Nonhuman Lentiviruses
David T. Evans
John H. Elder
Ronald C. Desrosiers
History
Use of the term “slow virus infections” and identification of the first lentivirus is generally credited to Sigurdsson et al.314,315,316 in Iceland. Twenty karakul sheep that were imported from Germany to Iceland in 1933 were responsible for the transmission of a chronic disease and death in massive numbers of Icelandic sheep over subsequent years. More than 100,000 Icelandic sheep died of the disease in the decades that followed. Sigurdsson et al.314,315,316 not only described the disease but also demonstrated that it was due to a transmissible agent. They used the term slow virus infections to refer to this disease as well as to what is now known as scrapie.313 The term has since been used to describe a wide variety of nonacute, persisting viral infections. In 1960, Sigurdsson et al.317 described the cultivation of the transmissible agent in tissue culture, and Gudnadottir and Palsson115 were able to reproduce the disease with the culture-grown virus. The diseases in the sheep were called maedi (Icelandic for dyspnea, that is, a lung disease resulting in difficulty breathing) and visna (Icelandic for a state of progressive apathy, a “fading away,” resulting from brain disease). Both disease states result from the same virus, now referred to as maedi/visna virus (MVV). MVV and related viruses are called lentiviruses, derived from the Latin lentus for slow. Approximately 600,000 sheep were slaughtered in Iceland in 1965 to eradicate MVV from the island.
The maedi/visna disease in Icelandic sheep, although the first specifically shown to be caused by a defined lentivirus, is probably not the first description of a lentiviral disease in the literature. Vallée and Carré described in 1904 the infectious nature of the disease equine infectious anemia,343 which is now known to be caused by a lentivirus (EIAV). Even for MVV, a chronic, progressive interstitial pneumonia had been described in South African sheep in 1915 and in Montana sheep in 1923.213 Work with EIAV was largely on a parallel track with that of MVV, and the first description of EIAV cultivation appeared in 1961.172 Only subsequently were EIAV and MVV shown to belong to the same lentivirus subfamily of retroviruses on the basis of morphologic criteria.111,253,363
The scenario of disease outbreak in a susceptible population leading to the identification of a new lentivirus has repeated itself dramatically on several occasions in more recent history. In 1964, an emerging infectious disease was first detected in Bali cattle in the Jembrana district of Bali.272,321 Bali cattle are the domesticated form of the wild banteng (Bos javanicus). Within 12 months, 26,000 of the 300,000 cattle on the island died of the disease. The cause of the disease was subsequently traced to a bovine lentivirus now called Jembrana disease virus (JDV), a distinct variant of bovine immunodeficiency virus (BIV).36,162 Bali cattle are particularly sensitive to disease caused by this virus, whereas other species are resistant.322,323,366 Outbreaks of immunodeficiency disease and lymphoma in captive colonies of macaque monkeys (Asian Old World primates) were subsequently traced to the introduction of a simian lentivirus or lentiviruses from African monkeys.18,61,201,208 Simian immunodeficiency viruses (SIVs) naturally infect African nonhuman primates without apparent disease, but Asian macaques appear to harbor no such virus naturally. Even the origins of human immunodeficiency virus (HIV) in people have followed a similar pattern. HIV-2 in people in western Africa clearly originated from SIV of sooty mangabey monkeys.41,70,99,129,184,214 The natural habitat of sooty mangabey monkeys is the same geographic region in western Africa where HIV-2 is endemic, and SIV from sooty mangabey monkeys groups phylogenetically with HIV-2 distinct from other lineages of primate lentiviruses. Sooty mangabey monkeys are naturally infected with SIV at high frequency without apparent disease. The origins
of HIV-1 in central Africa have similarly been traced to the chimpanzee Pan troglodytes.51,97,305 These examples illustrate the importance of studying animal viruses even when they are not apparently associated with any disease.
of HIV-1 in central Africa have similarly been traced to the chimpanzee Pan troglodytes.51,97,305 These examples illustrate the importance of studying animal viruses even when they are not apparently associated with any disease.
Table 51.1 Known Lentiviruses | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
The earliest descriptions of the isolation of HIV and its association with acquired immunodeficiency syndrome (AIDS) did not appreciate that the virus was a lentivirus.13,270 Only subsequently, through more careful examination of electron micrographs and through sequence analysis, did this become clear.112,227 At the time, study of lentiviruses was an obscure discipline with which many scientists were not familiar.
From a historical perspective, MVV317 and EIAV172 were discovered, isolated, and characterized long before the discovery of HIV; the field has retained use of the original designations for these viruses. Lentivirus groupings identified after the discovery of HIV have used a nomenclature similar to that for HIV (i.e., “immunodeficiency virus”). BIV, originally isolated from a cow with a chronic disease by Van Der Maaten et al. in 1972,345 received little attention until after the discovery of HIV. Subsequent to the discovery of HIV, lentiviruses were isolated from monkeys and cats (Table 51.1). Although discovery of MVV, EIAV, and BIV predates that of HIV, HIV has received such intense scrutiny that much more is known about it than all other lentiviruses in terms of the level of detailed knowledge. New information about other lentiviruses is thus usually compared with what is known for HIV.
Infectious Agent
Overview of General Properties
Related lentiviruses have been isolated from sheep, goats, horses, cattle, cats, monkeys, and humans (Table 51.1). Genetic analysis of the virus from goats (caprine arthritis encephalitis virus; CAEV) indicates that it clusters closely with MVV,342 therefore is placed in a single grouping with MVV. Based on host species and the genetic analyses described in more detail later in this chapter (Genome Organization and Composition section), five discrete evolutionary groupings of lentiviruses are generally recognized (Fig. 51.1A). It is likely that more remain to be identified. It is important to realize that even within a single grouping, discrete subgroupings based on host species, geography, and genetic distance can be defined. For example, among the nonhuman primate lentiviruses, distinctly different subgroupings exist for the SIVs from African green monkeys, sooty mangabey monkeys, Sykes’ monkeys, and L’hoesti monkeys (Fig. 51.1B). Even among African green monkeys, which inhabit virtually all of sub-Saharan Africa, discrete genetic sub-subgroupings of SIVagm can be defined that correlate with subspecies and natural geographic habitat (Table 51.2 and Fig. 51.1B).
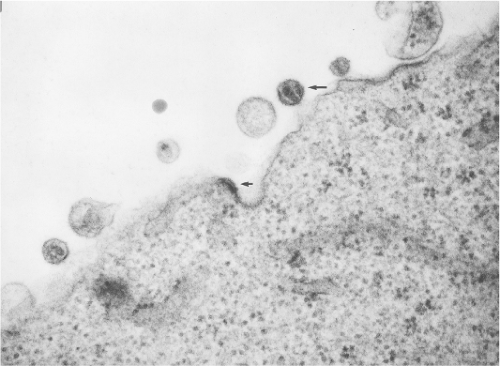
All lentiviruses have a common morphogenesis and morphology that distinguish them from other subgroups of retroviruses (see Chapter 47). Lentiviruses bud from the plasma membrane without a preformed nucleoid, and mature particles typically have a conical or rod-shaped nucleoid (Fig. 51.2). Classification into the lentivirus subgroup by morphologic criteria alone is entirely consistent with phylogenetic analysis of pol gene sequences. Viruses classified as lentiviruses have pol gene sequences more closely related to one another than to other retroviruses, and all have the characteristic morphogenesis/morphology. All lentiviruses have a propensity to replicate in macrophages, and all are associated with a chronic progressive disease course in susceptible hosts. The primate lentiviruses have acquired use of CD4 as one of two receptors used sequentially by virus for entry into cells (Table 51.3). FIV has been shown to use an analogous two-receptor sequential mechanism for entry. However, lentiviruses from nonprimates, including FIV, do not use CD4 as a receptor for entry. The chronic disease induced by the primate lentiviruses in susceptible hosts has an immunodeficiency component presumably because of the targeting of CD4+ lymphocytes by use of CD4 as a receptor. All lentiviruses have a number of auxiliary genes (i.e., genes in addition to gag, pol, and env not found in other, simpler retroviruses). Major distinguishing properties are summarized in Table 51.3.
Genome Organization and Composition
Widespread availability of DNA sequencing capabilities has greatly facilitated our understanding of phylogenetic relationships and the gene products of lentiviruses. Based on sequence analysis, five discrete groupings of lentiviruses are now recognized (see Fig. 51.1A). It is likely that other groupings remain to be discovered. Extensive diversity exists even within a grouping. For example, among the SIVs, distinct subgroups have been defined from the African green monkey, sooty mangabey monkey, L’hoesti monkey, and Sykes’ monkey (see Fig. 51.1B and Table 51.2). In all, 14 discrete phylogenetic lineages of primate lentiviruses have so far been defined (Fig. 51.1B and Table 51.2). Of the 69 species known to inhabit sub-Saharan Africa, SIV infection has been demonstrated in 40 of them, partial SIV sequence information is available from 32 species, and complete SIV genome sequences are available from 20 species.1 Since some species have not yet been surveyed, additional SIV lineages may still remain to be discovered. Extensive diversity has also been observed for FIV in wild and captive cat species.34,336 Even among the four subspecies of African green monkeys (vervet, grivet, tantalus, and sabeus), discrete sub-subgroupings of SIVagm have been defined (Table 51.2). These four subspecies naturally inhabit distinct or sometimes partially overlapping habitats that cover almost all of sub-Saharan Africa. JDV represents a distinct subgroup relative to the original BIV isolate, analogous to SIVagm versus SIVsm.36
The pol gene generally exhibits the greatest degree of sequence conservation. Thus, pol gene sequences are often used for the comparison of lentiviruses from different groups, subgroups, or sub-subgroups. Sequences from one subgroup of SIV (e.g., SIVsm) will typically exhibit only 55% to 60% identity in pol at the amino acid level when compared with sequences from another SIV subgroup (e.g., SIVagm). Diverse members within a subgroup may exhibit as little as 75% to 80% amino acid identity in pol, but the number is typically much higher. When different groups of lentiviruses are compared—for example, MVV with SIV—amino acid identity in pol is typically 35% or less. Relatedness in other genes is even less than that. Nonetheless, these lentiviral sequences are clearly more closely related to one another than to other retroviruses, justifying their classification as lentiviruses on the basis of morphogenesis/morphology and biologic properties.
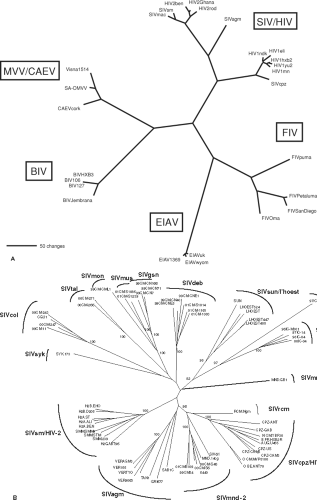 Figure 51.1. Phylogeny of the lentiviruses. A: Five discrete evolutionary groupings of lentiviruses. The unrooted tree depicts the phylogenetic relationships among the five recognized groups of lentiviruses. The tree is based on a 470-amino-acid alignment of reverse transcriptase sequences from representative members of each group, including the bovine (BIV, Jembrana), equine (EIAV), feline (FIV), ovine/caprine (visna, CAEV) and primate (SIV, HIV) lentiviruses. Maximum parsimony (shown) and neighbor-joining (not shown) analyses give trees of nearly identical topology. Branch lengths are proportional to the number of amino acid replacements. B: Phylogeny of primate lentiviruses. The 14 groupings of primate lentiviruses are shown. (See Table 51.2 for species codes.) [Adapted from Courgnaud V, Formenty P, Akoua-Koffi C, et al. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus). J Virol 2003;77:744–748.] |
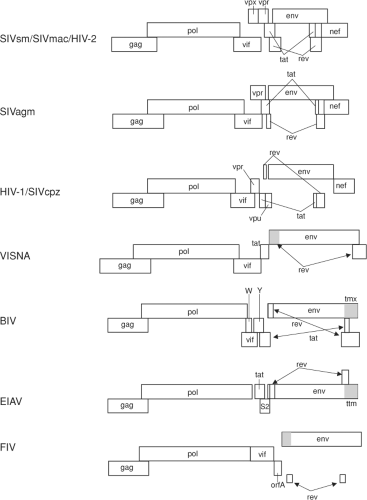
All known lentiviruses have at least three genes in addition to the standard gag, pol, and env genes that all replication-competent retroviruses possess (Tables 51.4 and 51.5, and Fig. 51.3). These extra genes likely contribute to a more complex life cycle for the lentiviruses, which includes persistent viral replication and strategies for immune evasion discussed in more detail later in this chapter (Immune Responses and Persistence section). A rev gene that encodes a protein responsible for controlling the relative level of full-length vs. multiply spliced viral messenger RNAs (mRNAs) is present in all lentiviruses, along with a downstream nucleotide sequence referred to as the Rev-Response Element (RRE). Interestingly, neither the sequence
nor location of the RRE is conserved, but the function appears to be the same in all lentiviruses. A vif gene, whose main role appears to be to counteract the cellular restriction factor APOBEC, is consistently present in five of the six lentivirus groupings. EIAV, however, stands alone in apparently lacking a vif gene. Whether another protein may contribute vif function for EIAV remains to be determined. A nef gene is found at the 3′ end of all primate lentiviruses (Table 51.5). None of the nonprimate lentiviruses contains a nef gene. However, cells infected with EIAV and BIV have been found to make spliced RNAs that predict protein products called Ttm and Tmx that correspond to the carboxy-terminal portion of transmembrane protein (TM).17,110 Whether these represent evolutionary precursors to nef or perform functions similar to those of nef is yet to be determined. All lentiviruses except FIV contain an unusually long (>120 amino acids) cytoplasmic domain of TM. In the intergenic region between vif and env, a number of other genes can be present, particularly in the primate lentiviruses. A vpr gene is present in this region in all primate lentiviruses that have been examined. The SIVsm/SIVmac/HIV-2, SIVrcm and SIVmnd2 subgroups of viruses contain an additional homolog of vpr called vpx (Table 51.5). The nonprimate lentiviruses also contain a gene between the vif and env genes with varied roles. BIV encodes a Tat protein from this location that acts on a downstream Tar element similar to the primate lentiviruses.197 A small protein is also encoded from this region of the ruminant lentiviruses that was originally thought to be a transcriptional transactivator but which may actually be more vpr-like in activity.354 Likewise, a small protein encoded in a similar region by FIV, termed OrfA, was originally thought to be a transactivator, but has recently been shown to downregulate CD134 receptor expression on the infected cell, similar to CD4 downregulation by HIV and SIV Nef.133 Only two of the 14 groups of primate lentiviruses that have been analyzed to date consistently have a gene called vpu: HIV-1/SIVcpz and SIVgsn/SIVmon/SIVmus (Table 51.5). SIVden from a pet Dent’s Mona monkey (Cercopithecus mona denti) was found to contain a vpu gene, although the virus clustered phylogenetically more closely to the SIV from DeBrazza monkeys, SIVdeb.62
nor location of the RRE is conserved, but the function appears to be the same in all lentiviruses. A vif gene, whose main role appears to be to counteract the cellular restriction factor APOBEC, is consistently present in five of the six lentivirus groupings. EIAV, however, stands alone in apparently lacking a vif gene. Whether another protein may contribute vif function for EIAV remains to be determined. A nef gene is found at the 3′ end of all primate lentiviruses (Table 51.5). None of the nonprimate lentiviruses contains a nef gene. However, cells infected with EIAV and BIV have been found to make spliced RNAs that predict protein products called Ttm and Tmx that correspond to the carboxy-terminal portion of transmembrane protein (TM).17,110 Whether these represent evolutionary precursors to nef or perform functions similar to those of nef is yet to be determined. All lentiviruses except FIV contain an unusually long (>120 amino acids) cytoplasmic domain of TM. In the intergenic region between vif and env, a number of other genes can be present, particularly in the primate lentiviruses. A vpr gene is present in this region in all primate lentiviruses that have been examined. The SIVsm/SIVmac/HIV-2, SIVrcm and SIVmnd2 subgroups of viruses contain an additional homolog of vpr called vpx (Table 51.5). The nonprimate lentiviruses also contain a gene between the vif and env genes with varied roles. BIV encodes a Tat protein from this location that acts on a downstream Tar element similar to the primate lentiviruses.197 A small protein is also encoded from this region of the ruminant lentiviruses that was originally thought to be a transcriptional transactivator but which may actually be more vpr-like in activity.354 Likewise, a small protein encoded in a similar region by FIV, termed OrfA, was originally thought to be a transactivator, but has recently been shown to downregulate CD134 receptor expression on the infected cell, similar to CD4 downregulation by HIV and SIV Nef.133 Only two of the 14 groups of primate lentiviruses that have been analyzed to date consistently have a gene called vpu: HIV-1/SIVcpz and SIVgsn/SIVmon/SIVmus (Table 51.5). SIVden from a pet Dent’s Mona monkey (Cercopithecus mona denti) was found to contain a vpu gene, although the virus clustered phylogenetically more closely to the SIV from DeBrazza monkeys, SIVdeb.62
Table 51.2 Detailed Listing of Primate Lentivirusesa | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 51.3 Properties of Lentiviruses | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 51.4 Auxiliary Genes in Nonprimate Lentiviruses | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||
Table 51.5 Auxiliary Genes Primate Lentiviruses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Except for BIV, nonprimate lentiviruses encode a deoxyuridine triphosphatase (dUTPase) activity from a distinct open reading frame located within the pol gene.85 dUTPase converts dUTP to a precursor of dTTP, thus keeping the concentration of dUTP in the cell at low levels. By doing so, dUTPase indirectly prevents misincorporation of uracil into viral DNA, ultimately reducing mutagenic G>A transitions. dUTPase is particularly relevant for virus replication in nondividing cells like macrophages in which endogenous dUTPase levels are low. Curiously, these dUTPase coding sequences are absent in the primate lentiviruses but present in the type D beta retroviruses. The explanation for this variable presence of dUTPase is not entirely clear. The type D retroviruses and lentiviruses may possibly have diverged from a common ancestor that possessed such a dUTPase reading frame, and this reading frame may then have been lost in the evolution of primate lentiviruses from the more primitive ones. The type D retroviruses and lentiviruses do share a common morphology of the mature particles (i.e., a cylindrical, rod-shaped, or conical nucleoid), which also may suggest some commonality. Alternatively, the presence of a dUTPase reading frame in nonprimate lentiviruses and in type D retroviruses could be an example of convergent evolution, with two distinct lineages acquiring these sequences independently by gene capture or by sequence duplication and point mutation. Another possibility is that one viral lineage may have acquired dUTPase sequences from the other viral
lineage by a recombination or capture event.12 The presence of a dUTPase in the endogenous SIV found in the germline of the prosimian lemur107 supports the notion that dUTPase was lost in the evolution of nonprimate to primate lentiviruses. The loss of dUTPase in the evolution from nonprimate to primate lentiviruses could relate at least in part to the predominant replication of nonprimate lentiviruses in cells with little potential to divide (macrophages) where endogenous dUTPase levels are low versus the predominant replication of primate lentiviruses in cells with much greater potential to divide (lymphocytes) in which high levels of endogenous dUTPase prevail. Primate lentiviruses incorporate uracil DNA glycosylase into virions through its binding to vpr,209 thus facilitating excision and subsequent repair of misincorporated uracil in newly synthesized viral DNA. The presence of the dUTPase or uracil DNA glycosylase in virions serves to reduce the mutation frequency in reverse transcription products.
lineage by a recombination or capture event.12 The presence of a dUTPase in the endogenous SIV found in the germline of the prosimian lemur107 supports the notion that dUTPase was lost in the evolution of nonprimate to primate lentiviruses. The loss of dUTPase in the evolution from nonprimate to primate lentiviruses could relate at least in part to the predominant replication of nonprimate lentiviruses in cells with little potential to divide (macrophages) where endogenous dUTPase levels are low versus the predominant replication of primate lentiviruses in cells with much greater potential to divide (lymphocytes) in which high levels of endogenous dUTPase prevail. Primate lentiviruses incorporate uracil DNA glycosylase into virions through its binding to vpr,209 thus facilitating excision and subsequent repair of misincorporated uracil in newly synthesized viral DNA. The presence of the dUTPase or uracil DNA glycosylase in virions serves to reduce the mutation frequency in reverse transcription products.
Table 51.6 Cell Substrates for Growing Lentiviruses | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Propagation
Propagation and Assay in Cell Culture
Primary isolates from all lentivirus groupings can be grown in normal macrophage cultures of the host species. Other types of cells may be more typically used for propagating the individual viruses in routine cell culture. The types of cells that can be used vary with the virus (Table 51.6) and almost certainly relate to the kinds of receptor that can be used in most cases. Isolates also can be adapted to replicate on particular cell substrates. Examples of the latter include growth of FIV on Crandell feline kidney cells and growth of SIVs and HIVs on human tumor T-cell lines (Table 51.6). It is important to note that the strong selective forces that allow replication in particular cells may make research convenient but may lead to results atypical of natural infection.
Host Range
The lentiviruses are typically restricted in their host range, but both natural and experimental cross-species infections have been documented. SIVs naturally infect a variety of African nonhuman primates, but a single example of natural infection of Asian Old World monkeys has never been documented. The SIVs from sooty mangabey monkeys and African green monkeys, when introduced into macaque monkeys (Asian Old World primates), can infect them and cause an AIDS-like disease.94,127,233 In fact, the SIV of sooty mangabey monkeys was accidentally introduced into macaque monkeys in captive United States colonies on at least one occasion and disseminated unknowingly into other macaques for more than a decade before it was discovered and eliminated.8,59,208 At least one clear case of laboratory-acquired infection of a human with SIVmac has been documented.166,325
Cross-species transmission of SIV to baboons in the wild has been documented. Baboons do not appear to harbor their own SIV naturally. Of 279 baboon sera taken from native habitats in Tanzania and Ethiopia, 277 were negative for the presence of antibodies to SIV. However, two gave strong reactivity to SIVagm antigens.173 One of these two was subsequently shown to harbor SIVagm sequences in stored plasma. The SIVagm sequences clustered with the vervet subtype, consistent with the known cohabitation of vervet monkeys in the same region where the baboon samples were taken.151 An SIVagm variant has similarly been detected in a chacma baboon of southern Africa.347 Baboons also can be infected experimentally with HIV-2.198 Evidence for cross-species transmission of SIV from West African green monkeys (Cercopithecus aethiops sabeus) to patas monkeys (Erythrocebus patas) and to white-crowned mangabeys (Cercocebus torquatus lunulatus) has also been presented.20,334 Transmission of SIV or HIV to nonprimates has not been documented.
The origins of both HIV-2 and HIV-1 in humans are now generally believed to be from cross-species transmission. HIV-2 is endemic in western Africa and has only slowly made its way to other regions of the world. The SIV from sooty mangabey monkeys is closely related to HIV-2, has the same genome organization as HIV-2, and groups phylogenetically with HIV-2, distinct from the other 13 groups of primate lentiviruses (Fig. 51.1B). The native habitat of the sooty mangabey monkey is the coastal forest regions of western African, the same region where HIV-2 is endemic. Thus, strong circumstantial evidence involving both viral sequences and geographic co-localization link the monkey and human lentiviruses in western Africa.42,70,99,129 Similarly, of the four subspecies of chimpanzees, at least two (Pan troglodytes troglodytes and Pan troglodytes schweinfurthii) naturally harbor lentiviruses in the HIV-1/SIVcpz group.51,97,305 HIV-1s from the three divergent HIV-1 groupings M, N, and O are each more closely related to the P. t. troglodytes SIVcpz than to the P. t. schweinfurthii SIVcpz. HIV-1 groups N and O are largely restricted to west equatorial Africa where P. t. troglodytes are found.51,97 Although more work needs to be done on the lentiviruses naturally harbored by the four subspecies of chimpanzees, the evidence to date provides
strong support for the introduction of a lentivirus from P.t. troglodytes (or possibly from gorilla,236) into the human population on three or more occasions for the origins of groups M, N, and O of HIV-1. Use of the hypodermic syringe, deforestation, massive migrations to urban centers in Africa, and vaccination campaigns have all been suggested as possible explanations for why these cross-species transmissions and disseminations have succeeded only recently in history.
strong support for the introduction of a lentivirus from P.t. troglodytes (or possibly from gorilla,236) into the human population on three or more occasions for the origins of groups M, N, and O of HIV-1. Use of the hypodermic syringe, deforestation, massive migrations to urban centers in Africa, and vaccination campaigns have all been suggested as possible explanations for why these cross-species transmissions and disseminations have succeeded only recently in history.
 Figure 51.4. Chimpanzee “Sagu” eating a leg of a red colobus monkey. (Photo by Sonja Metzger, Max-Planck-Institute for Evolutionary Anthropology. See Aghokeng et al (1) and Leendertz et al.183 (Courtesy of the Centers for Disease Control and Prevention, Atlanta, Georgia.) |
Detailed sequence analyses have revealed that SIVcpz is more closely related to SIVgsn than to other SIVs in the 3′ half of its genome and is more closely related to SIVrcm in the 5′ half of its genome.11 Furthermore, until recently, HIV-1/SIVcpz and SIVgsn/SIVmon/SIVmus were the only groups of primate lentiviruses with vpu genes. The natural range of the west-central African chimpanzee (P. t. troglodytes) overlaps that of both red-capped mangabeys and greater spot-nosed monkeys. In addition, chimpanzees are known to prey on and eat monkeys for food (Fig. 51.4). Thus, the origins of SIVcpz in chimpanzees may be connected directly or indirectly to greater spot-nosed monkeys and/or red-capped mangabeys. SIVden from C. mona denti has now also been found to contain a vpu gene, although overall it clusters more closely with the SIV from DeBrazza monkey, SIVdeb.62 Sorting out the origins of SIVcpz, and consequently HIV-1 in humans, is complicated by the sheer numbers of species likely to harbor phylogenetically distinct SIVs, the potential for cross-species transmission, and the potential for retroviral recombination events.305
Attempts to transmit HIV-1 experimentally to a variety of animal species have shown a restricted host range. HIV-1 is infectious for chimpanzees, and a chimpanzee-passaged HIV-1 has shown pathogenic potential.246,247 An early report described HIV-1 infection of pig-tail macaques (Macaca nemestrina),2 but this model has not proven sufficiently robust to date for general use.
The nonprimate lentiviruses are also restricted in their host range, limited to the same or closely related genera. BIV is a notable exception in that it has been reported to infect New Zealand white rabbits.264 Cross-species infections do occur, however. Cross-species infection of sheep from goats by the goat lentivirus has been observed,265 consistent with the high degree of genetic relatedness between the caprine arthritis-encephalitis virus (CAEV) of goats and the sheep lentivirus MVV. Studies have also shown that domestic cats can be infected by puma lentivirus (FIVpco) and by lion lentivirus (FIVple).349,350 No overt disease symptoms have been associated with the latter infections, but long-term studies have not been performed.
Restriction
Cellular proteins have been defined in recent years that directly interfere with different stages of virus replication. These proteins, termed restriction factors, constitute an important defense against lentivirus infection at the cellular level, collectively referred to as “intrinsic immunity.” There are presently three lentiviral restriction factors that have been extensively studied: APOBEC3G/F, TRIM5α, and tetherin/BST-2, discovered in 2002,307 2004,328 and 2008,238,344 respectively. Although their mechanisms of antiviral activity are diverse, they are linked to innate immunity through induction by type I and type II interferons. Lentiviruses have, in turn, evolved countermeasures to each of these restriction factors to facilitate persistent replication in their respective hosts. As a consequence of this ongoing evolutionary conflict, APOBEC3G/F, TRIM5α, and tetherin have acquired species-specific differences that represent important host range determinants of lentiviral infection.222,291,292
APOBEC3G and 3F are incorporated into virus particles in the absence of Vif and result in a producer cell–dependent block to virus infectivity.96,357 These cytidine deaminases introduce C-to-U mutations in minus-strand DNA during the process of reverse transcription, which are then copied into G-to-A transitions upon plus-strand DNA synthesis.119,377 APOBEC3G and 3F preferentially deaminate the 3′ dC in CC and TC dinucleotides respectively.14,21,38,193,365 The antiviral activity of APOBEC3G and 3F is a direct result of the accumulation of extensive mutations in the proviral genome, a phenomenon known as hypermutation.120,182,206,383
HIV, SIV, and other lentiviruses are resistant to the APOBEC proteins of their respective hosts by virtue of the ability of their Vif proteins to prevent APOBEC incorporation into virions. Vif does this by serving as an adaptor to recruit the cullin-5-elongin B/C-Rbx ubiquitin ligase complex to APOBEC3G and 3F, which leads to the polyubiquitylation and subsequent proteasomal degradation of these proteins in infected cells.49,211,306,378 By depleting cytosolic pools of APOBEC3G and 3F, Vif significantly reduces their encapsidation into virions.
The sensitivity of APOBEC3G to Vif-mediated degradation is often species specific.318 HIV-1 Vif can degrade the APOBEC3G proteins of humans and chimpanzees, but not African green monkeys or rhesus macaques.210 Similarly, SIVagm Vif can degrade African green monkey APOBEC3G, but not human APOBEC3G.210 This specificity is governed by a single amino acid difference at position 128 (aspartic acid in humans versus lysine in Old World monkeys) that is critical for binding to Vif.22,207,298 The failure of HIV-1 Vif to counteract the APOBEC3G proteins of Old World monkeys in part accounts for the inability of HIV-1 to replicate in these species. Likewise, the inability of SIVagm Vif to counteract human APOBEC3G may pose a significant barrier to the zoonotic transmission of this group of primate lentiviruses to humans.
TRIM5α imposes a post-entry block that represents a major host range determinant for lentiviruses as well as other types of retroviruses. This block occurs before reverse transcription and susceptibility/resistance is determined by sequences in the capsid protein.124,132,311 Evolutionary analyses have revealed that TRIM5 has been co-evolving with retroviral pathogens for tens of millions of years, perhaps since the radiation of eutherian mammals.290 Homologs of the TRIM5 gene have been found in the genomes of primates, cows, pigs, dogs, rabbits, rats, and mice,290 and anti-viral activity has been demonstrated for the TRIM5α proteins of various species of Old and New World primates121,287,324 as well as for the related TRIM5 proteins of rabbits and cows.294,376 A common theme is that these TRIM5 proteins do not block infection by retroviral pathogens of their host, but exhibit variable patterns of restriction against retroviruses of other species.121 The underlying basis for this differential restriction is sequence variation in the B30.2 domain.190,235,294,368,375 One of the more peculiar twists in TRIM5 evolution is the independent emergence of a TRIM5-cyclophilin A (TRIM5Cyp) fusion protein in at least two different primate lineages. Owl monkey cells exhibit a potent post-entry block to HIV-1 infection due to a TRIM5Cyp fusion protein resulting from the LINE-1-mediate retrotransposition of an open reading frame for CypA into the intron between exons 7 and 8 of TRIM5.243,293 A similar TRIM5 Cyp fusion was identified in Old World primate species of the macaque genus, including rhesus macaques, pig-tailed macaques, and cynomolgous macaques.27,191,241,355,369 In this case, the CypA insertion occurred in the 3′ UTR of TRIM5 and is linked to a second mutation in the splice acceptor site of exon 7 favoring splicing of the mRNA transcript to the downstream open reading frame for CypA. Macaque TRIM5Cyp poorly restricts HIV-1, but does block infection by other lentiviruses, including SIVagm, HIV-2 and FIV.355,369 The functional blocks to lentiviral infection imposed by these TRIM5Cyp proteins, which clearly represent distinct retrotransposition events, are believed to be the result of convergent evolution.
Sequence analyses have also revealed extensive TRIM5 polymorphism in rhesus macaques and sooty mangabeys.240 Clusters of synonymous and nonsynonymous nucleotide substitutions were identified in regions of the gene coding for the CC and B30.2 domains with multiple alleles present at high frequencies in both species.240 Moreover, a number of specific polymorphisms were found in the TRIM5 genes of both rhesus macaques and sooty mangabeys, despite an estimated divergence time of over 8 million years.240 Sequence variation in the B30.2 domains of these species’ TRIM5α proteins was also shown to result in the differential restriction of a number of lentiviruses, including HIV-1, HIV-2, FIV, and EIAV.240,368 These observations imply that balancing selection has acted over millions of years to preserve ancient TRIM5α polymorphisms with functional diversity in their ability to block virus infection in Old World monkeys.
TRIM5 polymorphisms have been found to account for the highly variable course of infection for SIVsmE543-3 in rhesus macaques. In contrast to SIVmac239, which consistently results in high viral loads with minimal animal-to-animal variation, viral loads in SIVsmE543-3-infected rhesus macaques are highly variable. Kirmaier et al. found that this variation is strongly associated with differences in TRIM5 genotype.170 The resurgence of SIVsmE543-3 replication in animals with restrictive TRIM5 alleles was also associated with an arginine-to-serine change at position 97 in capsid (R97S) corresponding to the residue present at this position in SIVmac239.170 Thus, while SIVmac239 has had sufficient time to adapt to rhesus macaque TRIM5 protein, SIVsmE543-3 evidently has not. These observations illustrate the influence of TRIM5 polymorphisms on the differential suppression of a virus that has not fully adapted to its host.
Efforts to determine the role of the HIV-1 Vpu protein in virus release led to the identification of tetherin (BST-2 or CD317) as an interferon-inducible host-cell factor that interferes with the detachment of virus particles from infected cells.237,238,344 Tetherin is a type II integral membrane protein with a topology that allows both ends of the molecule to be anchored in lipid membranes.174 It has an N-terminal cytoplasmic domain, a transmembrane domain, an extracellular coiled-coil domain and a C-terminal glycosyl-phosphatidylinositol anchor.174 Tetherin is upregulated in response to interferon and becomes incorporated into virus particles as they attempt to bud from the surface of infected cells.89,116,260 Captured virions are then internalized and routed for lysosomal degradation.226,260
The primate lentiviruses have evolved at least three different viral gene products to overcome restriction by tetherin. Whereas HIV-1 Vpu and HIV-2 Env antagonize human tetherin,180,238,344 most SIVs, including members of the phylogenetically diverse SIVcpz, SIVagm, and SIVsmm lineages, use Nef to counteract the tetherin proteins of their nonhuman primate
hosts.149,289,382 An exception are the SIVs of Old World monkeys that contain a vpu gene (SIVgsn, SIVmon, SIVmus and SIVden), which use Vpu rather than Nef to counteract the tetherin proteins of their respective hosts.289 Tetherin antagonism by HIV-1 Vpu depends upon a physical interaction between the membrane-spanning domains of Vpu and tetherin147,281 and in part on the recruitment of the βTrCP-2 component of the Skp1-Cullin1-F-box ubiquitin ligase complex, nonlysine ubiquitylation of tetherin, and ESCRT-mediated trafficking of tetherin for lysosomal degradation.79,148,205,225,333 In contrast, tetherin antagonism by HIV-2 Env involves a physical interaction between the extracellular domains of Env and tetherin, and the internalization and sequestration of tetherin within the trans-Golgi network, without degradation, by a pathway that depends on a conserved tyrosine-based endocytosis motif in the cytoplasmic tail of gp41.122,180 The mechanism of antagonism by Nef remains to be defined, but it appears to involve the downregulation of tetherin from the surface of infected cells.149,302
hosts.149,289,382 An exception are the SIVs of Old World monkeys that contain a vpu gene (SIVgsn, SIVmon, SIVmus and SIVden), which use Vpu rather than Nef to counteract the tetherin proteins of their respective hosts.289 Tetherin antagonism by HIV-1 Vpu depends upon a physical interaction between the membrane-spanning domains of Vpu and tetherin147,281 and in part on the recruitment of the βTrCP-2 component of the Skp1-Cullin1-F-box ubiquitin ligase complex, nonlysine ubiquitylation of tetherin, and ESCRT-mediated trafficking of tetherin for lysosomal degradation.79,148,205,225,333 In contrast, tetherin antagonism by HIV-2 Env involves a physical interaction between the extracellular domains of Env and tetherin, and the internalization and sequestration of tetherin within the trans-Golgi network, without degradation, by a pathway that depends on a conserved tyrosine-based endocytosis motif in the cytoplasmic tail of gp41.122,180 The mechanism of antagonism by Nef remains to be defined, but it appears to involve the downregulation of tetherin from the surface of infected cells.149,302
In accordance with a now familiar theme, resistance to tetherin is species dependent. HIV-1 Vpu counteracts human, chimpanzee, and gorilla tetherin, but is ineffective against the tetherin orthologs of Old World monkeys.149,222,289 Conversely, the Vpu proteins of SIVgsn, SIVmon and SIVmus counteract the tetherin proteins expressed by various species of Old World monkeys, but are unable to counteract great ape tetherin.289 Likewise, the Nef proteins of SIVcpz, SIVsmm/mac and SIVagm counteract the tetherin proteins of their respective hosts, and with varying efficiency, the tetherin proteins of other nonhuman primates.149,289,382 However, these Nef proteins are universally ineffective against human tetherin.149,289,382 The specificity of Vpu reflects amino acid variation in the transmembrane domain of tetherin and corresponding variation in the transmembrane domain of Vpu.149,222,289 In contrast, the specificity of Nef for nonhuman primate tetherin is dependent on a five amino acid sequence in the cytoplasmic domain (G/D14DIWK18 in chimpanzee, rhesus macaque, and sooty mangabey tetherin) that is missing from human tetherin.149,382
The absence of sequences in human tetherin that confer susceptibility to Nef has had a profound effect on the evolution of HIV-1 and HIV-2. Since HIV-1 arose from the cross-species transmission of SIVcpz from chimpanzees to humans, and SIVcpz uses Nef to antagonize tetherin, the absence of sequences in human tetherin that confer susceptibility to Nef explains why HIV-1 Vpu acquired this activity in humans. However, this function appears only to have been acquired by the Vpu proteins of the pandemic HIV-1 group M viruses, but not by the nonpandemic HIV-1 group N or O viruses.289 It has therefore been suggested that antagonism of tetherin by Vpu may have contributed to the global spread of HIV-1 group M.289 A similar scenario may explain the role of HIV-2 Env in the antagonism of tetherin. Since HIV-2 arose from the cross-species transmission of SIVsmm from sooty mangabeys to humans, and this virus does not have a vpu gene, the inability of Nef to antagonize human tetherin may also account for the selective pressure for HIV-2 Env to acquire this activity.
Compensatory changes in the cytoplasmic tail of gp41 that restore resistance to tetherin were recently identified in a nef-deleted strain of SIV that acquired a pathogenic phenotype after serial passage in rhesus macaques. Similar to HIV-2 Env antagonism of human tetherin, these changes result in a physical interaction with rhesus tetherin and facilitate virus release by a mechanism that depends on a conserved tyrosine-based endocytosis motif in the gp41 tail.302 However, unlike HIV-2 Env, these changes afford resistance to rhesus tetherin, but not to human tetherin, by stabilizing a selective physical interaction that depends on residues in the cytoplasmic domain of rhesus tetherin.302 These observations are analogous to the adaptation of HIV-2 Env for antagonism of human tetherin, and imply that antagonism of tetherin is important for virus replication in vivo and ultimately for lentiviral pathogenesis.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree