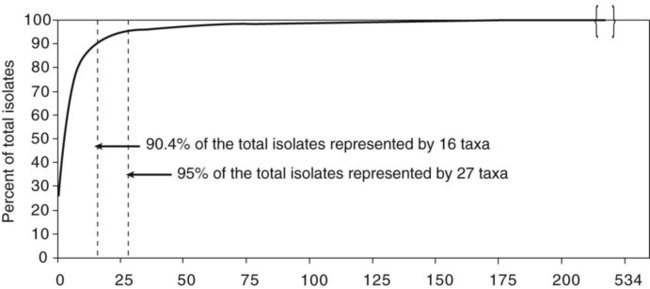
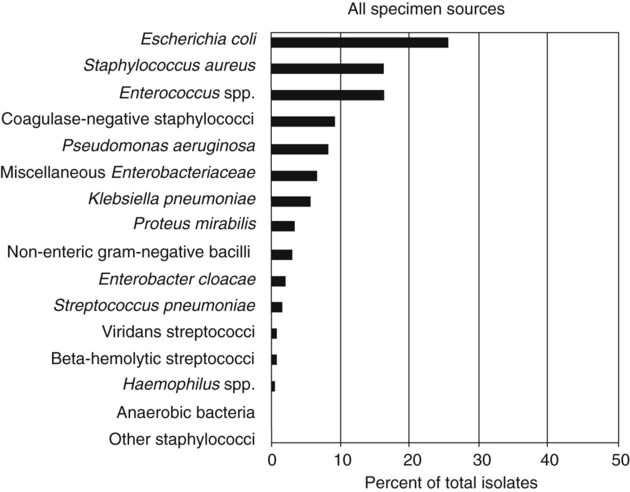
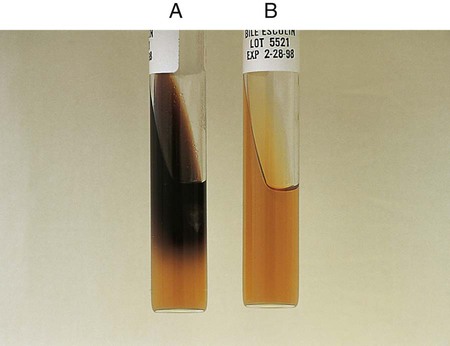
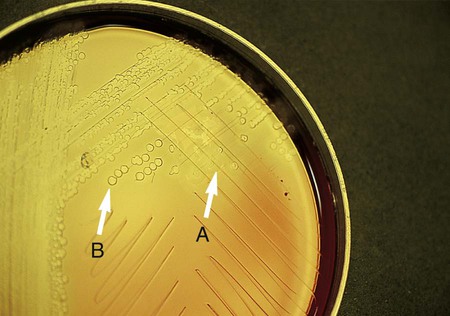
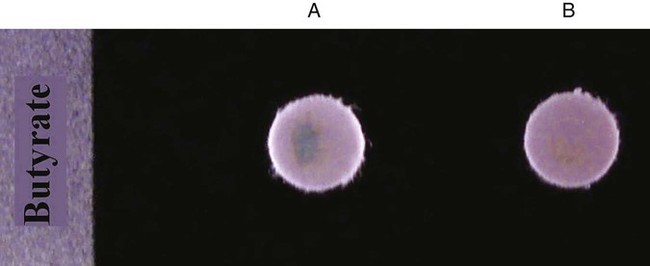
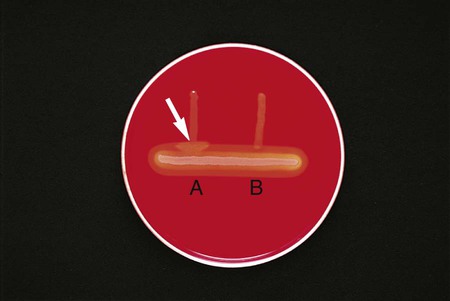
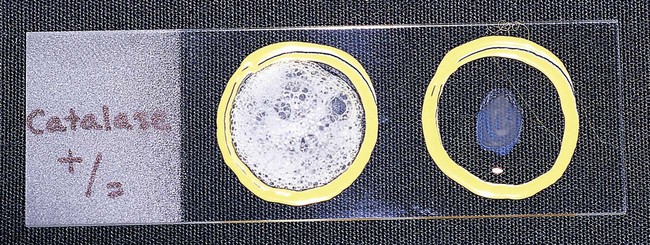
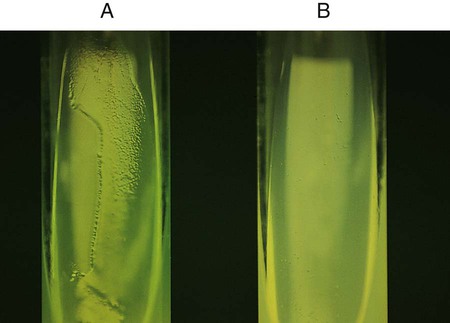
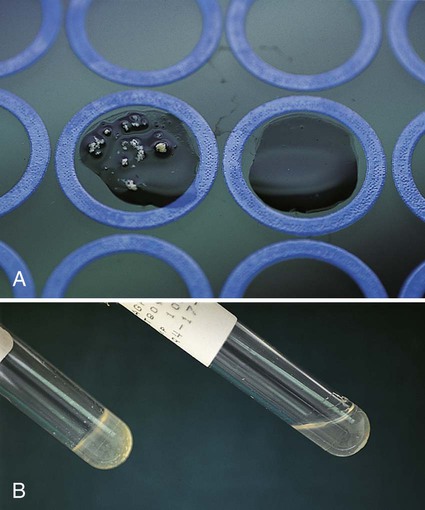
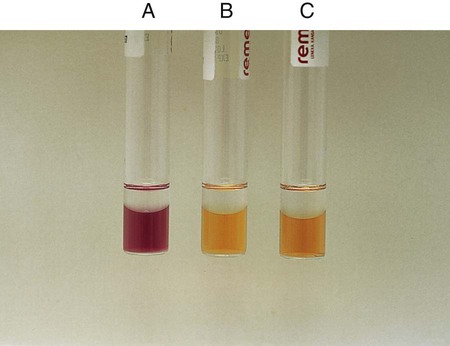
Chapter 13 1. State the specific diagnostic purpose for each test methodology. 2. Briefly describe the test principle associated with each test methodology. 3. Outline limitations and explain ways to trouble-shoot or report results in the event the test result indicates a false positive or false negative or is equivocal. 4. State the appropriate quality control organisms and results used with each testing procedure. It is challenging to determine how most effectively to present and teach diagnostic microbiology in a way that is sufficiently comprehensive and yet not excessively cluttered with rare and seldom-needed facts about bacterial species uncommonly encountered. Approximately 530 different bacterial species or taxa are reported by clinical microbiology laboratories across the United States (Figure 13-1). Yet 95% of the bacterial identifications reported are distributed across only 27 of these taxa. This is an indication of how infrequently the other 500 or more taxa are identified and reported. Therefore, although the chapters in Part III, Bacteriology, are intended to be comprehensive in terms of the variety of bacterial species presented, it is helpful to keep in perspective which taxa are most likely to be encountered in the clinical environment. The relative frequencies with which the common bacterial species and organism groups are reported in clinical laboratories are presented in Figure 13-2. To meet the challenges of bacterial identification processes beyond what can be portrayed in flow charts, the chapters in Part III have been arranged to guide the student through the entire workup of a microorganism, beginning with initial culture of the specimen. In most instances, the first information a microbiologist uses in the identification process is the macroscopic description of the colony, or colony morphology. This includes the type of hemolysis (if any), pigment (if present), size, texture (opaque, translucent, or transparent), adherence to agar, pitting of agar, and many other characteristics (see Chapter 7). After careful observation of the colony, the Gram stain is used to separate the organism into a variety of broad categories based on Gram stain reaction and the cellular morphology of gram-positive or gram-negative bacteria (e.g., gram-positive cocci, gram-negative rods; see Chapter 6). For gram-positive organisms, the catalase test should follow the Gram stain, and testing on gram-negative organisms should begin with the oxidase test. These simple tests, plus growth on MacConkey agar, if the isolate is a gram-negative rod or coccobacillus, help the microbiologist assign the organism to one of the primary categories (organized here as subsections). Application of the various identification methods and systems outlined in this chapter generate the data and criteria discussed in each chapter for the definitive identification of clinically relevant bacteria. Most of the procedures described in the following chapters can be found at the end of this chapter. In this chapter, each procedure includes a photograph of positive and negative reactions. Chapter 6 includes photographs of some commonly used bacteriologic stains. In addition, Table 13-1 lists several commonly used commercial identification systems for a variety of the microorganisms discussed in the following pages. TABLE 13-1 Examples of Commercial Identification Systems for Various Organisms *Durham, N.C.: www.bioMerieux-Vitek.com †Sparks, Md.: www.bectondickinson.com ‡Lenexa, Kan.: www.remelinc.com §West Sacramento, Calif.: www.dadebehring.com
Overview of Bacterial Identification Methods and Strategies
Rationale for Approaching Organism Identification
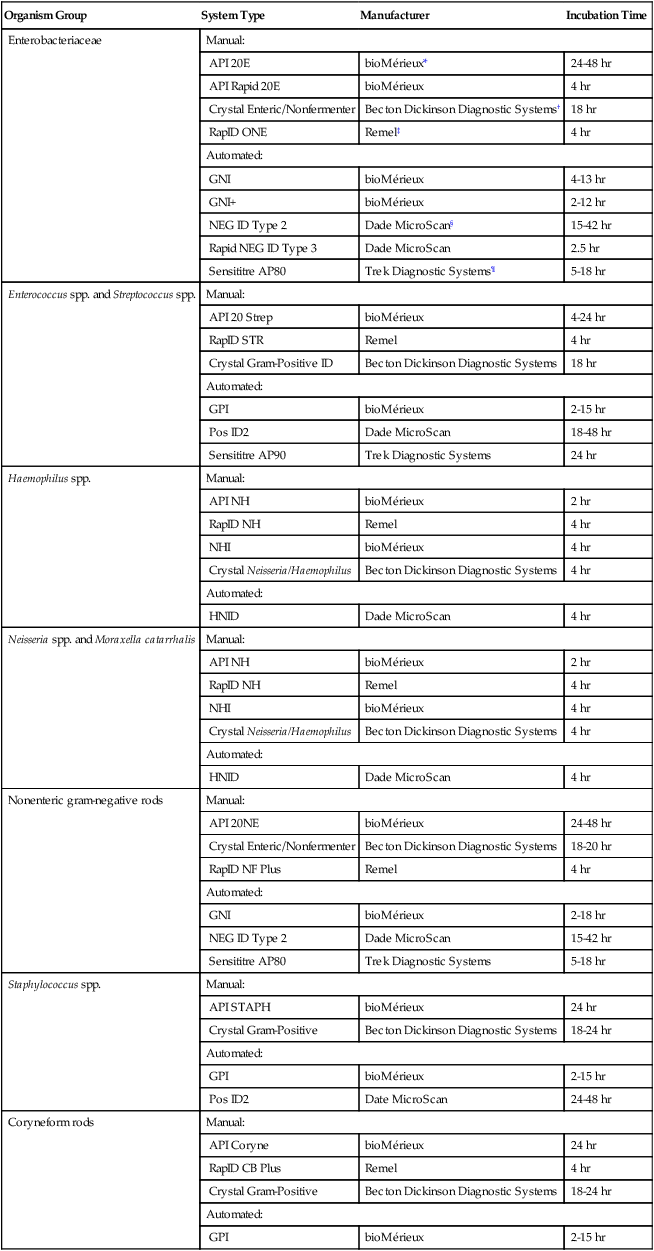
Organism Group
System Type
Manufacturer
Incubation Time
Enterobacteriaceae
Manual:
API 20E
bioMérieux*
24-48 hr
API Rapid 20E
bioMérieux
4 hr
Crystal Enteric/Nonfermenter
Becton Dickinson Diagnostic Systems†
18 hr
RapID ONE
Remel‡
4 hr
Automated:
GNI
bioMérieux
4-13 hr
GNI+
bioMérieux
2-12 hr
NEG ID Type 2
Dade MicroScan§
15-42 hr
Rapid NEG ID Type 3
Dade MicroScan
2.5 hr
Sensititre AP80
Trek Diagnostic Systems¶
5-18 hr
Enterococcus spp. and Streptococcus spp.
Manual:
API 20 Strep
bioMérieux
4-24 hr
RapID STR
Remel
4 hr
Crystal Gram-Positive ID
Becton Dickinson Diagnostic Systems
18 hr
Automated:
GPI
bioMérieux
2-15 hr
Pos ID2
Dade MicroScan
18-48 hr
Sensititre AP90
Trek Diagnostic Systems
24 hr
Haemophilus spp.
Manual:
API NH
bioMérieux
2 hr
RapID NH
Remel
4 hr
NHI
bioMérieux
4 hr
Crystal Neisseria/Haemophilus
Becton Dickinson Diagnostic Systems
4 hr
Automated:
HNID
Dade MicroScan
4 hr
Neisseria spp. and Moraxella catarrhalis
Manual:
API NH
bioMérieux
2 hr
RapID NH
Remel
4 hr
NHI
bioMérieux
4 hr
Crystal Neisseria/Haemophilus
Becton Dickinson Diagnostic Systems
4 hr
Automated:
HNID
Dade MicroScan
4 hr
Nonenteric gram-negative rods
Manual:
API 20NE
bioMérieux
24-48 hr
Crystal Enteric/Nonfermenter
Becton Dickinson Diagnostic Systems
18-20 hr
RapID NF Plus
Remel
4 hr
Automated:
GNI
bioMérieux
2-18 hr
NEG ID Type 2
Dade MicroScan
15-42 hr
Sensititre AP80
Trek Diagnostic Systems
5-18 hr
Staphylococcus spp.
Manual:
API STAPH
bioMérieux
24 hr
Crystal Gram-Positive
Becton Dickinson Diagnostic Systems
18-24 hr
Automated:
GPI
bioMérieux
2-15 hr
Pos ID2
Date MicroScan
24-48 hr
Coryneform rods
Manual:
API Coryne
bioMérieux
24 hr
RapID CB Plus
Remel
4 hr
Crystal Gram-Positive
Becton Dickinson Diagnostic Systems
18-24 hr
Automated:
GPI
bioMérieux
2-15 hr