomipramine, nortriptyline, and amitriptyline), β-adrenoreceptor antagonists (metoprolol, propranolol, bupranolol, and carvedilol), opioids (codeine and tramadol), and the estrogen receptor antagonist tamoxifen.122,123,129 Since CYP2D6 is not inducible, genetic polymorphisms greatly contribute to the interindividual variations in enzyme activity. Five of the prototypic substrates for CYP2D6 activity assessment are sparteine,129 debrisoquine,130–132 dextromethorphan,133–135 bufuralol,129,136,137 and tramadol.138–140 Among the five probe substrates, bufuralol and dextromethorphan are commonly used substrates in in vitro studies. The phenotype of CYP2D6 metabolism can be categorized based on genotypes such as UM (more than two functional alleles), EM (at least one or two normal functional alleles), IM (two reduced functional alleles or one null allele and the other allele with reduced function), and PM (two nonfunctional alleles). The phenotype of CYP2D6 metabolism shows ethnic differences; while UMs are mainly found in North Africa and Oceania, IMs are mainly from Asia, and high incidences of PMs are found in whites.75,122,123,129,141
Functional Polymorphism of CYP2D6
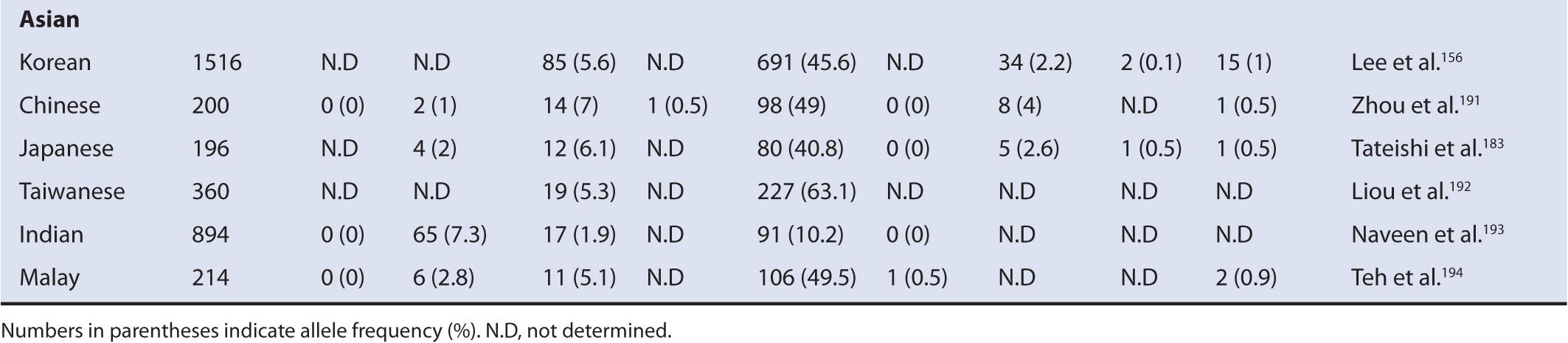
Currently, 81 CYP2D6 variant alleles were assigned to the Human Cytochrome P450 Allele Nomenclature Committee. Among them, the null alleles of CYP2D6 include *3, *4, *5, *6, *7, *8, *11, *12, *13, *14, *15, *16, *18, *19, *20, *21, *38, *40, *42, *44, *56, *60, and *62.126,142–170 Since the null allele has no activity, its clinical impact is greater than any of the other functional alleles and therefore most of the null alleles are related to the PM phenotype. Among these null alleles, the most frequently studied alleles that are responsible for most of the CYP2D6 PM phenotype are CYP2D6*3 (frameshift), CYP2D6*4 (splice variant), CYP2D6*5 (gene deletion), and CYP2D6*6 (frameshift). The most common CYP2D6 variants having reduced function include CYP2D6*10, *17, and *41. Some of these CYP2D6 variants exhibit a clear ethnic disposition. For example, CYP2D6*4 is found at a frequency of about 15% in whites171; however, this variant occurs at a frequency of about <5% in blacks172,173 and <1% in Asians.174 This varying frequency of CYP2D6*4 is the major reason for the ethnically different PM distributions, accounting for 5-10% PMs in whites,61,75,175 0-19% PM in blacks,75,176,177 and <1% PMs in Asians.75,141,178 The CYP2D6*10 allele is a reduced functional allele, most commonly found in Asians at up to the frequency of 63%.156,174,179–183 This allele contributes to a great extent of IM phenotype in Asians compared with the other ethnic populations. The highest prevalence of UM phenotype is found in Northeast Africa and the Oceania regions where individuals carrying duplication of CYP2D6*1 or *2 are high. For example, the frequency of CYP2D6*2xN is about 28% in the Mozabite population in Algeria and about 22% of CYP2D6*1xN has been found in Oceania,122,184,185 whereas the frequency of CYP2D6*5 is similar in different ethnic populations ranging from 4% to 7%.122,123,141 Ethnic differences in the frequency of functional CYP2D6 alleles are summarized in Table 6A–3. Genetic mutations of CYP2D6 can cause altered enzyme activity through various ways, which include changes in substrate recognition, enzyme affinity for substrates, protein stability, and regulation of expression. CYP2D6 alleles with reduced activity include CYP2D6*10, *14, *17, *18, *36, *41, *47, *49, *50, *51, *54, *55, *57, *59, *62, and *72 (http://www.cypalleles.ki.se/cyp2d6.htm). Alleles with similar activity to the wild-type include *27, *39, and *48.195 Most of these variant alleles are characterized in different labs with different protocols. Therefore, absolute determination of expressed activity through a quantitative manner is currently difficult. CYP2D6 gene exhibits copy number variations. For example, carriers of CYP2D6*2xN or CYP2D6*1xN show extremely high activity compared with the wild-type. Individuals having duplication or multiplication of the CYP2D6 active gene are expected to express significantly increased CYP2D6 substrate drug clearance, resulting in a lack of therapeutic effect or adverse drug reaction in some cases due to the significantly increased metabolism of the precursor into the active form of drug.
TABLE 6A–3 Frequencies of major CYP2D6 alleles in different ethnic populations.
Clinical Relevance of CYP2D6 Genetic Polymorphism
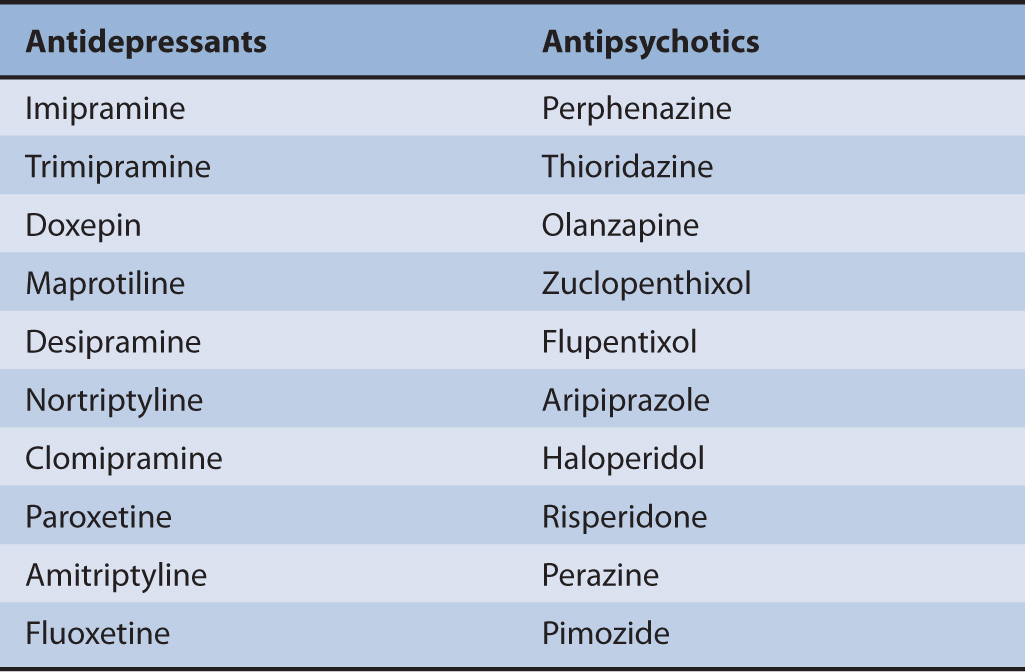
Various systems have been developed to categorize CYP2D6 activity based on its genotype, such as UM, EM, IM, PM, and activity scores.99,141,196 Many clinical drugs are greatly influenced by CYP2D6 genetic polymorphisms. First, antidepressants and antipsychotics exhibit significant differences in plasma concentrations, depending on CYP2D6 polymorphisms.197–199 Several studies have shown that CYP2D6 PMs exhibit a higher incidence of undesirable side effects when taking CYP2D6-dependent antidepressants,200–208 antipsychotic drugs,209–216 and selective serotonin reuptake inhibitors (SSRIs).217–220 Similarly, multiple studies have found that CYP2D6 UMs experience diminished response to these drugs.198,202,221,222 A list of the top 10 antidepressants and antipsychotic drugs whose plasma concentrations are greatly influenced by CYP2D6 polymorphisms is presented in Table 6A–4. Several articles suggest that physicians might consider reducing the dose of tricyclic antidepressants by about 50% of the normally prescribed dose for PMs and 140-180% for UMs.141,198,223 However, routine testing for CYP2D6 genotype has not been approved by the FDA or endorsed by an expert panel, since the clinical significance and cost-effectiveness of genotyping have not been demonstrated.
TABLE 6A–4 List of the top 10 antidepressants and antipsychotic drugs that are greatly influenced by CYP2D6 genotypes.
Tamoxifen metabolism by CYP2D6 into its active agent is critical for its antitumor activity in breast cancer patients.224–226 Data from numerous studies support that the relationship between CYP2D6 genotype and tamoxifen response is clinically relevant.227–230 The CYP2D6 “PM” genotype has been associated with poor clinical outcome with tamoxifen.225,231–236 Tamoxifen-treated patients homozygous for CYP2D6*4 exhibited a shorter disease-and relapse-free survival rate.233,234,237,238 Similarly, patients carrying the CYP2D6 “poor” and “intermediate metabolizer” genotypes showed a significantly increased incidence of recurrence, shorter relapse-free survival, and shorter event-free survival rates.229,239,240 In addition, patients having CYP2D6 UM genotype were associated with significantly longer relapse-free time, less breast cancer recurrences, and higher event-free survival compared with CYP2D6 EM carriers.229,237,239,241
Increased codeine metabolism by CYP2D6 to morphine has been shown to cause serious morphine toxicity.242,243 CYP2D6 UM genotype exhibited about a 50% higher concentration of morphine compared with the EM genotype. However, individuals having defective alleles of CYP2D6 exhibited very low or undetectable morphine concentrations with a lack of analgesia.244–246 A case report that a breastfed infant died from toxic levels of morphine strongly suggested an association between CYP2D6 genotype and morphine toxicity.243 Both the mother and infant were CYP2D6 UM genotypes, resulting in high concentrations of morphine in the breast milk and blood due to the rapid and extensive conversion of codeine to morphine.
Tramadol is metabolized by CYP2D6 to generate a pharmacologically active O-desmethyltramadol and CYP2D6 genotype is shown to be linked to the concentration of O-desmethyltramadol, resulting in the different efficacy of tramadol treatment.247–249 Individuals with the CYP2D6 PM genotypes exhibited lesser analgesic effect than the EM genotypes.250–252 Patients carrying CYP2D6 UM genotype have higher concentrations of O-desmethyltramadol and better pain control than those carrying the EM genotype, but higher incidences of the side effects (i.e., nausea, respiratory depression) have been reported.253,254 Therefore, CYP2D6 UMs might be at greater risk from regular tramadol doses and might benefit from a lower dose of tramadol.
Pharmacokinetic parameters for beta-adrenergic receptor antagonists, such as carvedilol, metoprolol, propranolol, and timolol, have been reported to be linked to CYP2D6 genetic polymorphisms.123,255 Metoprolol consists of a racemic mixture of S– and R-metoprolol with the S-metoprolol exhibiting about a 500-fold greater affinity for beta1-adrenergic receptors than the R-form.256 Alpha-hydroxylation of metoprolol is converted by CYP2D6 and O-demethylation is metabolized by CYP2D6 and CYP3A4.257,258 Among the several metabolic pathways of carvedilol, 4′- and 5′-hydroxylations of carvedilol are mainly catalyzed by CYP2D6 with a minor contribution of these reactions catalyzed by CYP2E1 and CYP2C9.259
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree